Biosensors and Bio-Bar Code Assays Based on Biofunctionalized Magnetic Microbeads
Abstract
:1. Introduction
2. Use of magnetic microbeads for affinity biosensors
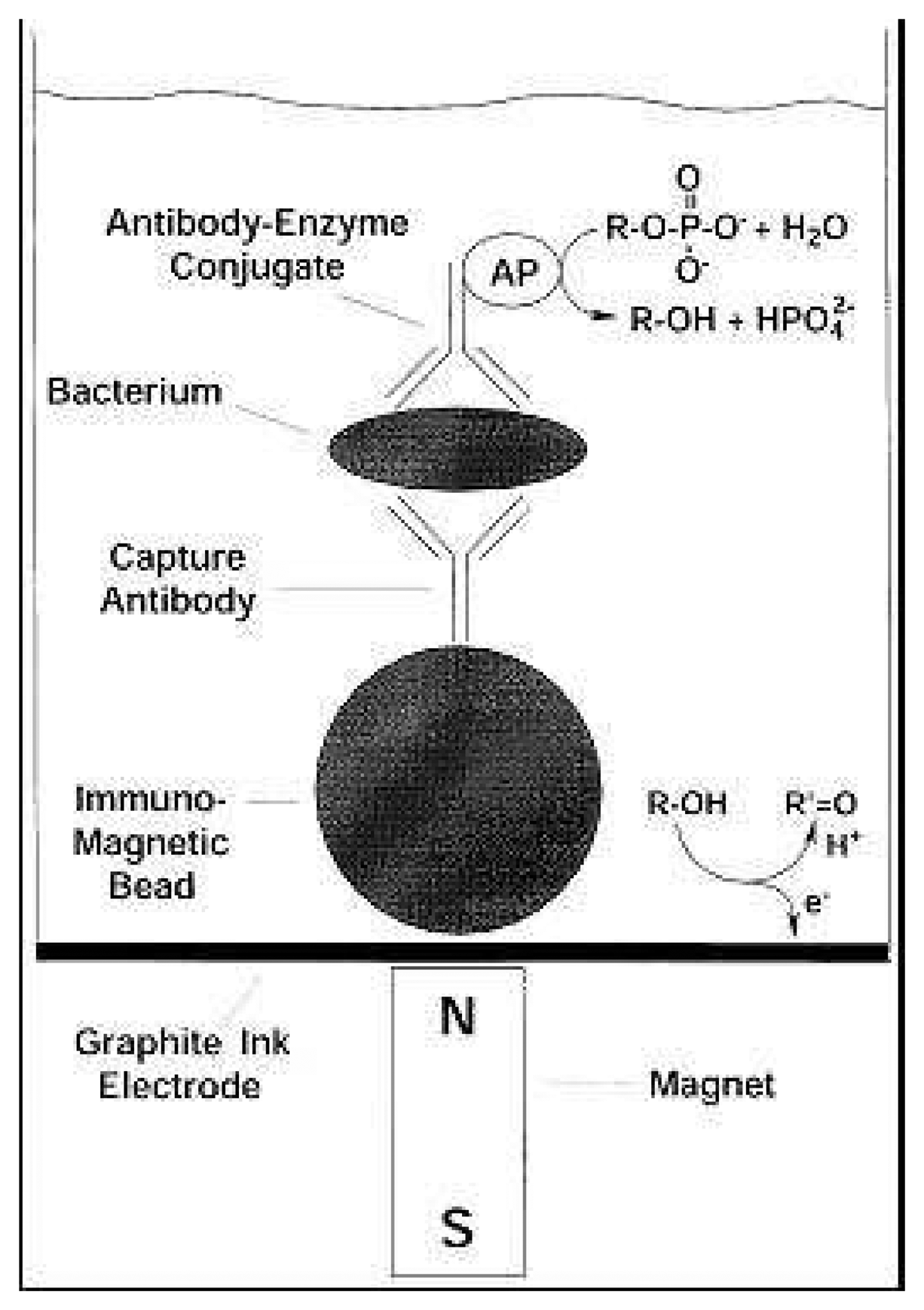
2.1. Immunomagnetic electrochemical sensors (ELIME)
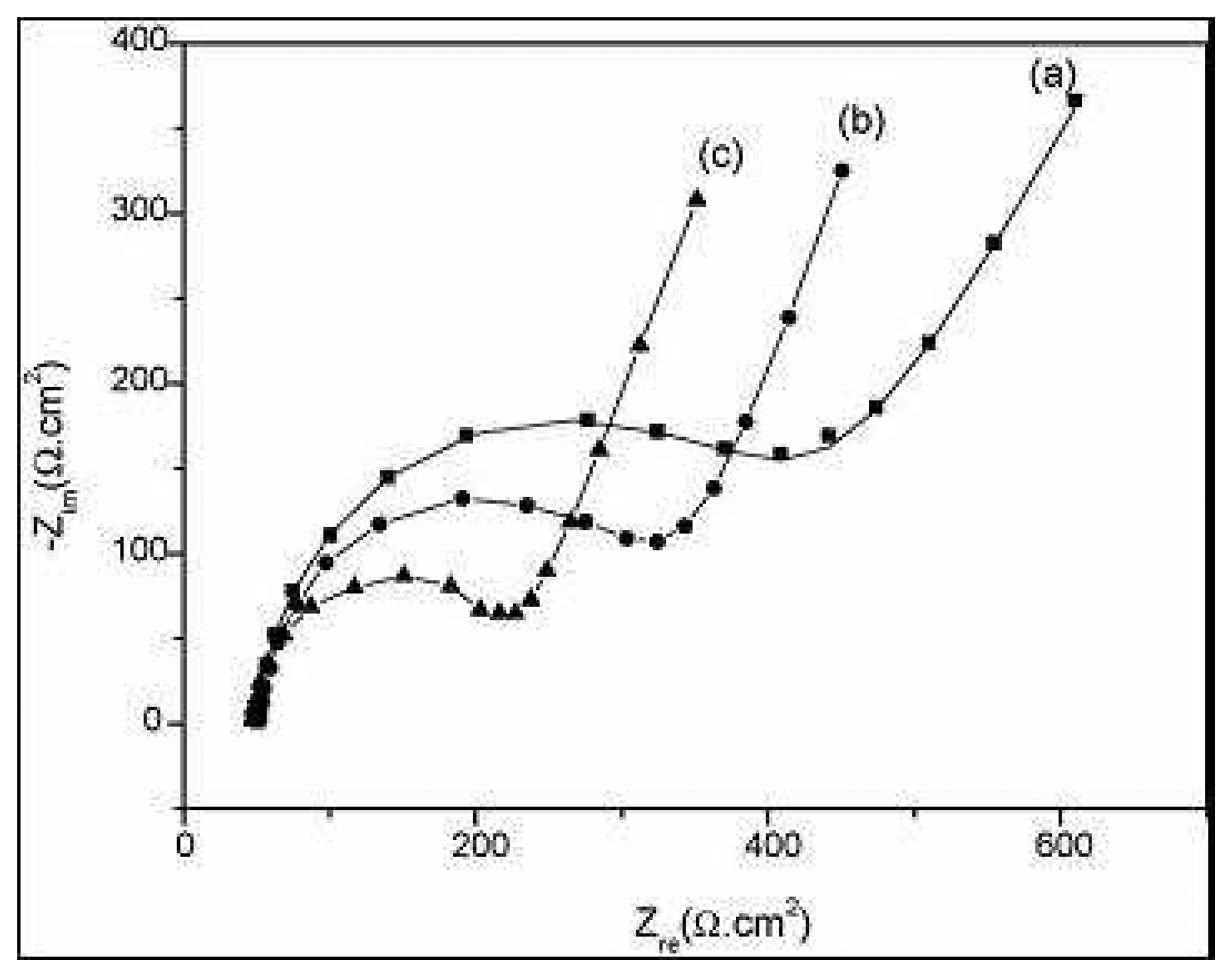
2.2. Label-free immunomagnetic impedancemetric sensors [7]
2.3. Piezoimmunosensors
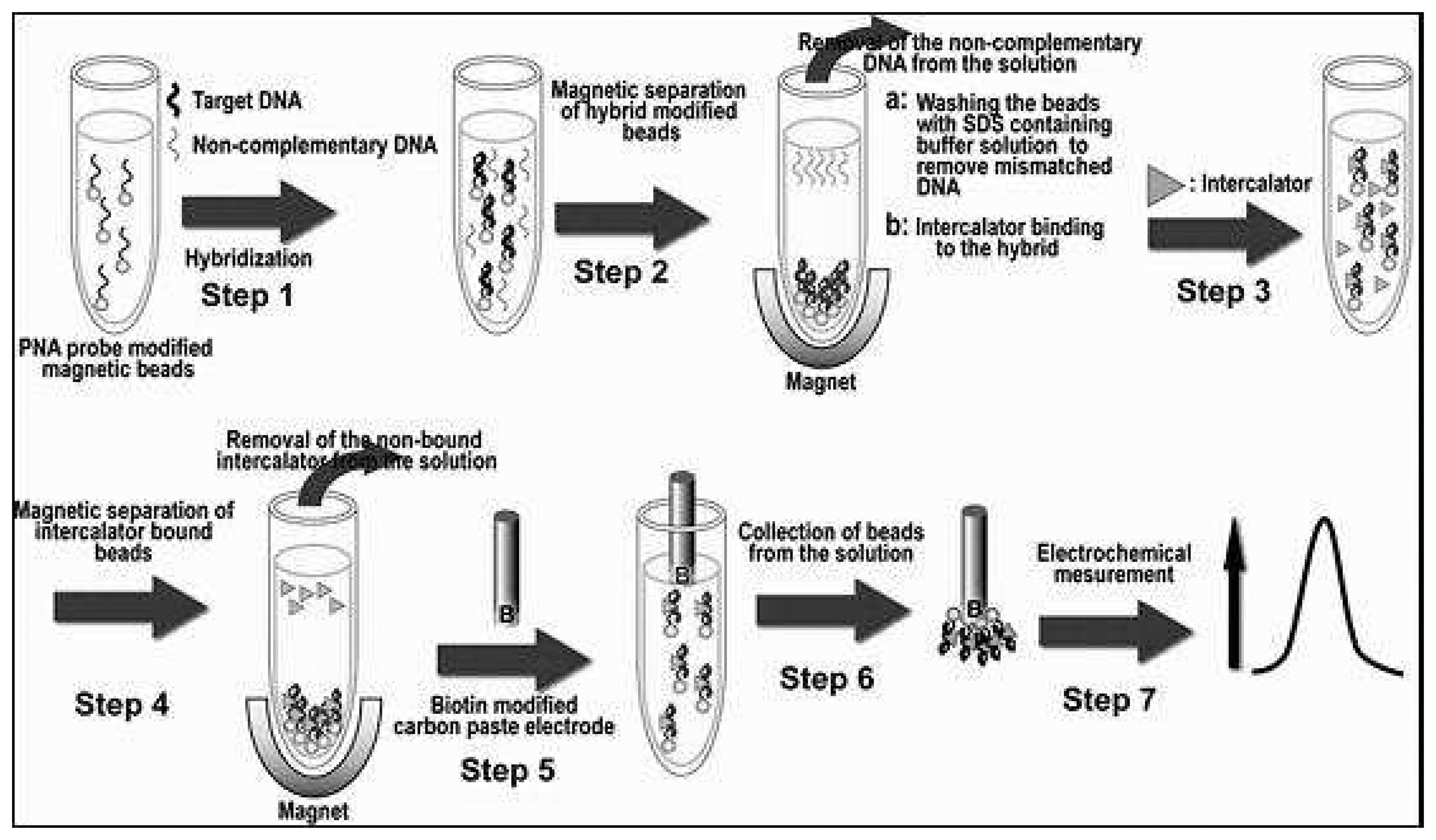
2.4. Genomagnetic electrochemical assays
2.5. Magnetic detection
2.5.1. Magnetic Permeability measurements
2.5.2. Frequency mixing
2.5.3. Magnetic AFM [25]
2.5.4. Magnetic Resonance Imaging (MRI)
3. Enzymatic biosensors based onto magnetic microbeads
- enzyme immobilization onto magnetic microbeads, having high specific area, allows a high loading of the sensitive matrix
- such a matrix provides a good macroenvironment for retention of the bioactivity
- the most decisive advantage is a good control of the localisation of the sensitive material through the use of magnets or of magnetised transducing parts allowing enzyme reactions to occur close to the detection device
- the sensitive transducing part can be easily renewed
4. Bio-bar code
- Magnetic Micro Beads (MMB) bearing the biological probe (DNA or Antibody, Figure 18) for target recognition.
4.1. Nanoparticles (NP)
4.2. Magnetic Microbeads (MMB)
4.3. Detection
4.4. Biological applications
4.5. Conclusion
References
- Hermanson, G. T.; Mallia, A. K; Smith, P. K. Immobilised affinity ligand techniques.; Academic Press, 1992. [Google Scholar]Hermanson, G. T. Bioconjugate Techniques.; Academic Press, 1996. [Google Scholar]
- Georganopoulou, D. G.; Chang, L.; Nam, J-M.; Thaxton, C. S.; Mufson, E. J.; Klein, W. L.; Mirkin, C. A. Fluorescent and scanometric ultrasensitive detection technologies with the biobar code assay for alzheimer's disease diagnosis. PNAS 2005, 102, 2273–2276. [Google Scholar]
