Voltammetric Detection of Antioxidative Properties of Flavonoids Using Electrically Heated DNA Modified Carbon Paste Electrode
Abstract
:Introduction
Experimental
Apparatus and Reagents
Construction of CPE
Preparation of the DNA Biosensor
Procedures at The Evaluation of Antioxidative Properties
Results and Discussion
Voltammetric properties of the flavonoids at DNA/CPE
Antioxidative Effects of Flavonoids
Conclusions
Acknowledgments
References
- Haliwell, B. Food Sci. Agric. Chem. 1999, 1, 67.
- Labuda, J.; Bučková, M.; Jantová, S.; Štepánek, I.; Surugiu, I.; Danielsson, B.; Mascini, M. J. Anal. Chem. 2000, 367, 364.
- Mascini, M.; Palchetti, I.; Marrazza, G. Fres. J. Anal. Chem. 2001, 369, 15.
- Erdem, A.; Ozsoz, M. Electroanalysis 2002, 14, 965.
- Oliveira-Brett, A. M.; Vivan, M.; Fernandes, I. R.; Piedade, J. A. P. Talanta 2002, 56, 959.
- Wang, J.; Kawde, A. N. Anal. Chim. Acta 2001, 431, 219.
- Palecek, E.; Fojta, M. Anal. Chem. 2001, 73, 74A.
- Kubičárová, T.; Fojta, M.; Vidic, J.; Havran, L.; Paleček, E. Electroanalysis 2000, 12, 1422.
- Labuda, J.; Bučková, M.; Vaníčková, M.; Matusch, J.; Wenrich, R. Electroanalysis 1999, 11, 101.
- Bučková, M.; Labuda, J.; Šandula, J.; Križková, L.; Štěpánek, I.; Ďuračková, Z. Talanta 2001, 56, 939.
- Labuda, J.; Buckova, M.; Heilerova, L.; Caniova-Ziakova, A.; Brandsteterova, E.; Mattusch, J.; Wennrich, R. Sensors 2002, 2, 1.
- Gründler, P.; Kirbs., A. Electroanalysis 1999, 11, 223.
- Gründler, P. Fresenius J. Anal. Chem. 2000, 367, 324.
- Korbut, O.; Bučková, M.; Tarapčik, P.; Labuda, J.; Griindler, P. J. Electroanal. Chem. 2001, 506, 143.
- Puppo, A. Phytochemistry 1992, 31, 85.
- Ferguson, L. R. Mut. Res. 2001, 475, 89.
- Ohsima, H.; Yoshie, Y.; Auriol, S.; Gilibert, I. Free Rad Bio Med 1998, 25, 1057.
- Yamashita, N.; Tanemura, H.; Kawanishi, S. Mutat Res 1999, 425, 107.
- Johnson, M. K.; Loo, G. Mutat. Res. 2000, 459, 211.
- Anderson, R. F.; Fisher, L. J.; Hara, Y.; Harris, T.; Mak, W. B.; Melton, L. D.; Packer, J. E. Carcinogenesis 2001, 22, 1189.
- Griindler, P. Fres. J. Anal. Chem. 1998, 362, 180.
- Dollimore, L. S.; Gillerd, R. D. J. Chem. Soc. 1973, 933.
- Oliver, A. Electrochim. Acta 1992, 37, 1945.
- Zerihun, T.; Gruendler, P. J. Electroanal. Chem. 1996, 404, 243.
- Wang, J.; Rivas, G.; Luo, D.; Cai, X.; Valera, F. S.; Dontha, N. Anal. Chem. 1996, 68, 4365.
- Pang, D.-W.; Abruna, H.D. Anal. Chem. 1998, 70, 3162.
- Labuda, J.; Bučková, M.; Heilerová, L.; Šilhár, S.; Štepánek, I. Anal. Bioanal. Chem. submitted.
- Sample Availability: Available from the authors.
