A Non-Label and Enzyme-Free Sensitive Detection Method for Thrombin Based on Simulation-Assisted DNA Assembly
Abstract
:1. Introduction
2. Experimental Details
2.1. Materials and Reagents
2.2. Apparatus
2.3. Procedure for Thrombin Assay
2.4. Gel Electrophoresis Analysis
3. Results and Discussion
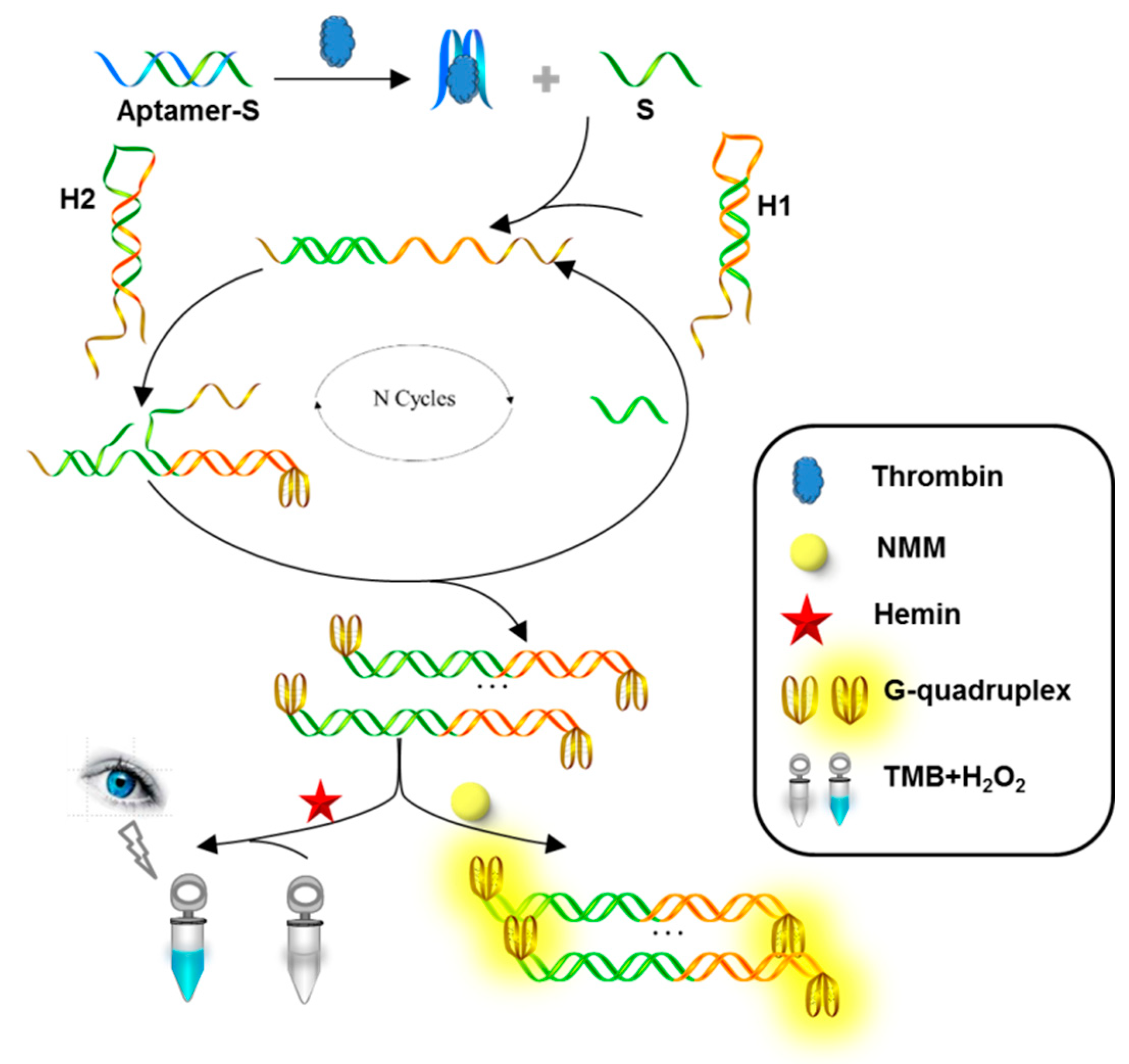
3.1. Principle of Thrombin Detection
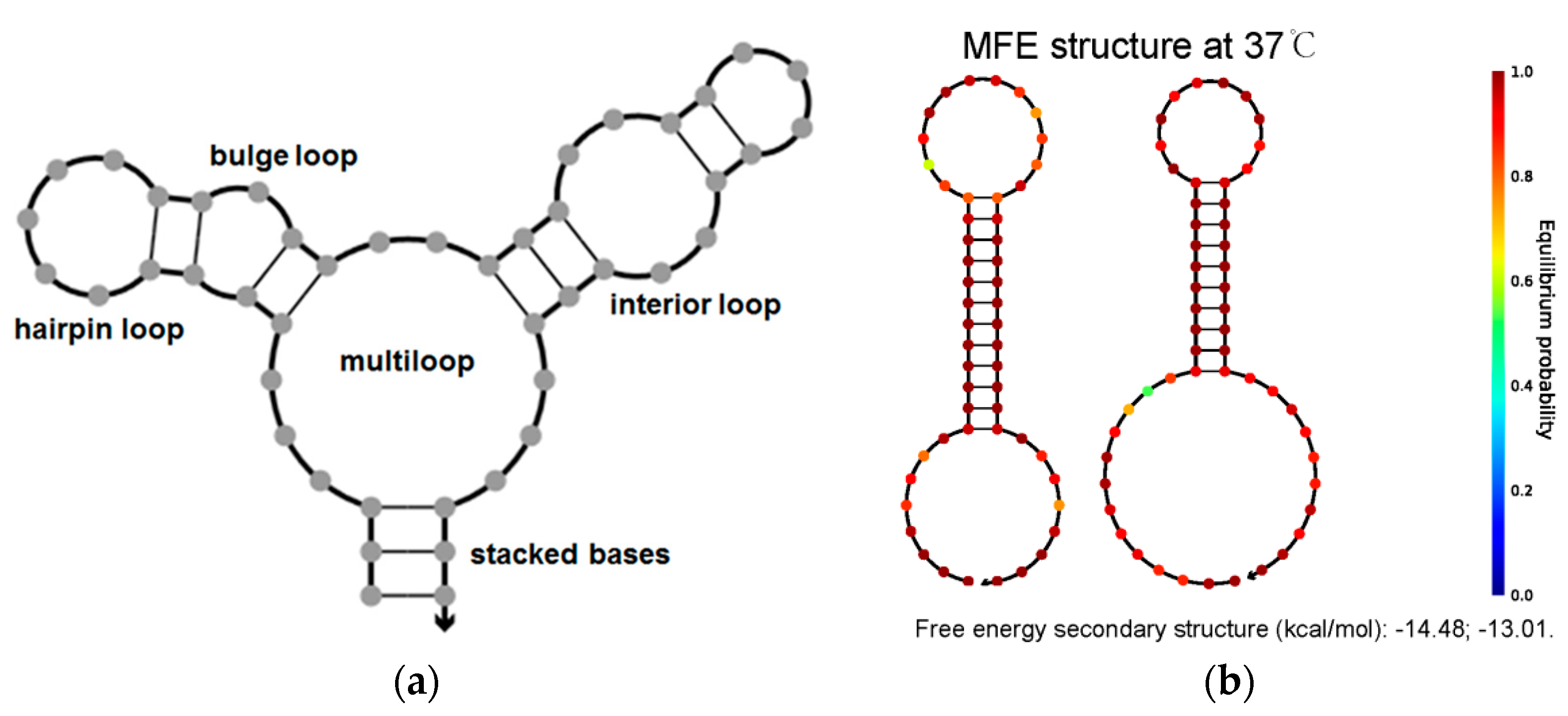
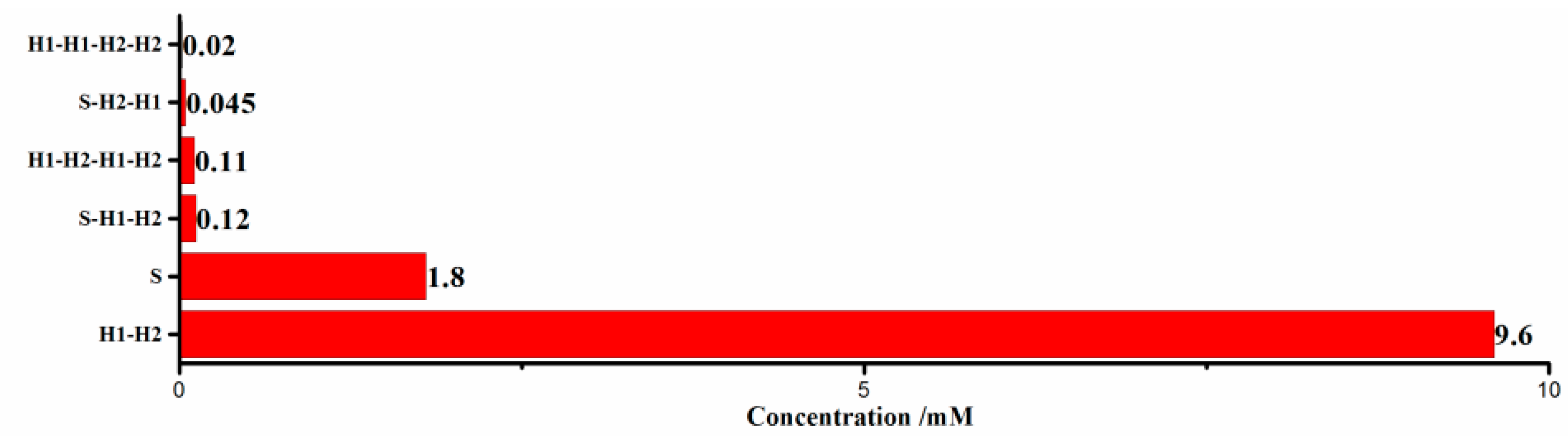
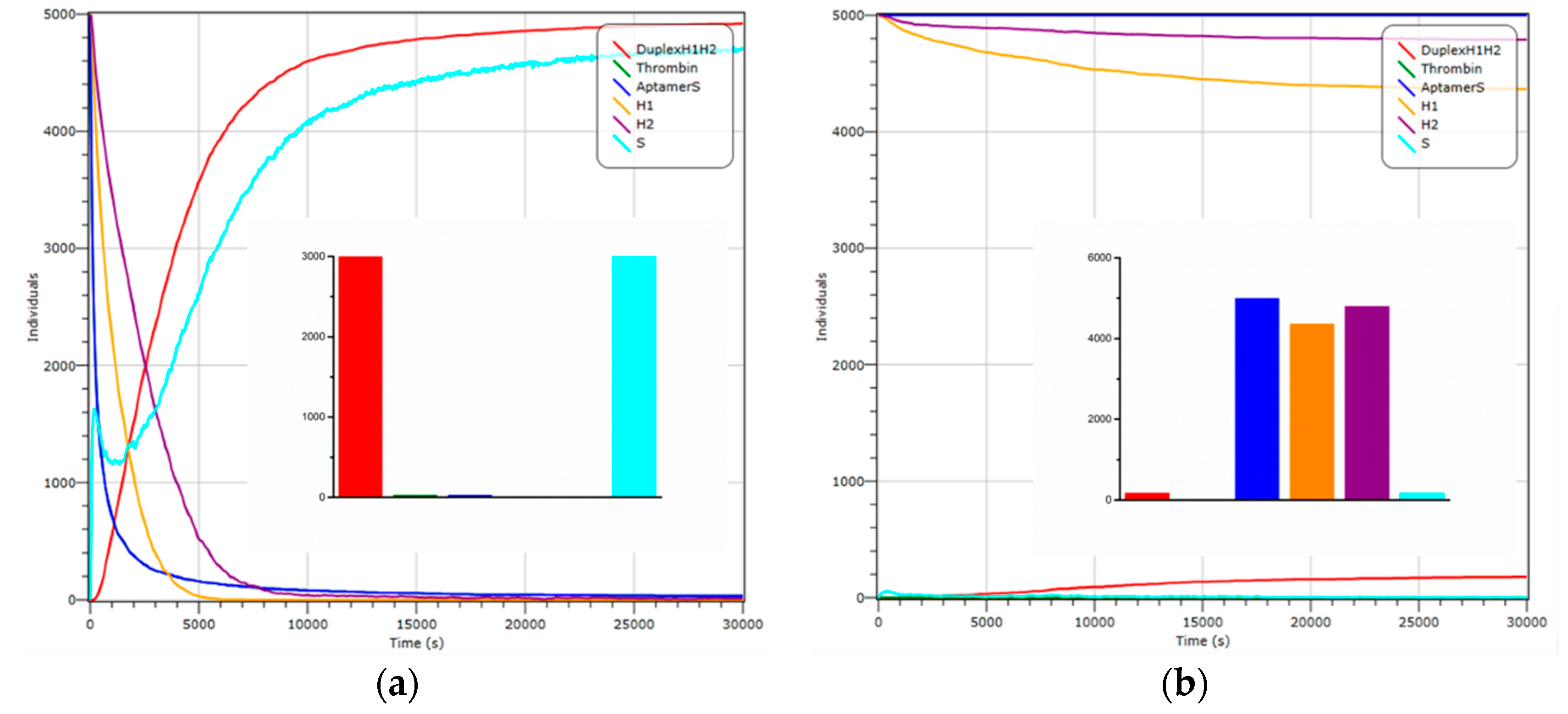
3.2. Simulation Experiment
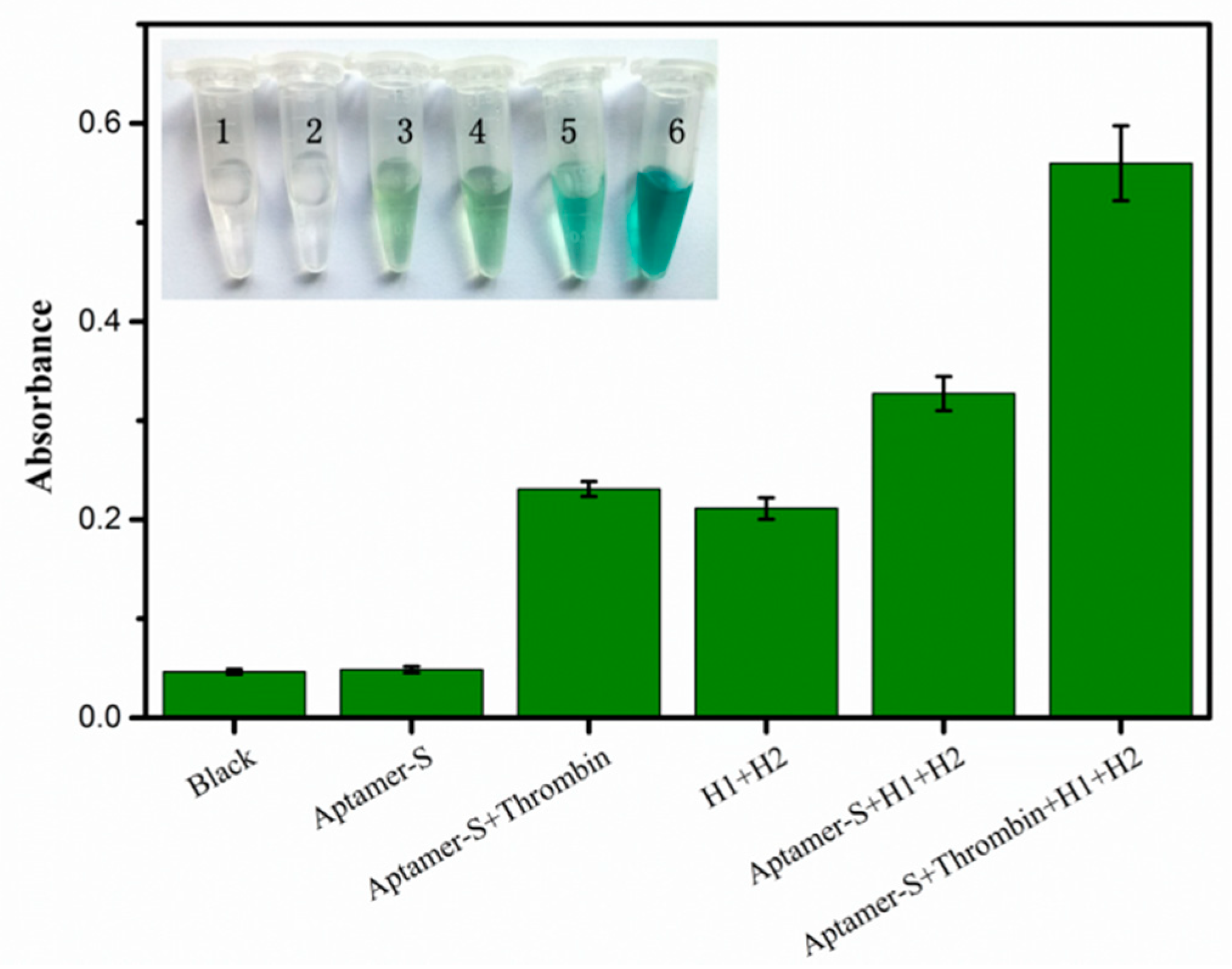
3.3. Feasibility of CHA Amplification Approach for Thrombin Analysis
3.4. Optimization of Detection Condition
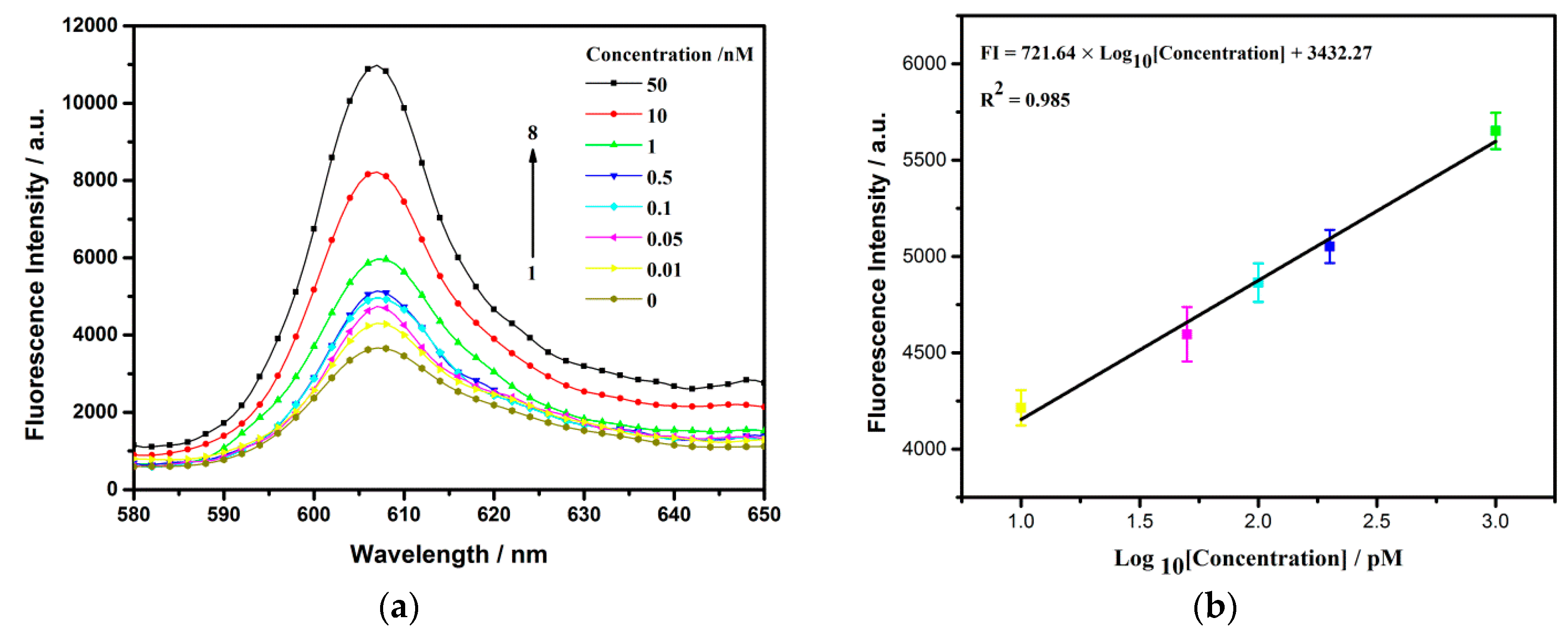
3.5. Sensitive Detection of Thrombin with Fluorescence as Signal Report
3.6. Detection of Thrombin in Bovine Serum
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lowe, C.R. As introduction to the concepts and technology of biosensors. Biosensors 1985, 1, 3–16. [Google Scholar] [CrossRef]
- Scheller, F.W.; Hintsche, R.; Pfeiffer, D.; Schubert, F.; Riedel, K.; Kindervater, R. Biosensors: Fundamentals, applications and trends. Sens. Actuators B Chem. 1991, 4, 197–206. [Google Scholar] [CrossRef]