- Liu, Z.; Liu, Y.; Yang, H.; Yang, Y.; Shen, G.; Yu, R. A phenol biosensor based on immobilizing tyrosinase to modified core-shell magnetic nanoparticles supported at a carbon paste electrode. Analytica Chimica Acta 2005, 533(1), 3–9. [Google Scholar]
- Stoeva, S. I.; Lee, J-S.; Thaxton, S.; Mirkin, C. A. Multiplexed DNA detection with biobarcoded Nanoparticle probes. Angewandte Chemie 2006, 45, 3303–3306. [Google Scholar]
- Nam, J.; Stoeva, S. I.; Mirkin, C. A. Bio-Bar-code-Based DNA Detection with PCR like sensitivity. J. Am. Chem. Soc. 2004, 126, 5932–5933. [Google Scholar]
- Nam, J.; Taxton, C. S.; Mirkin, C. A. Nanoparticle-based Bio-barcodes for the ultrasensitive detection of proteins. Science. 2003, 301, 1884–1886. [Google Scholar]
- Helali, S.; Martelet, C.; Abdelghani, A.; Maaref, M.A.; Jaffrezic-Renault, N. A disposable immunomagnetic electrochemical sensor based on functionalised magnetic beads on gold surface for the detection of atrazine. Electrochimica Acta 2006, 51(24), 5182–5186. [Google Scholar]
- Santandreu, M.; Sole, S.; Fabregas, E.; Alegret, S. Development of electrochemical immunosensing systems with renewable surfaces. Biosensors Bioelectronics 1998, 13, 7–17. [Google Scholar]
- Gehring, A.G.; Brewster, J.D.; Irwin, P.L.; Tu, S.I.; Van Houten, L. J. 1-Naphtyl phosphate as an enzymatic substrate for enzyme-linked immunomagnetic electrochemistry. J. Electroanal. Chem. 1999, 469, 27–33. [Google Scholar]
- Sole, S.; Alegret, S.; Cespedes, F.; Fabregas, E. ; Flow injection immunoanalysis based on a magnetoimmunosensor system. Anal. Chem. 1998, 70, 1462–1467. [Google Scholar]
- Dequaire, M.; Degrand, C.; Limoges, B. An immunomagnetic electrochemical sensor based on a perfluorosulfonate-coated screen-printed electrode for the determination of 2,4-dichlorooxyacetic acid. Anal. Chem. 1999, 71, 2571–2577. [Google Scholar]
- Liu, Z.M.; Yang, H.F.; Li, Y.F.; Liu, Y.L.; Shen, G.L.; Yu, R.Q. Core-shell magnetic nanoparticles applied for immobilization of antibody on carbon paste electrode and amperometric immunosensing. Sensors and Actuators B 2006, 113, 956–962. [Google Scholar]