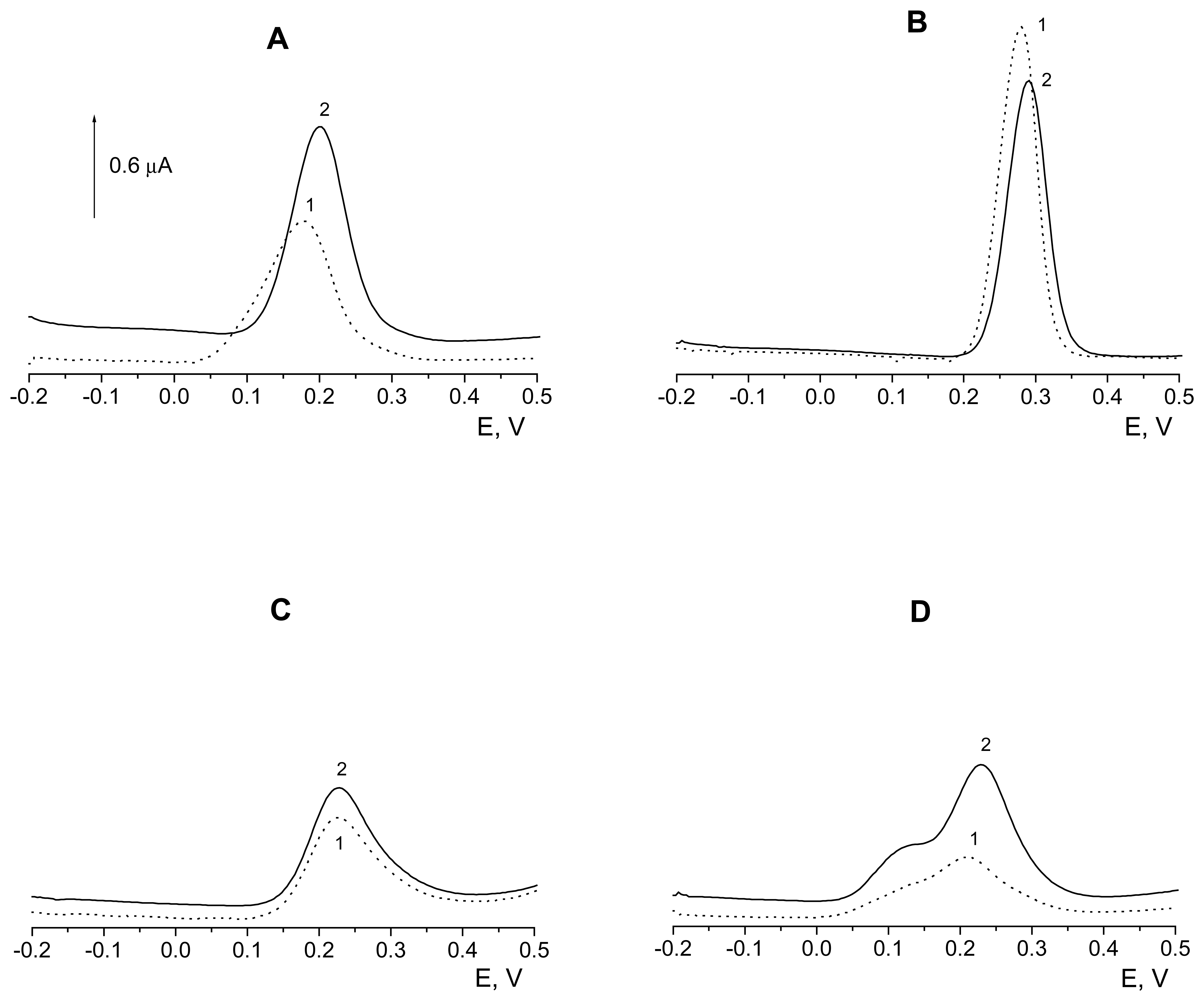
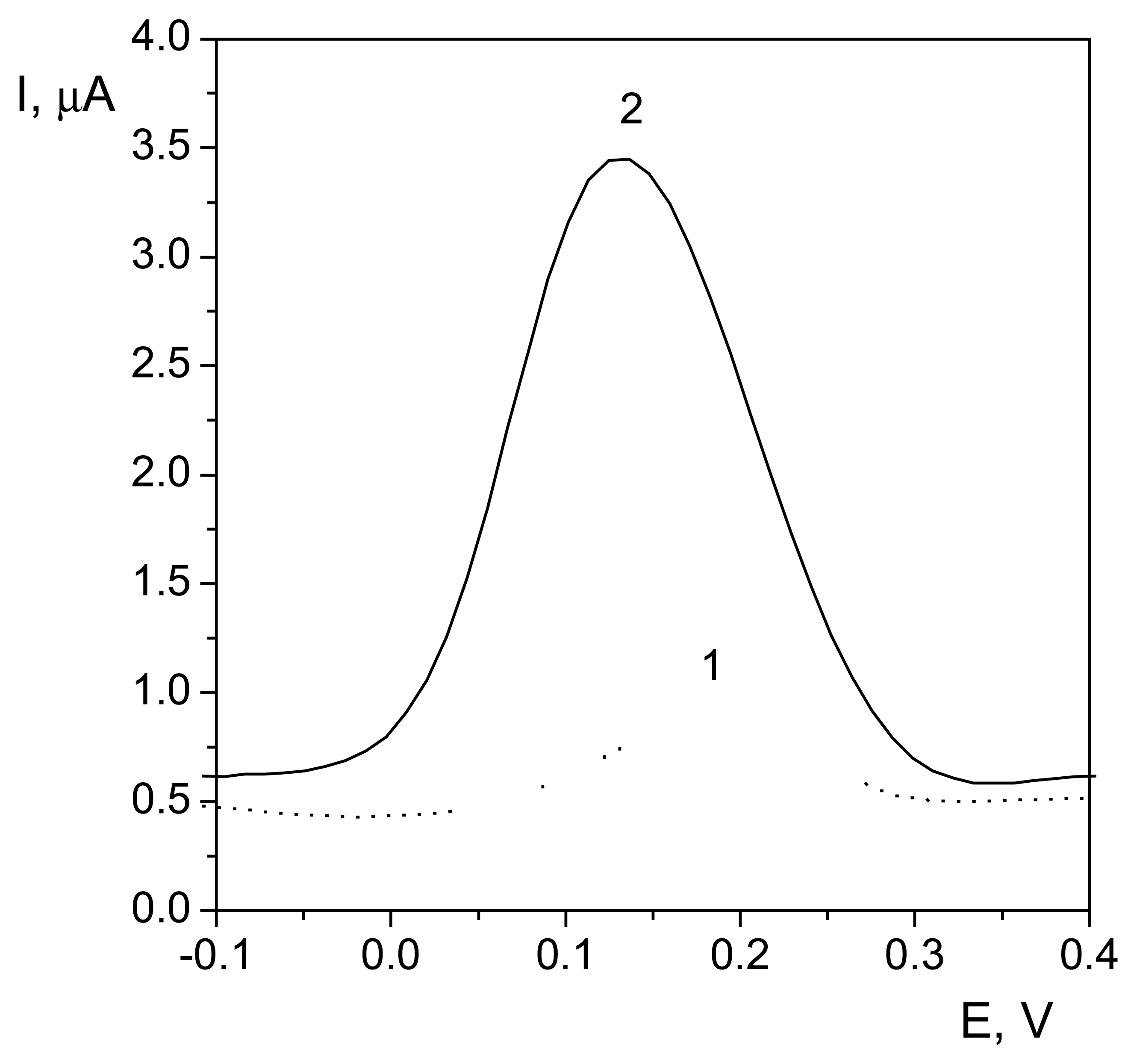
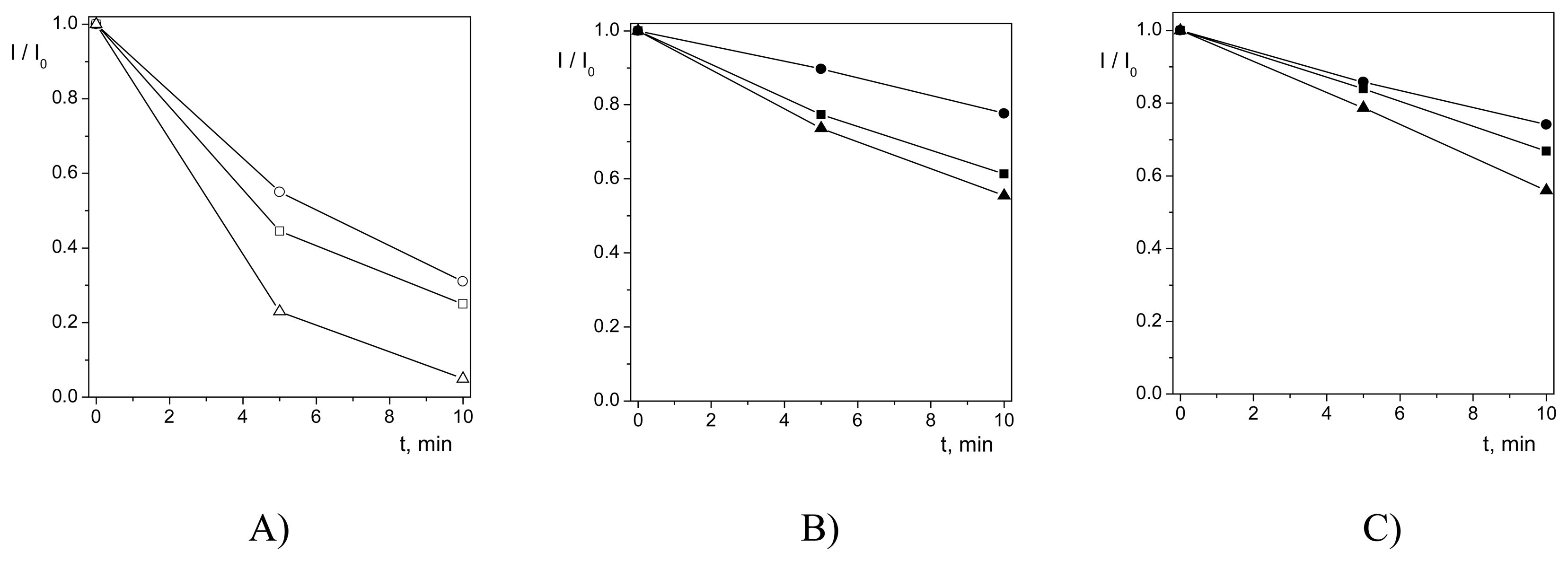
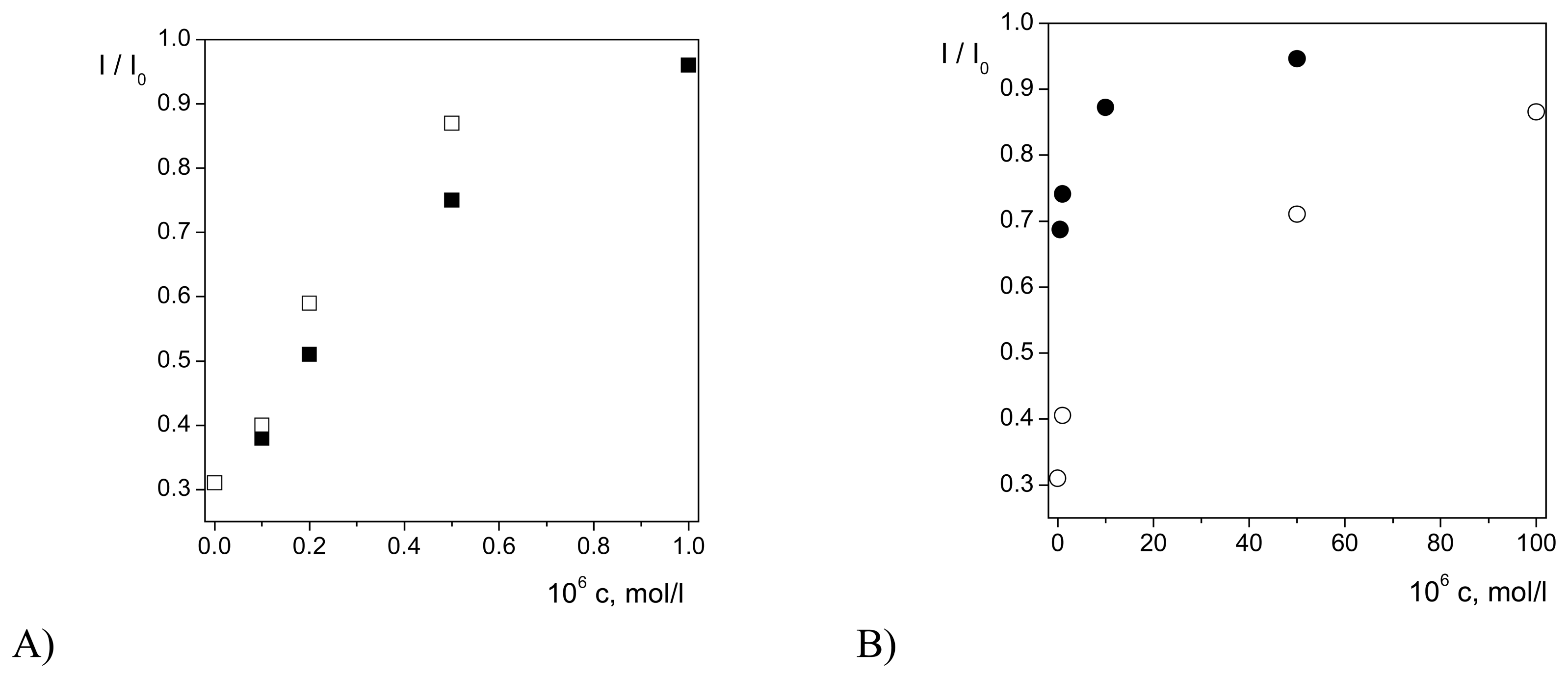

| Flavonoid | Unheated electrode | Heated electrode (t = 38 °C) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CPE | DNA/CPE | CPE | DNA/CPE | ||||||
| Ip, μA | Ep,mV | Ip, μA | Ep,mV | Ip, μa | Ep,mV | Ip, μA | Ep,mV | ||
| 5× l0-7 mol/l Quercetin | A | 0.825 | 0.177 | 1.233 | 0.199 | 0.763 | 0.177 | 1.488 | 0.196 |
| B | 0.259 | 0.177 | 0.593 | 0.199 | 0.240 | 0.188 | 0.450 | 0.199 | |
| C | 0.072 | 0.199 | 0.171 | 0.210 | 0.096 | 0.203 | 0.188 | 0.214 | |