- Tan, Y.; Guo, Q.; Zhao, X.; Yang, X.; Wang, K.; Huang, J.; Zhou, Y. Proximity-dependent protein detection based on enzyme-assisted fluorescence signal amplification. Biosens. Bioelectron. 2014, 51, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.C.; Spencer, F.A. Thrombin: Structure, Biochemistry, Measurement, and Status in Clinical Medicine. J. Thromb. Thrombolysis 1998, 5, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Xu, J.; Xu, Y.; Xiang, Y.; Yuan, R.; Chai, Y. A universal and label-free aptasensor for fluorescent detection of ATP and thrombin based on SYBR Green I dye. Biosens. Bioelectron. 2013, 42, 193. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, X.; Xie, Y.; Zhao, R.; Tan, C.; Jiang, Y. Label-free fluorescent assays based on aptamer-target recognition. Analyst 2012, 137, 2309. [Google Scholar] [CrossRef] [PubMed]
- Zheng, A.X.; Wang, J.R.; Li, J.; Song, X.R.; Chen, G.N.; Yang, H.H. Enzyme-free fluorescence aptasensor for amplification detection of human thrombin via target-catalyzed hairpin assembly. Biosens. Bioelectron. 2012, 36, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Zhou, X.; Da, X. Sensitive and Homogeneous Protein Detection Based on Target-Triggered Aptamer Hairpin Switch and Nicking Enzyme Assisted Fluorescence Signal Amplification. Anal. Chem. 2012, 84, 3507–3513. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Zhou, X.; Xing, D. Highly sensitive protein detection based on aptamer probe and isothermal nicking enzyme assisted fluorescence signal amplification. Chem. Commun. 2010, 46, 7373–7375. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Qu, L.; Zhang, Q.; Lin, J. Fluorescence detection of thrombin using autocatalytic strand displacement cycle reaction and a dual-aptamer DNA sandwich assay. Anal. Biochem. 2012, 421, 362. [Google Scholar] [CrossRef] [PubMed]
- Assicot, M.; Bohuon, C. Presence of two distinct catechol-O-methyltransferase activities in red blood cells. Biochimie 1971, 53, 871–874. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, M.T.; Wan, Q.H.; Boulet, C.A.; Le, X.C. Competitive immunoassay for staphylococcal enterotoxin A using capillary electrophoresis with laser-induced fluorescence detection. J. Chromatogr. A 1999, 853, 545–553. [Google Scholar] [CrossRef]
- Baker, K.N.; Rendall, M.H.; Patel, A.; Boyd, P.; Hoare, M.; Freedman, R.B.; James, D.C. Rapid monitoring of recombinant protein products: A comparison of current technologies. Trends Biotechnol. 2002, 20, 149–156. [Google Scholar] [CrossRef]
- Yin, P.; Choi, H.M.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.S.; Yoon, M.; Baeg, J.O.; Kim, J. Label-free dual assay of DNA sequences and potassium ions using an aptamer probe and a molecular light switch complex. Chem. Commun. 