- Li, J.; He, X.; Wu, Z.; Wang, K.; Shen, G.; Yu, R. Piezoelectric based on magnetic nanoparticles with simple immobilization procedures. Analytica Chimica Acta 2003, 481, 191–198. [Google Scholar]
- Kim, G.H.; Rand, A.G.; Letcher, S.V. Impedance characterisation of a piezoelectric immunosensor. Part I: antibody coating and buffer solution. Biosensors Bioelectronics 2003, 18, 83–89. [Google Scholar]
- Kim, G.H.; Rand, A. G.; Letcher, S.V. Impedance characterization of a piezoelectric immunosensor. Part II: Salmonella typhinium detection using magnetic enhancement. Biosensors Bioelectronics 2003, 18, 91–99. [Google Scholar]
- Palacek, E.; Fojta, M; Jelen, F. New approaches in the developement of DNA sensors: hybridization and electrochemical detection of DNA and RNA at two different surfaces. Bioelectrochemistry 2002, 56, 85–90. [Google Scholar]
- Wang, J.; Kawde, A.N.; Erdem, A.; Salazar, M. Magnetic bead-based label-free elecrochemical detection of DNA hybridization. Analyst 2001, 126, 2020–2024. [Google Scholar]
- Kerman, K.; Matsubara, Y.; Morita, Y.; Takamura, Y. Peptide nucleic acid modified magnetic beads for intercalator based electrochemical detection of DNA hybridization. Science and technology of Advanced Materials 2004, 5, 351–357. [Google Scholar]
- Erdem, A.; Pividori, M.I.; Lermo, A.; Bonanni, A.; Del Valle, M.; Alegret, S. Genomagnetic assay based on label-free electrochemical detection using magneto-composite electrodes. Sensors and Actuators B 2006, 114, 591–598. [Google Scholar]
- Wang, J.; Xu, D.; Erdem, A.; Polsky, R.; Salazar, M. A. Genomagnetic electrochemical assays of DNA hybridization. Talanta 2002, 56, 931–938. [Google Scholar]
- Kriz, K.; Gehrke, J.; Kriz, D. Advancements toward magneto immunoassays. Biosensors Biolectronics 1998, 13, 817–823. [Google Scholar]
