Partial Least Square Discriminant Analysis Based on Normalized Two-Stage Vegetation Indices for Mapping Damage from Rice Diseases Using PlanetScope Datasets
Abstract
:1. Introduction
2. Materials and Methods
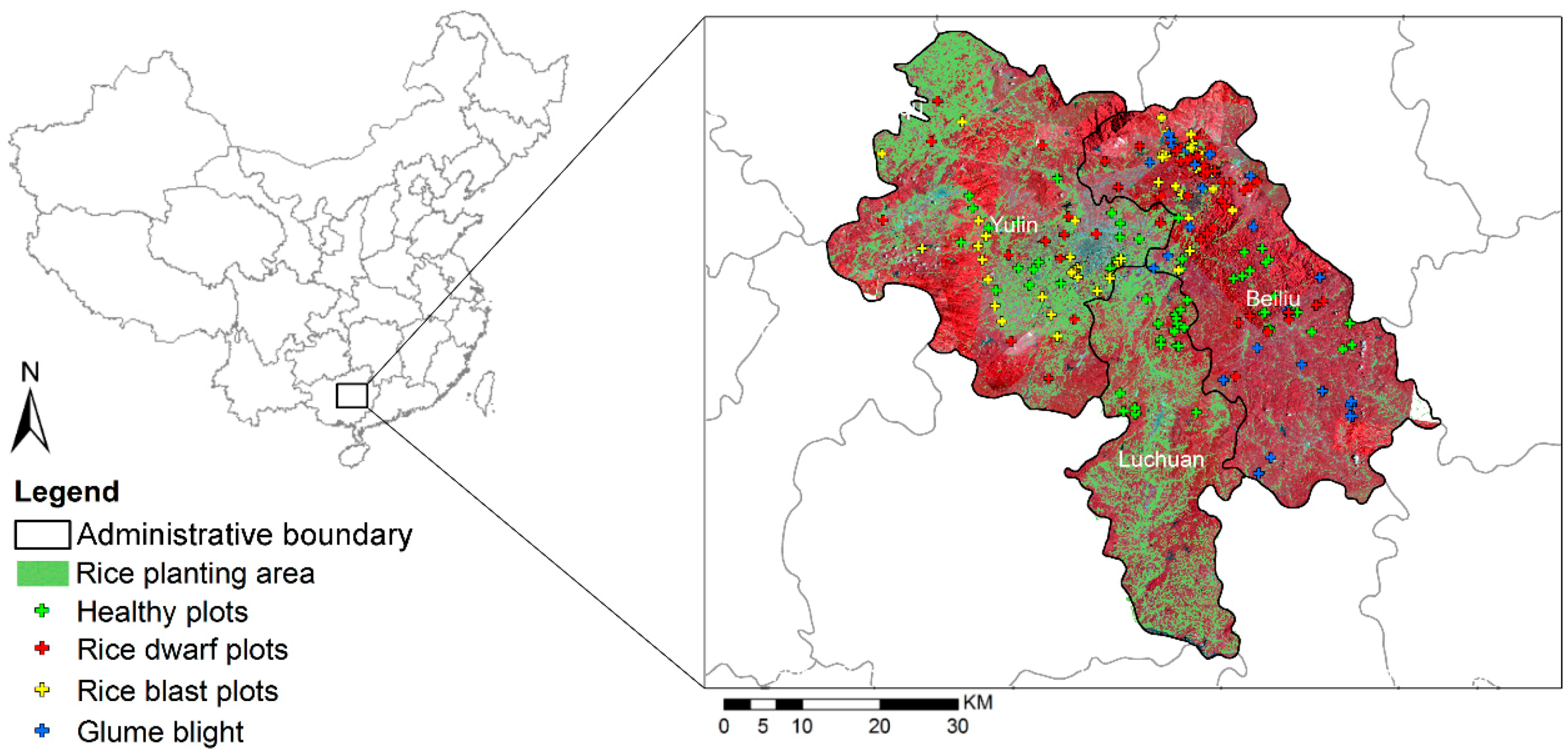
2.1. Study Area
2.2. Satellite Data
2.3. Field Investigation
2.4. Mapping Various Rice Disease Infested Areas
2.4.1. Spectral Features for Mapping Diseases
2.4.2. Normalized Two-Stage Vegetation Indices
2.4.3. The Sensitivity of the Identified Spectral Features to Rice Diseases
2.4.4. Diseases Occurrence Mapping Using Partial Least Squares Discriminant Analysis (PLS-DA)
3. Results
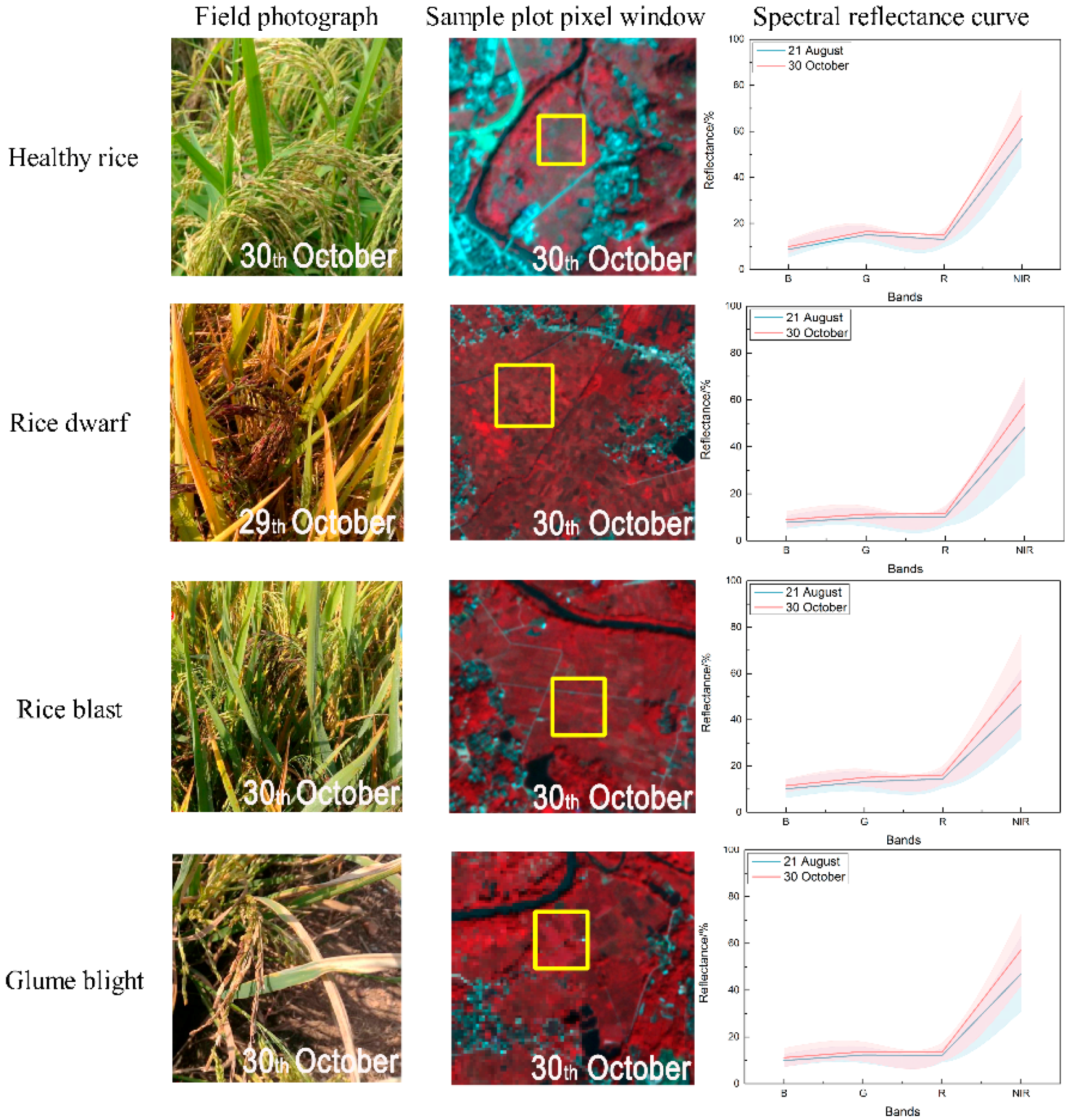
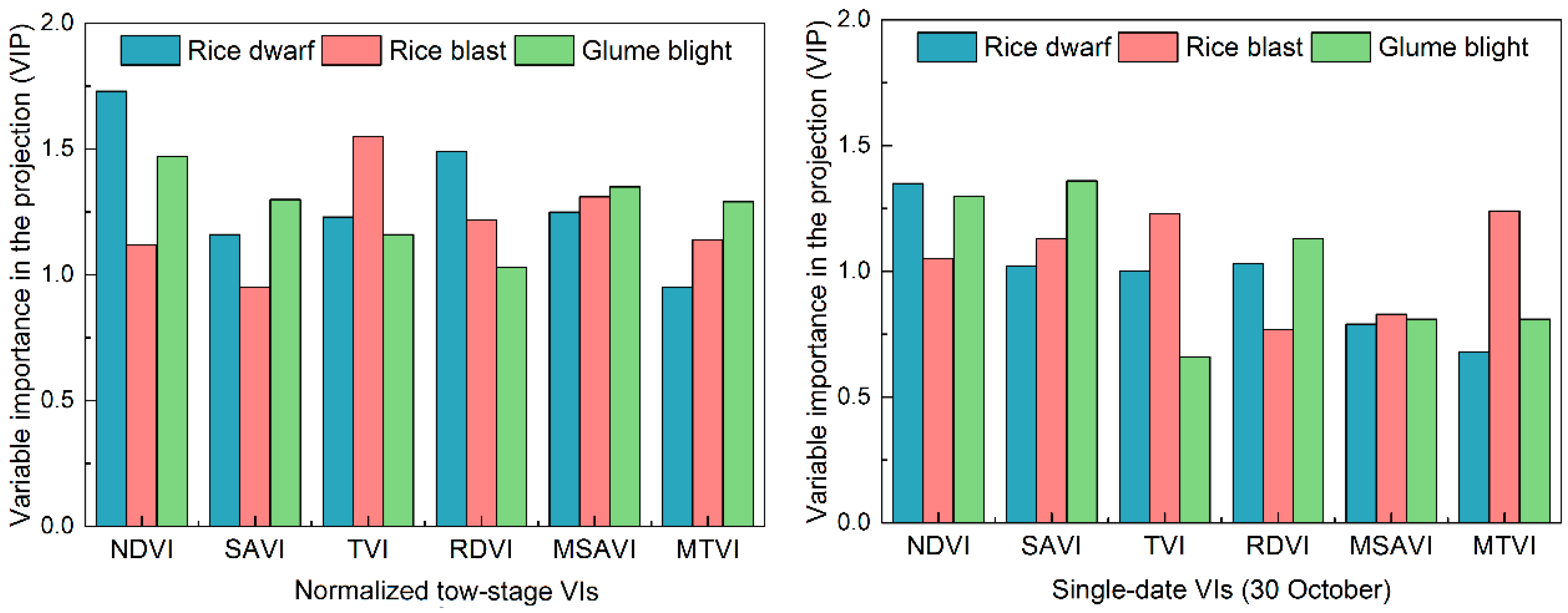
3.1. Responses of Spectral Features to Different Infestations
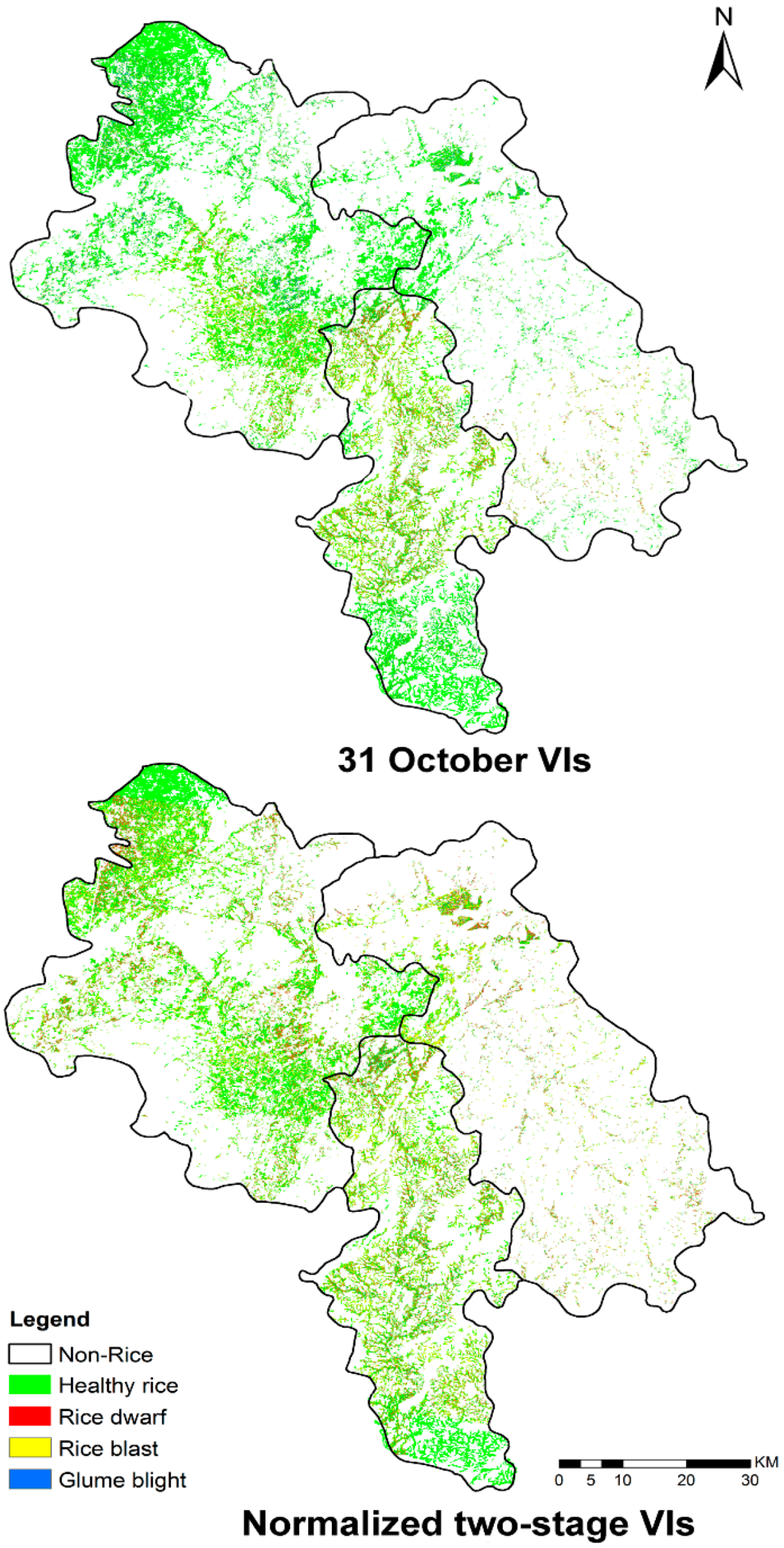
3.2. Mapping Disease Infestations with PL Satellite Imagery
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greer, C.A.; Webster, R.K. Occurrence, distribution, epidemiology, cultivar reaction, and management of rice blast disease in California. Plant Dis. 2007, 85, 1096–1102. [Google Scholar] [CrossRef]
- Jin, X.H.; Guo, Y.X.; Zheng, W.; Tai, L.M.; Zhang, Y.L. The occurrence characteristic and analysis of the trends in rice blast disease in 2007 in Heilongjiang province. North Rice 2007, 2, 57–61. [Google Scholar]