2009, 47, 7419–7421. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Cao, J.; Jiang, W.; Wang, L. A novel enzyme-free and label-free fluorescence aptasensor for amplified detection of adenosine. Biosens. Bioelectron. 2013, 44, 52. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, W.; Zhou, M.; Xiang, Y.; Yuan, R.; Chai, Y. Toehold strand displacement-driven assembly of G-quadruplex DNA for enzyme-free and non-label sensitive fluorescent detection of thrombin. Biosens. Bioelectron. 2015, 64, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Guo, Z.; Wang, J.; Wang, E. G-quadruplex as signal transducer for biorecognition events. Curr. Pharm. Des. 2012, 18, 2076–2095. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wang, W.; Wang, M.; Kang, T.S.; Lu, J.J.; Chen, X.P.; Han, Q.B.; Leung, C.H.; Ma, D.L. A luminescent G-quadruplex-selective iridium(III) complex for the label-free detection of lysozyme. J. Mater. Chem. B 2016, 4, 2407–2411. [Google Scholar] [CrossRef]
- Lu, L.; Mao, Z.; Kang, T.S.; Leung, C.H.; Ma, D.L. A versatile nanomachine for the sensitive detection of platelet-derived growth factor-BB utilizing a G-quadruplex-selective iridium(III) complex. Biosens. Bioelectron. 