- Abrahamsson, D.; Kriz, K.; Lu, M.; Kriz, D. A preliminary sutdy on DNA detection based on relative magnetic permeability measurements and histone H1 conjugated superparamagnetic nanoparticles as magnetic tracers. Biosensors Bioelectronics 2004, 19, 1549–1557. [Google Scholar]
- Lu, M.; Ibraimi, F.; Kriz, D.; Kriz, K. A combination of magnetic permeability detection with nanometer-scaled puperparamagnetic tracer and its application for one-step detection of human urinary albumin in undiluted urine. Biosensors Bioelectronics 2006, 21, 2248–2254. [Google Scholar]
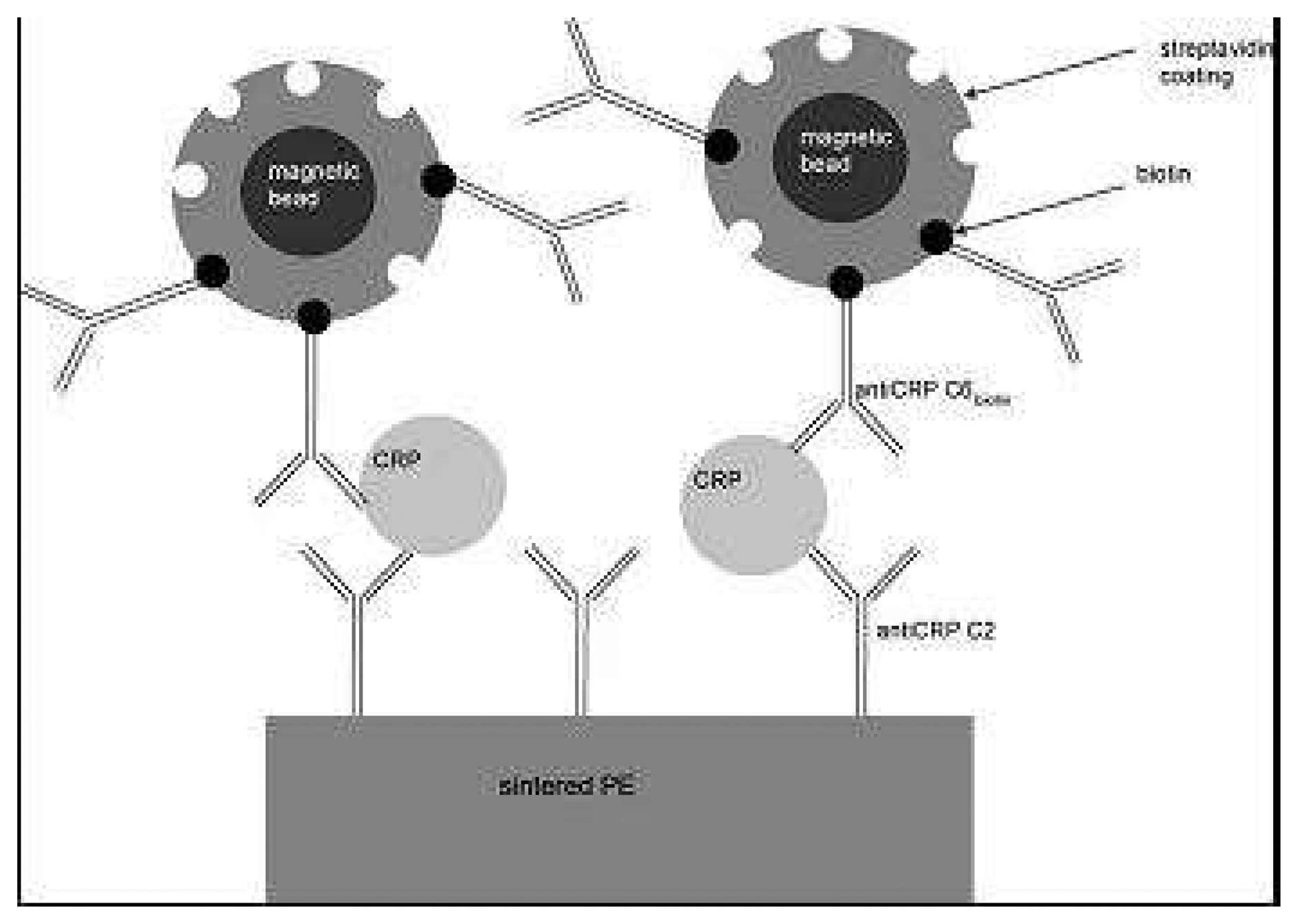
- Meyer, M.H.F.; Hartmann, M.; Krause, H.J; Blankenstein, G.; Mueller-Chorus, B.; Oster, J.; Miethe, P.; Keusgen, M. CRP determination based on a novel magnetic biosensor. Biosensors Bioelectronics 2007, 22(6), 973–979. [Google Scholar]
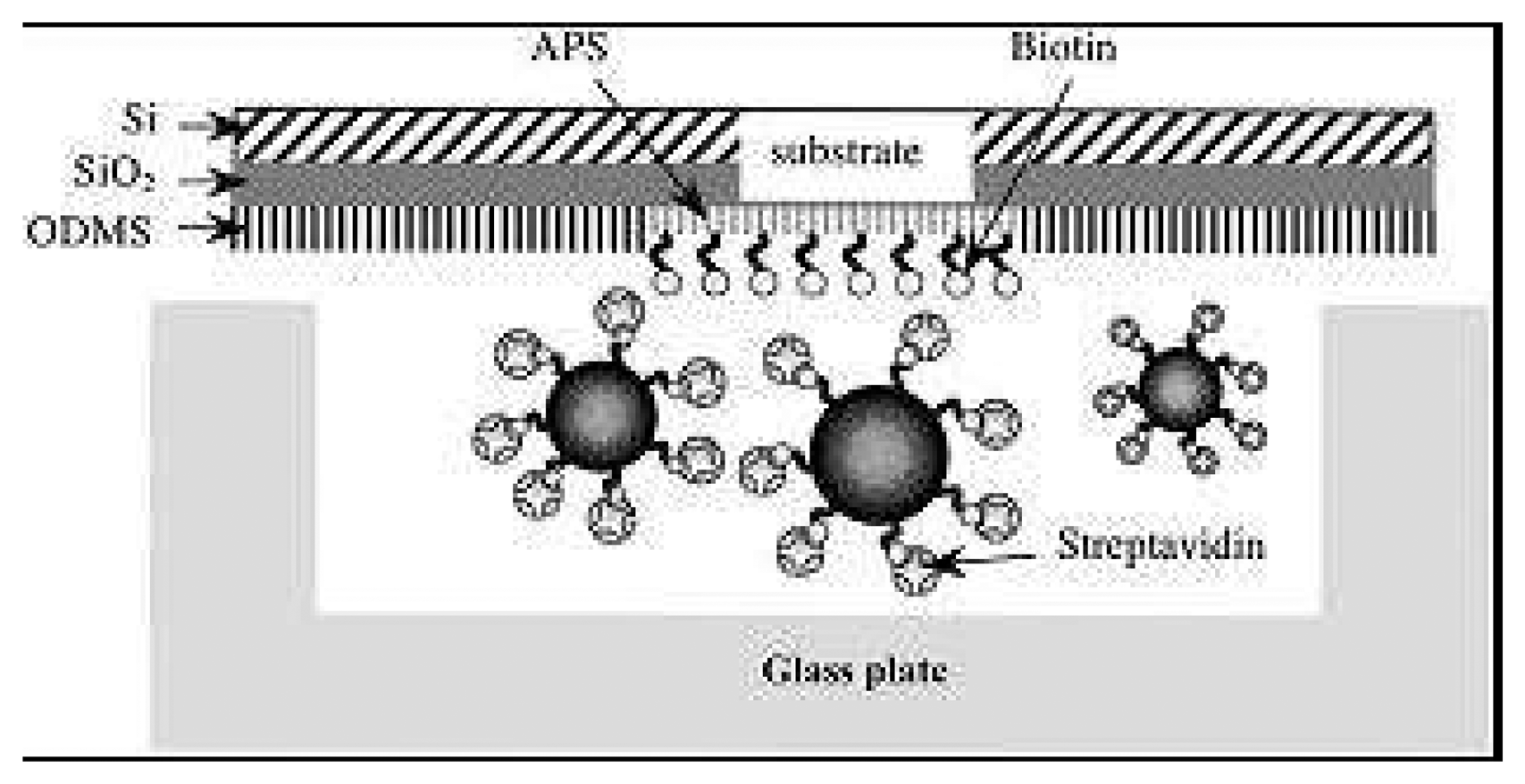
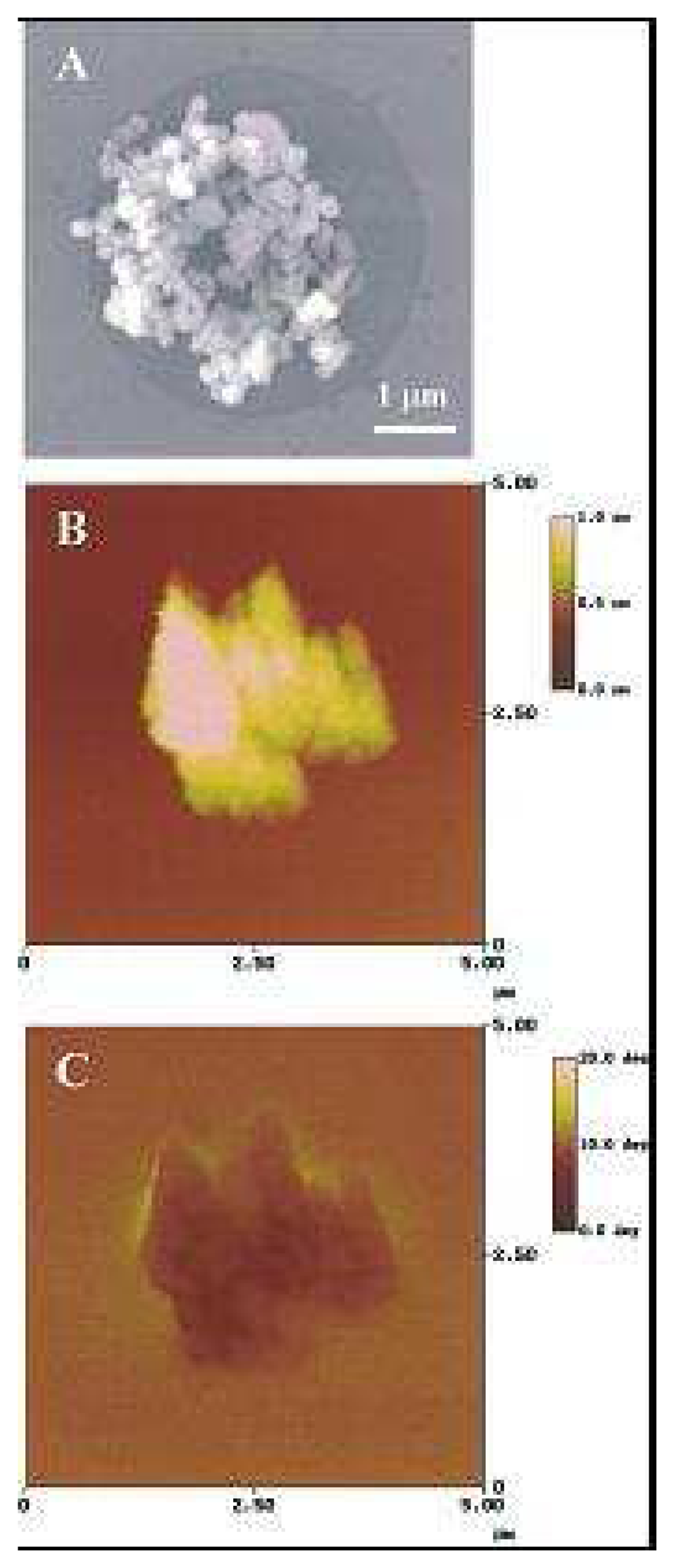
- Arakaki, A.; Hideshima, S.; Nakagawa, T; Niwa, D.; Tanaka, T.; Matsunaga, T. Detection of biomolecular interaction between biotin and streptavidin on a self-assembled monolayer using magnetic nanoparticles. Biotechnology & Bioengineering 2004, 88, 543–546. [Google Scholar]
- Perez, J. M.; Josephson, L.; O'Loughlin, T.; Högemann, D.; Weissleder, R. Magnetic relaxation switches capable of sensing molecular interactions. Nature Biotechnology 2002, 20, 816–820. [Google Scholar]
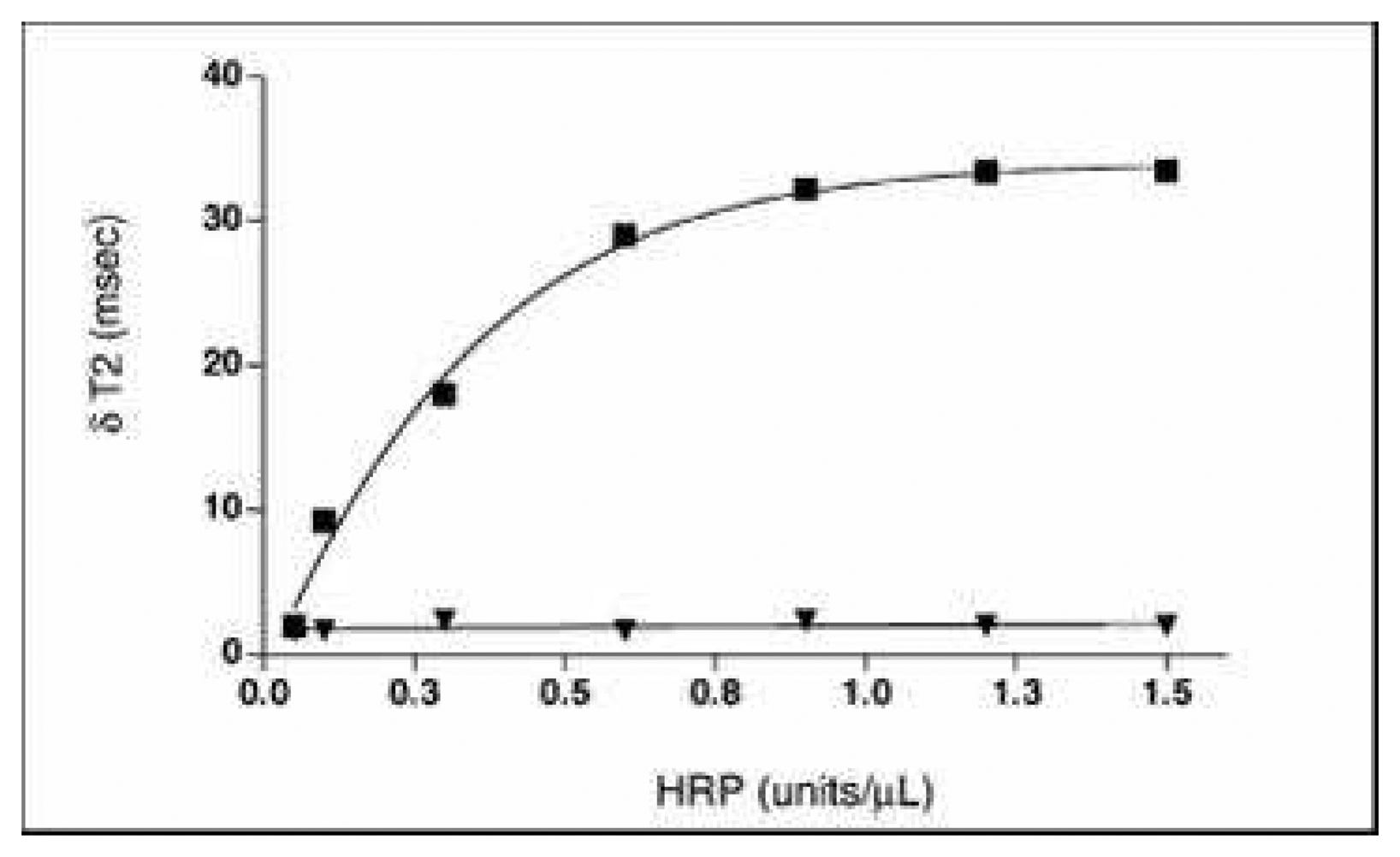
- Perez, J. M.; Simeone, F. J.; Tsourkas, A.; Josephson, L.; Weissleder, R. Peroxidase Substrate Nanosensors for MR imaging. Nanoletters 2004, 4, 119–122. [Google Scholar]
- Perez, J. M.; Simeone, F. J.; Saeki, Y.; Josephson, L.; Weissleder, R. Viral-Induced Self-Assembly of Magnetic Nanoparticles Allows the Detection of Viral Particles in Biological Media. JACS 2003, 125, 10192–10193. [Google Scholar]
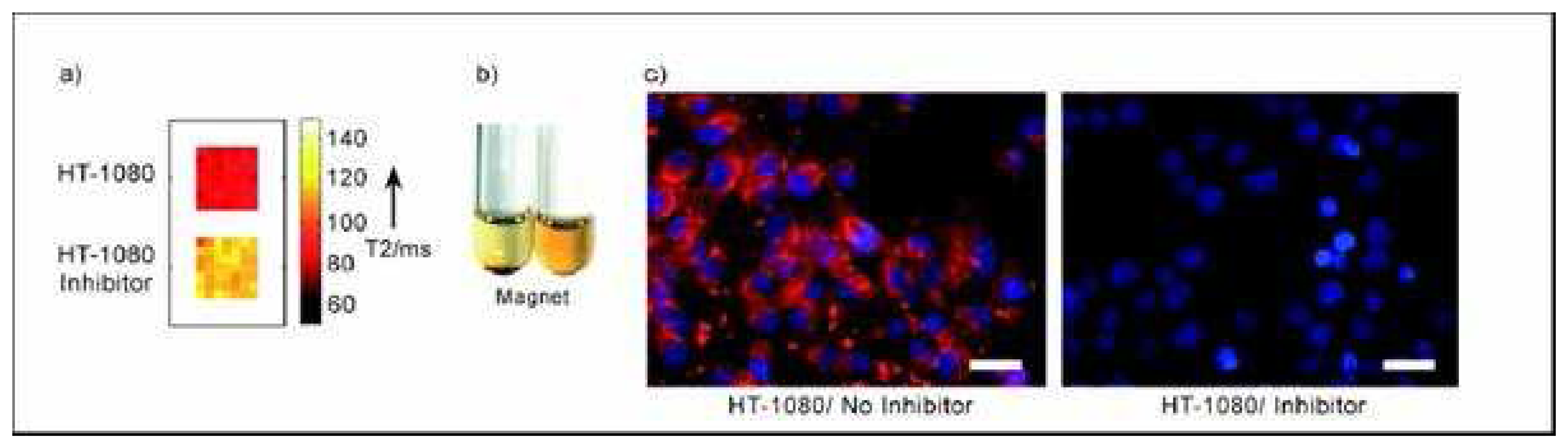
- Harris, T. J.; von Maltzahn, G.; Derfus, A. M.; Ruoslahti, E.; Bhatia, S. N. Proteolytic actuation of Nanoparticles Self assembly. Angew.Chem. Int. Ed. 2006, 45, 3161–3165. [Google Scholar]