- Wang, H.D.; Chen, J.P.; Wang, A.G.; Jiang, X.H.; Adams, M.J. Studies on the epidemiology and yield losses from rice black-streaked dwarf disease in a recent epidemic in Zhejiang province, China. Plant Pathol. 2009, 58, 815–825. [Google Scholar] [CrossRef] [Green Version]
- Khush, G.S.; Pollard, L.R. Multiple disease and insect resistance for increased yield stability in rice. In Progress in Irrigated Rice Research; FAO: Rome, Italy, 1989; pp. 79–92. [Google Scholar]
- Bonman, J.M.; Estrada, B.A.; Kim, C.K.; Lee, E.J. Assessment of blast disease and yield loss in susceptible and partially resistant rice cultivars in two irrigated lowland environments. Plant Dis. 1991, 75, 462. [Google Scholar] [CrossRef]
- Hammond, N.E.B.; Hardie, D.; Hauser, C.E.; Reid, S.A. Can general surveillance detect high priority pests in the Western Australian Grains Industry? Crop Prot. 2016, 79, 8–14. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.; Richard, K.; Landmann, T.; Thiel, M.; Nyasani, J.; Le Ru, B.P. Maize pest and disease mapping using RapidEye-based vegetation indices and machine learning methods. In Proceedings of the 11th International Conference of the African Association of Remote Sensing of the Environment (AARSE), Kampala, Uganda, 24–28 October 2016; pp. 24–28. [Google Scholar]