| D | 0.036 | 0.203 | 0.081 | 0.214 | 0.068 | 0.203 | 0.121 | 0.217 | |
| 5× 10-7 mol/l Rutin | A | 1.965 | 0.280 | 1.623 | 0.291 | 2.727 | 0.269 | 2.153 | 0.280 |
| B | 1.897 | 0.276 | 1.534 | 0.291 | 2.415 | 0.276 | 1.768 | 0.287 | |
| C | 0.936 | 0.283 | 0.840 | 0.294 | 1.697 | 0.283 | 1.160 | 0.294 | |
| D | 0.539 | 0.283 | 0.639 | 0.294 | 1.147 | 0,287 | 0.879 | 0.298 | |
| 5× 10-6 mol/l Epigallocatechin gallate | A | 0.341 | 0.210 | 0.794 | 0.228 | 0.381 | 0.196 | 0.892 | 0.214 |
| B | 0.077 | 0.225 | 0.145 | 0.239 | 0.109 | 0.221 | 0.185 | 0.232 | |
| C | 0.033 | 0.243 | 0.035 | 0.254 | 0.026 | 0.243 | 0.044 | 0.250 | |
| D | 0.022 | 0.245 | 0.020 | 0.256 | 0.012 | 0.244 | 0.021 | 0.252 | |
| 5×10-5 mol/l Catechin | A | 0.558 | 0.225 | 0.670 | 0.228 | 0.891 | 0.214 | 1.112 | 0.214 |
| B | 0.343 | 0.225 | 0.429 | 0.225 | 0.454 | 0.217 | 0.604 | 0.217 | |
| C | 0.248 | 0.225 | 0.313 | 0.228 | 0.312 | 0.228 | 0.410 | 0.232 | |
| D | 0.208 | 0.228 | 0.275 | 0.228 | 0.269 | 0.228 | 0.364 | 0.232 | |
2003 by MDPI ( http://www.mdpi.net). Reproduction is permitted for noncommercial purposes.
Share and Cite
Korbut, O.; Bučková, M.; Labuda, J.; Gründler, P. Voltammetric Detection of Antioxidative Properties of Flavonoids Using Electrically Heated DNA Modified Carbon Paste Electrode. Sensors 2003, 3, 1-10. https://doi.org/10.3390/s30100001
Korbut O, Bučková M, Labuda J, Gründler P. Voltammetric Detection of Antioxidative Properties of Flavonoids Using Electrically Heated DNA Modified Carbon Paste Electrode. Sensors. 2003; 3(1):1-10. https://doi.org/10.3390/s30100001
Chicago/Turabian StyleKorbut, O., M. Bučková, J. Labuda, and P. Gründler. 2003. "Voltammetric Detection of Antioxidative Properties of Flavonoids Using Electrically Heated DNA Modified Carbon Paste Electrode" Sensors 3, no. 1: 1-10. https://doi.org/10.3390/s30100001