- Varlan, A.R.; Sansen, W.; Van Loey, A.; Hendrickx, M. Covalent enzyme immobilization on paramagnetic polyacrolein beads. Biosensors Bioelectronics 1996, 11(4), 443–448. [Google Scholar]
- Miyabayashi, A.; Mattiasson, B. An enzyme electrode based on electromagnetic entrapment of the biocatalyst bound to magnetic beads. Analytica Chimica Acta 1988, 213, 121–130. [Google Scholar]
- Varlan, A.R.; Suls, J.; Jacobs, P.; Sansen, W. A new technique of enzyme entrapment for planar biosensors. Biosensors Bioelectronics 1995, 10(8), XV-XIX. [Google Scholar]
- Solé, S.; Merkoci, A.; Alegret, S. New materials for electrochemical sensing III. Beads. TrAC Trends in Analytical Chemistry 2001, 20(2), 102–110. [Google Scholar]
- Liao, M-H.; Guo, J-C.; Chen, W.C. A disposable amperometric ethanol biosensor based on screen-printed carbon electrodes mediated with ferricyanide-magnetic nanoparticle mixture. Journal of Magnetism and Magnetic Materials 2006, 304(1), e421–e423. [Google Scholar]
- Lu, B-W.; Chen, W.C. A disposable glucose biosensor based on drop-coating of screen-printed carbon electrodes with magnetic nanoparticles. Journal of Magnetism and Magnetic Materials 2006, 304(1), e400–e402. [Google Scholar]
- Elyacoubi, A.; Zayed, S.; Blankert, B.; Kauffmann, J-M. Development of an Amperometric Enzymatic Biosensor Based on Gold Modified Magnetic Nanoporous Microparticles. Electroanalysis 2006, 18, 345–350. [Google Scholar]
- Yu, D.; Blankert, B.; Bodoki, E.; Bollo, S.; Viré, J-C; Sandulescu, R.; Nomura, A.; Kauffmann, J-M. Amperometric biosensor based on horseradish peroxidase-immobilised magnetic microparticles. Sensors and Actuators B 2006, 113(2), 749–754. [Google Scholar]
- Kauffmann, J-M.; Yu, D.; El Yacoubi, A.; Blankert, B. Magnetic nanoporous microparticles for biosensors and bioreactors. LabPlus international 2006, 20(3), 6–8. [Google Scholar]
- Willner, I.; Willner, B. Functional nanoparticles architectures for sensoric, optoelectronic and bioelectronic applications. Pure Appl. Chem. 2002, 74(9), 1773–1783. [Google Scholar]