2016, 85, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Huang, R.; Zhou, Y.; Zhang, M.; Deng, M.; Wang, X.; Yan, S. Aptamer-based turn-on fluorescent four-branched quaternary ammonium pyrazine probe for selective thrombin detection. Chem. Commun. 2011, 47, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Ji, D.; Shang, J.; Hu, Y.; Gao, J.; Lin, Z.; Ge, J.; Li, Z. A rapid biosensor for highly sensitive protein detection based on G-quadruplex-Thioflavin T complex and terminal protection of small molecule-linked DNA. Sens. Actuators B Chem. 2017, 252, 1146–1152. [Google Scholar] [CrossRef]
- Li, H.; Ren, J.; Liu, Y.; Wang, E. Application of DNA machine in amplified DNA detection. Chem. Commun. 2014, 50, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ren, J.; Fan, D.; Liu, Y.; Wang, E. Integration of graphene oxide and DNA as a universal platform for multiple arithmetic logic units. Chem. Commun. 2014, 50, 14390. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, Y.; Zheng, J.; Zhong, W.; Shen, G.; Yang, R.; Tan, W. Aptamer degradation inhibition combined with DNAzyme cascade-based signal amplification for colorimetric detection of proteins. Chem. Commun. 2013, 49, 6137–6139. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Z.; Lin, H.; Nie, Z.; Chen, J.; Xu, X.; Yao, S. Label-Free Colorimetric Assay for Methyltransferase Activity Based on a Novel Methylation-Responsive DNAzyme Strategy. Anal. Chem. 2010, 82, 1935–1941. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, K.; Liu, Y.; Wang, H.; Wu, J.; Zhu, F.; Zou, P. Binding-induced and label-free colorimetric method for protein detection based on autonomous assembly of hemin/G-quadruplex DNAzyme amplification strategy. Biosens. Bioelectron. 2015, 64, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Kosman, J.; Juskowiak, B. Peroxidase-mimicking DNAzymes for biosensing applications: A review. Anal. Chim. Acta 2011, 707, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhu, J.; Li, T.; Wang, E. Bifunctional colorimetric oligonucleotide probe based on a G-quadruplex DNAzyme molecular beacon. Anal. Chem. 2011, 83, 8871–8876. [Google Scholar] [CrossRef] [PubMed]
- McCaskill, J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990, 29, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.M.; Bois, J.S.; Schaeffer, J.M.; Winfree, E.; Pierce, N.A. Thermodynamic Analysis of Interacting Nucleic Acid Strands. Siam Rev. 2007, 49, 65–88. [Google Scholar] [CrossRef] [Green Version]
- Lakin, M.R.; Simon, Y.; Filippo, P.; Stephen, E.; Andrew, P. Visual DSD: a design and analysis tool for DNA strand displacement systems. Bioinformatics 2011, 27, 3211–3213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded-dna molecules that bind and inhibit human thrombin nature. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Jang, H.; Park, K.S.; Park, H.G. A label-free and enzyme-free signal amplification strategy for a sensitive RNase H activity assay. Nanoscale 2017, 9, 16149–16153. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, I.; Borer, P.N.; Dengler, B.; Levine, M.D.; Uhlenbeck, O.C.; Crothers, D.M.; Gralla, J. Improved Estimation of Secondary Structure in Ribonucleic Acids. Nature 1971, 230, 362. [Google Scholar] [CrossRef] [PubMed]
- Waterman, M.S. Secondary Structure of Single-Stranded Nucleic Acidst. Stud. Found. Comb. Adv. Math. Suppl. Stud. 1978, 1, 167–212. [Google Scholar]
- Dirks, R.M.; Pierce, N.A. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. 2008, 25, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.M.; Pierce, N.A. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 2003, 24, 1664–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, B.R.; Pierce, N.A. Sequence Design for a Test Tube of Interacting Nucleic Acid Strands. ACS Synthetic Biol. 2015, 4, 1086–1100. [Google Scholar] [CrossRef] [PubMed]
- Gallian, J.A. Contemporary Abstract Algebra, 2nd ed.; Cengage Learning Emea: London, UK, 1998; Volume 24, pp. 2309–2316. [Google Scholar]



| Name | Sequences (5′–3′) |
|---|---|