- Yuan, L.; Bao, Z.; Zhang, H.; Zhang, Y.; Liang, X. Habitat monitoring to evaluate crop disease and pest distributions based on multi-source satellite remote sensing imagery. Opt. Int. J. Light Electron Opt. 2017, 145, 66–73. [Google Scholar] [CrossRef]
- Silva, J.R.M.D.; Damásio, C.V.; Sousa, A.M.O.; Bugalho, L.; Pessanha, L.; Quaresma, P. Agriculture pest and disease risk maps considering MSG satellite data and land surface temperature. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Guo, N.; Sha, S.; Die, H.U.; Wang, W.; Deng, Z.; Liu, W. Effect of Mixed Pixel on Monitoring Ability of Remote Sensing Drought Index. J. Arid Meteorol. 2016, 16, 407–426. [Google Scholar]
- Savage, S.L.; Lawrence, R.L.; Squires, J.R. Predicting relative species composition within mixed conifer forest pixels using zero-inflated models and Landsat imagery. Remote Sens. Environ. 2015, 171, 326–336. [Google Scholar] [CrossRef]
- Maglione, P. Very High Resolution optical satellites: An overview of the most commonly used. Am. J. Appl. Sci. 2016, 13, 91–99. [Google Scholar] [CrossRef]
- Houborg, R.; Mccabe, M.F. High-Resolution NDVI from Planet’s Constellation of Earth Observing Nano-Satellites: A New Data Source for Precision Agriculture. Remote Sens. 2016, 8, 768. [Google Scholar] [CrossRef]
- Kääb, A.; Altena, B.; Mascaro, J. Coseismic displacements of the 14 November 2016 Mw 7.8 Kaikoura, New Zealand, earthquake using the Planet optical cubesat constellation. Nat. Hazards Earth Syst. Sci. 2017, 17, 1–18. [Google Scholar] [CrossRef]
- Baloloy, A.; Blanco, A.; Candido, C.; Argamosa, R.; Dumalag, J.; Dimapilis, L.; Paringit, E. Estimation of mangrove forest aboveground biomass using multispectral bands, vegetation indices and biophusical variables derived from optical satellite imageries: Repideyes, PlanetScope, and Sentinel-2. ISPRS Ann. Photogramm. Remote Sens Spat. Inf. Sci. 2018, 4, 41–45. [Google Scholar]
- Marta, A.D.; Grifoni, D.; Mancini, M.; Orlando, F.; Guasconi, F.; Orlandini, S. Durum wheat in-field monitoring and early-yield prediction: Assessment of potential use of high resolution satellite imagery in a hilly area of Tuscany, Central Italy. J. Agric. Sci. 2015, 153, 68–77. [Google Scholar] [CrossRef]
- Helmholz, P.; Rottensteiner, F.; Heipke, C. Semi-automatic verification of cropland and grassland using very high resolution mono-temporal satellite images. ISPRS J. Photogramm. Remote Sens. 2014, 97, 204–218. [Google Scholar] [CrossRef]