- Willner, I; Katz, E. Magnetic control of electrocatalytic and bioelectrocatalytic process. Angew. Chem. Int. Ed 2003, 42, 4576–4588. [Google Scholar]
- Li, Y.; Hong Cu, Y.T.; Luo, D. Multiplexed detection of pathogen DNA with DNA-bases fluorescence nanobarcodes. Nature Biotechnology 2005, 23(7), 885–889. [Google Scholar]
- Oh, B-K.; Nam, J.M.; Lee, S. W; Mirkin, C. A. A fluorophore-base Bio-barcode Amplification Assay for Proteins. Small 2006, 2, 103–108. [Google Scholar]
- Thaxton, C. S.; Hill, H. D.; Georganopoulou, D. G.; Stoeva, S. I.; Mirkin, C. A. A Bio-Bar Code Assay Based upon Dithiothreitol-Induced Oligonucleotide Release. Anal. Chem 2005, 77, 8174–8178. [Google Scholar]
- Khan, S.; Klein, W.; Mirkin, C. A.; Chang, L.; Georganopoulou, D. Fluorescent and scanometric ultrasensitive detection technologies with the bio-bar code assay for alzheimer's disease diagnosis. Nanoscape 2005, 2(1), 7–15. [Google Scholar]
- Schweitzer, B.; Wiltshire, S.; Lambert, J.; O'Malley, S.; Kukanskis, K.; Zhu, Z.; Kingsmore, S. F.; Lizardi, P. M. Ward. D. C. Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. PNAS 2000, 97(18), 10113–10119. [Google Scholar]
- Lee, A.; Mirkin, C.; Georganopoulos, D. Real-time polymerase chain reaction completed detection of cardiac tropin I with the bio-bar code assay. Nanoscape 2005, 2(1), 17–25. [Google Scholar]








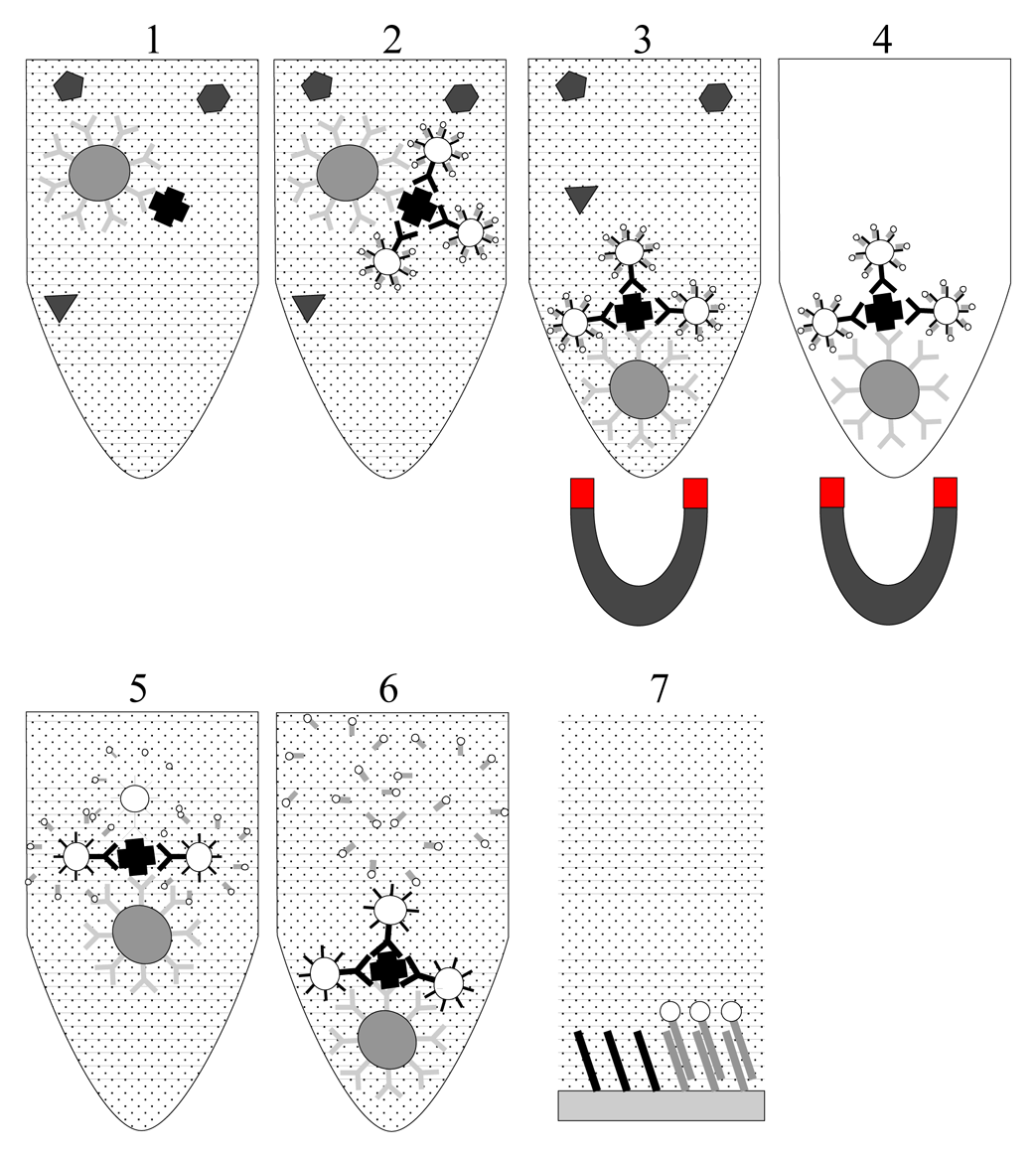
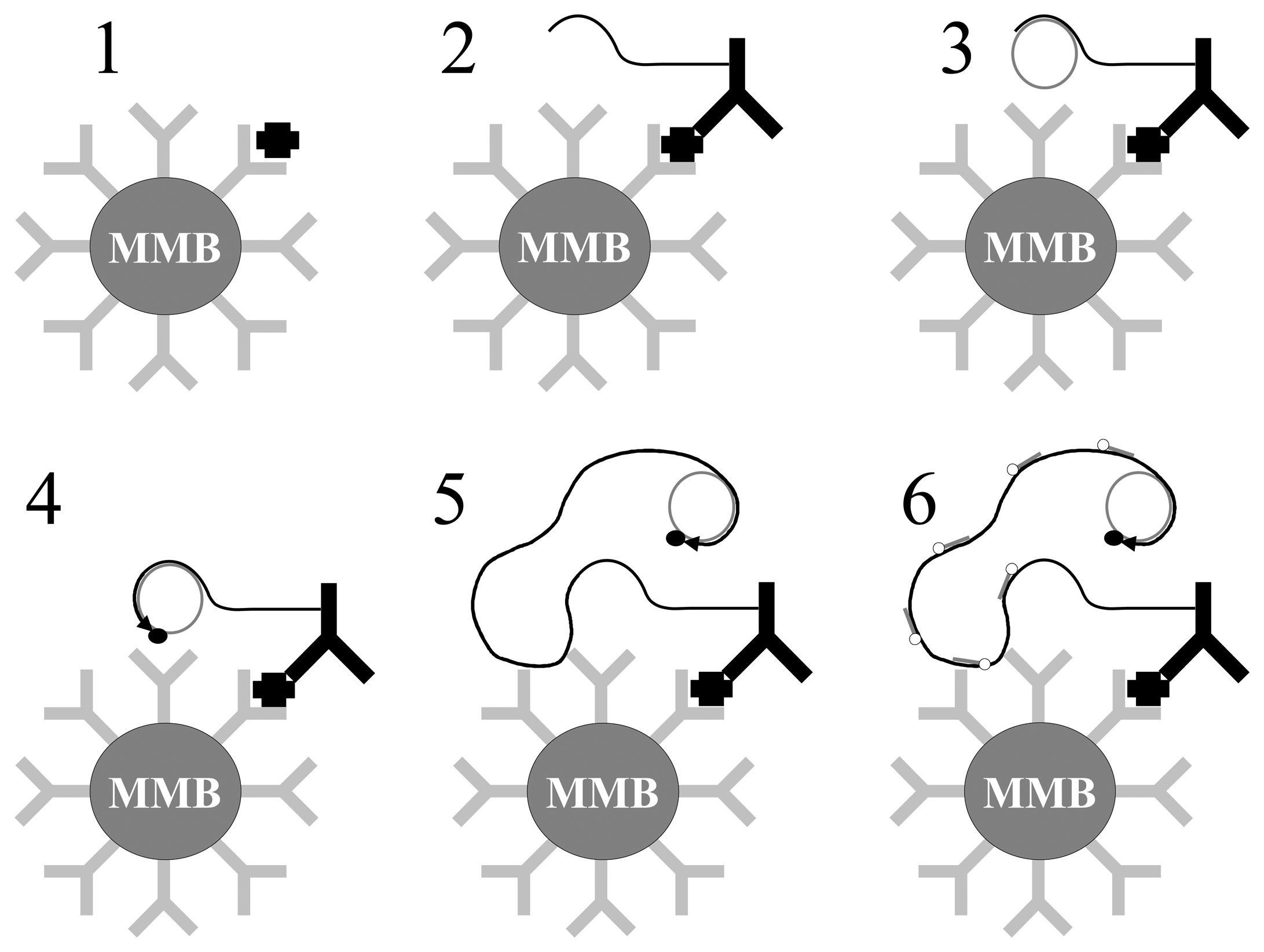
- 1
- Interaction between MMB and target.
- 2
- Recognition between target and particles in complex biological medium : sandwich MMB/target/NP
- 3
- Magnetic separation of MMB
- 4
- Removal of biological medium. Only sandwiches MMB/target/NP and MMB are kept in the tube
- 5
- Redispersion of sandwiches in distilled water causes dehybridation of biobar-codes
- 6
- Removal and analysis of bio–bar-codes using DNA microarray or other methods
- 1
- Interaction between MMB and target.
- 2