| Aptamer | GGT TGG TGT GGT TGG AAT AGT C |
| S | AGT CAC ACG GAC TAT TCC AA CC |
| H1 | GGG TAA TAG TCC GTG TGA CTA TGG ACT ATA GGA GTC ACA CGG GCG GGT AGG G |
| H2 | GGG TTG TAT GAC TCC TAT AGT CCA TAG TCA CAC GGA CTA TTG GGC GGG TAG GG |
| Analytical Method | Dynamic Range | Detection Limit | References |
|---|---|---|---|
| Fluorescence detection | 0–500 nM | 1.1 nM | [5] |
| Fluorescence detection | 0.02–2 nM | 20 pM | [7] |
| Fluorescence detection | 0–1 nM | 100 pM | [8] |
| Colorimetric detection | 2.5–2500 pM | 1.9 pM | [29] |
| Fluorescence detection | 0.01–50 nM | 2.4 pM | Our method |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, L.; Wang, Y.; Dong, Y. A Non-Label and Enzyme-Free Sensitive Detection Method for Thrombin Based on Simulation-Assisted DNA Assembly. Sensors 2018, 18, 2179. https://doi.org/10.3390/s18072179
Zhang Y, Wang L, Wang Y, Dong Y. A Non-Label and Enzyme-Free Sensitive Detection Method for Thrombin Based on Simulation-Assisted DNA Assembly. Sensors. 2018; 18(7):2179. https://doi.org/10.3390/s18072179
Chicago/Turabian StyleZhang, Yingying, Luhui Wang, Yanan Wang, and Yafei Dong. 2018. "A Non-Label and Enzyme-Free Sensitive Detection Method for Thrombin Based on Simulation-Assisted DNA Assembly" Sensors 18, no. 7: 2179. https://doi.org/10.3390/s18072179