- Recognition between target and particles in complex biological medium : sandwich MMB/target/NP
- 3
- Magnetic separation of MMB
- 4
- Removal of biological medium. Only sandwiches MMB/target/NP and MMB are kept in the tube
- 5
- Redispersion of sandwiches in distilled water causes dehybridation of biobar-codes
- 6
- Removal and analysis of bio–bar-codes using DNA microarray or other methods
| Surface function | Immobilisation protocole | Biomolecules | Advantage | Disavantage | Reference |
|---|---|---|---|---|---|
| Amine | Active Ester | ||||
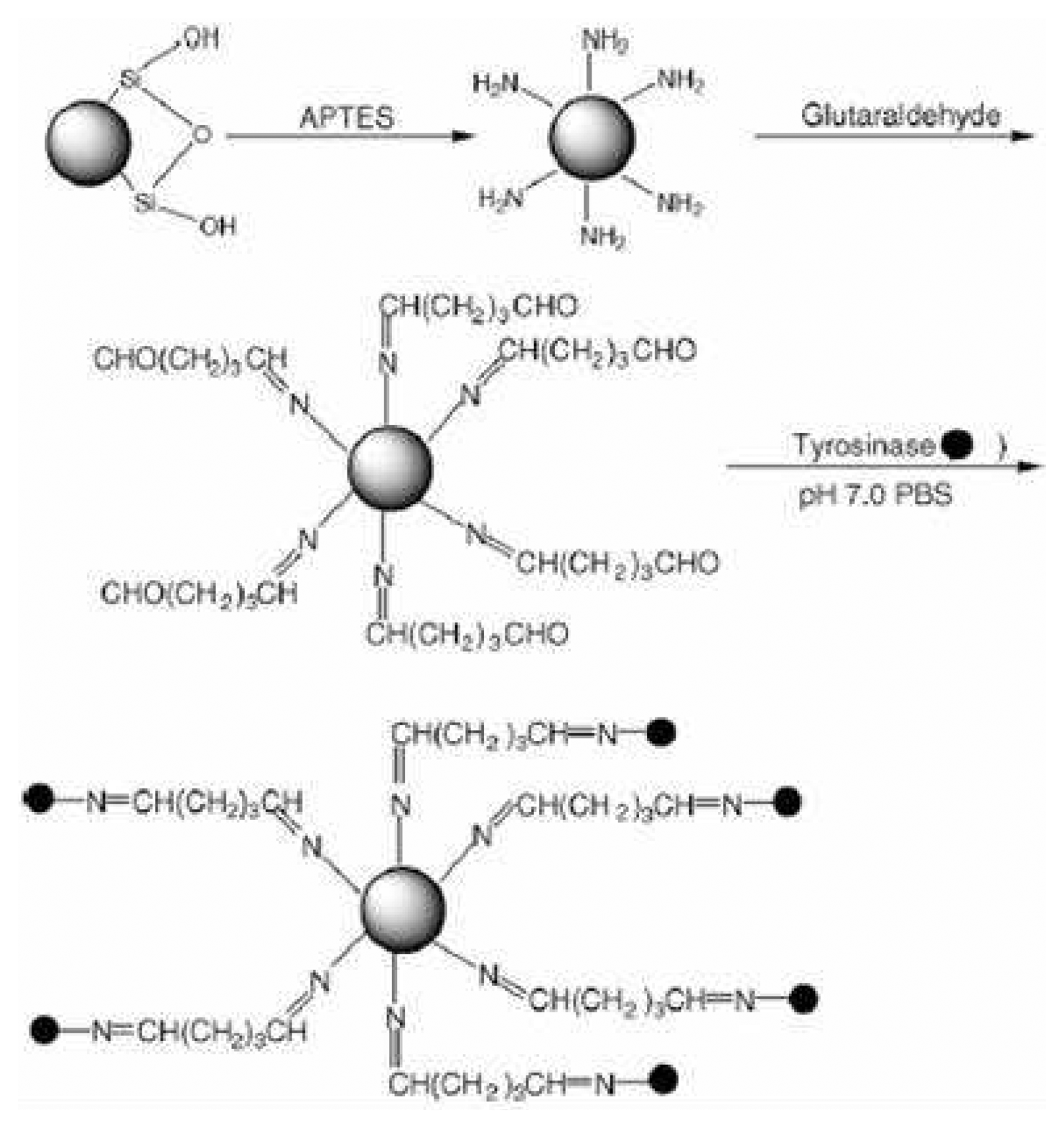
| Amine | Glutaraldehyde | Antibody (Ab) Enzyme | Easy Yield | Non reproducible Non Oriented | [2], [3] |
| Amine | Maleimido groups: SMPB sulfo-SMCC | Thiol derivatives | [4], [5] | ||
| Gold | Thiol | Thio-DNA | Simple | Limited to thiol containing molecules | [6] |
| Active ester | Amine | ||||
| Avidin, Streptavidin | Biotinylated biomolecules | Antibody | Simple | Need biotinylated biomolecules | [7] |
| Analyte | Transducer | Enzyme | Detection Limit | Dynamic range | Refs |
|---|---|---|---|---|---|
| Rabbit IgG | pH-ISFET | urease | 8nM | 0 – 2.07μM | [10] |
| Rabbit IgG | Graphite composite electrode | HR Peroxidase | 9×10-6μg.l-1 | 0 – 0.26μM | [8] |
| E. coli 0157:H7 | Graphite ink electrode | Alkaline Phosphatase | 4.7×103 cells.ml-1 | 0 – 105cells.ml-1 | [9] |
| 2,4-D herbicide | Nafion-SPE | Alkaline phosphatase | 0.01 μg.l-1 | 0.01 - 100μg.l-1 | [11] |
| Human IgG | Carbon paste electrode | HR Peroxidase | 0.18μg.ml-1 | 0.51 – 30.17 μg.ml-1 | [12] |
| Analyte | Electrochemical Transducer | Label | Detection Limit | Dynamic range | Refs |
|---|---|---|---|---|---|
| Breast-cancer BRCA1 gene | Potentiometric stripping measurements Graphite pencil electrode | No | 100ng.ml-1 (ppb) | 100 ppb-20ppm | [17] |
| Breast-cancer BRCA1 gene | Differential pulse voltammetry | Alkaline phosphatase | 100ppb | 0.25-1.0ppm (20 min Hybridization) | [20] |
| 21 mer DNA | Square wave voltammetry | Meldola's blue intercalator | 2pM | 2 – 20pM | [18] |
| DNA Sequence from Salmonella | Differential pulse voltammetry Graphite composite electrodes | No | 9.68pM | [19] |
| Analyte | Magnetic beads | Transducer | Enzyme | Sensitivity | Dynamic range | Ref. |
|---|---|---|---|---|---|---|
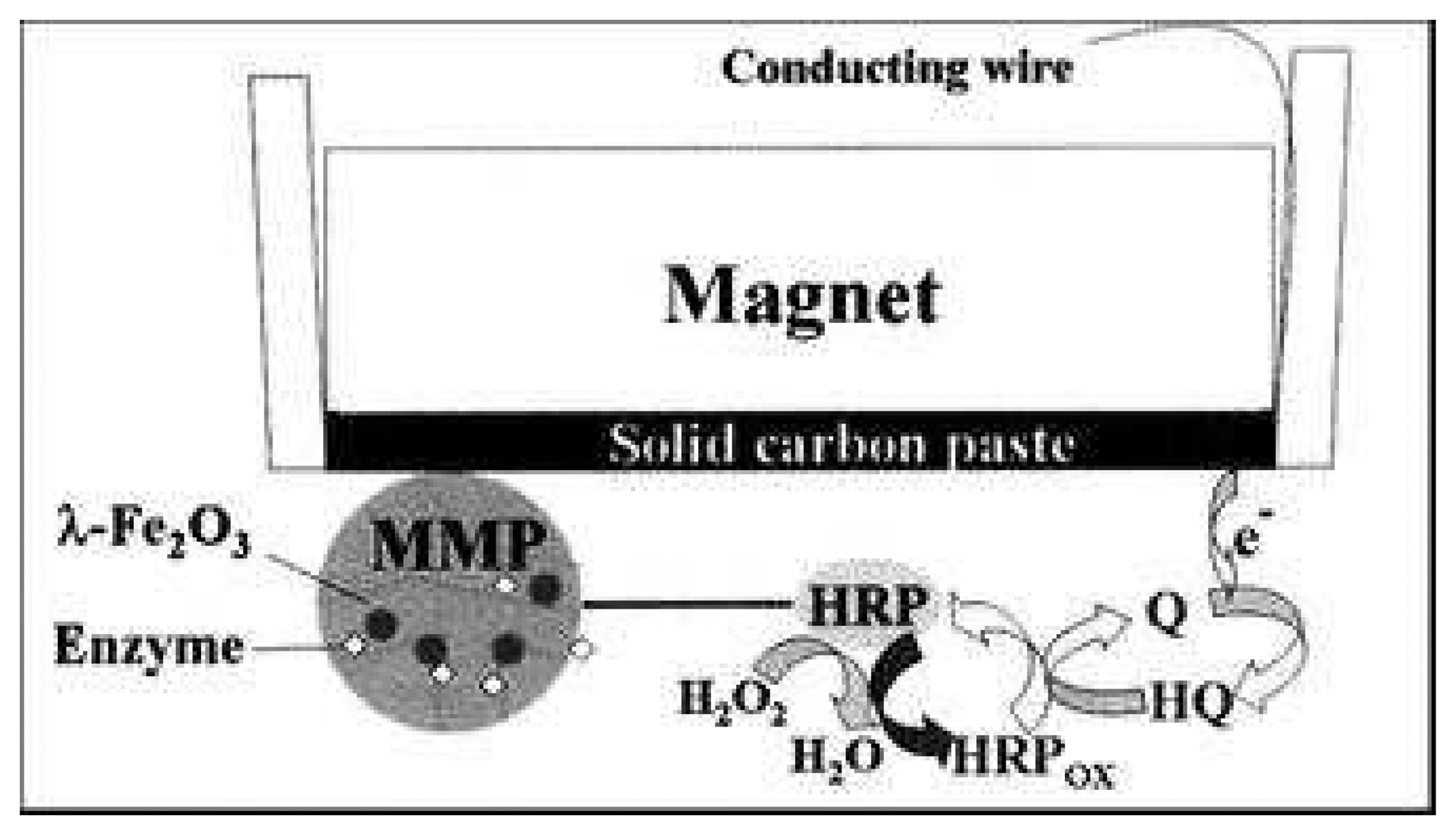
| Phenol | MgFe2O4silica coated ϕ = 120nm | C paste | Tyrosinase | 54,2μA.mM-1 | 10-6 – .5 10-4 M | [3] |
| Ethanol | Precipitated Fe3O4 ϕ = 9.8nm | Screen printed C | Yeast YADH/NAD+ | 0,61μA.mM-1 | 1-9 mM | [34] |
| Glucose | Precipitated Fe3O4 ϕ = 9.8nm | Screen printed C | Glucose oxydase | 1.74 mA mM-1 | 0-33 mM | [35] |
| # | Target | Recognition event | Detection | Lower detection limit | Authors |
|---|---|---|---|---|---|
| 1 | PSA (Prostate Specific Antigen) | Ab/Ag | Scanometric | 30 aM | [6] |
| 2 | PSA | Ab/Ag | Fluorescence | 300 aM | [42] |
| 3 | Cardiac troponin I (cTnI) | Ab/Ag | RT-PCR | 500 fM | [46] |
| 4 | ADDL (Amyloid Beta Derived Diffusible Ligands) | Ab/Ag | Scanometric | 10 aM | [2] |
| 5 | Hepatitis B, Variolas virus, Ebola, HIV | DNA/DNA | Scanometric | 500 fM | [4] |
| 6 | Anthrax lethal factor | DNA/DNA | Scanometric | 500 zM | [5] |
| 7 | Mock RNA | RNA/DNA | Scanometric | 700 aM | [43] |
© 2007 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Jaffrezic-Renault, N.; Martelet, C.; Chevolot, Y.; Cloarec, J.-P. Biosensors and Bio-Bar Code Assays Based on Biofunctionalized Magnetic Microbeads. Sensors 2007, 7, 589-614. https://doi.org/10.3390/s7040589
Jaffrezic-Renault N, Martelet C, Chevolot Y, Cloarec J-P. Biosensors and Bio-Bar Code Assays Based on Biofunctionalized Magnetic Microbeads. Sensors. 2007; 7(4):589-614. https://doi.org/10.3390/s7040589
Chicago/Turabian StyleJaffrezic-Renault, Nicole, Claude Martelet, Yann Chevolot, and Jean-Pierre Cloarec. 2007. "Biosensors and Bio-Bar Code Assays Based on Biofunctionalized Magnetic Microbeads" Sensors 7, no. 4: 589-614. https://doi.org/10.3390/s7040589