- Ursani, A.A.; Kpalma, K.; Lelong, C.C.D.; Ronsin, J. Fusion of Textural and Spectral Information for Tree Crop and Other Agricultural Cover Mapping with Very-High Resolution Satellite Images. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 225–235. [Google Scholar] [CrossRef]
- Shen, J.; Liu, J.; Lin, X.; Zhao, R.; Xu, S. Cropland Extraction from Very High Spatial Resolution Satellite Imagery by Object-Based Classification Using Improved Mean Shift and One-Class Support Vector Machines. Sens. Lett. 2011, 9, 997–1005. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, W.; Luo, J.; Huang, L.; Zhou, X. Detection and discrimination of pests and diseases in winter wheat based on spectral indices and kernel discriminant analysis. Comput. Electron. Agric. 2017, 141, 171–180. [Google Scholar] [CrossRef]
- Whiteside, T.; Bartolo, R. Mapping Aquatic Vegetation in a Tropical Wetland Using High Spatial Resolution Multispectral Satellite Imagery. Remote Sens. 2015, 7, 11664–11694. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Huang, Y.; Yuan, L.; Yang, G.; Chen, L.; Zhao, C. Using satellite multispectral imagery for damage mapping of armyworm (Spodoptera frugiperda) in maize at a regional scale. Pest Manag. Sci. 2016, 72, 335. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.A.; Galvão, L.S.; Santos, J.R. Relationships between Hyperion-derived vegetation indices, biophysical parameters, and elevation data in a Brazilian savannah environment. Remote Sens. Lett. 2010, 1, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.; Meng, J.; Shang, J.; Liu, J.; Wu, B.; Huffman, T. Modified vegetation index for estimating crop fraction of absorbed photosynthetically active radiation. Int. J. Remote Sens. 2015, 36, 3097–3113. [Google Scholar] [CrossRef]
- Houborg, R.; Soegaard, H.; Boegh, E. Combining vegetation index and model inversion methods for the extraction of key vegetation biophysical parameters using Terra and Aqua MODIS reflectance data. Remote Sens. Environ. 2007, 106, 39–58. [Google Scholar] [CrossRef]
- Raper, T.B.; Varco, J.J.; Hubbard, K.J. Canopy-Based Normalized Difference Vegetation Index Sensors for Monitoring Cotton Nitrogen Status. Agron. J. 2013, 105, 1345. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Zhang, J.; Nie, C.; Wei, L.; Yang, G.; Wang, J. Selection of Spectral Channels for Satellite Sensors in Monitoring Yellow Rust Disease of Winter Wheat. Intell. Autom. Soft Comput. 2013, 19, 501–511. [Google Scholar] [CrossRef]
- Lehnert, L.W.; Meyer, H.; Wang, Y.; Miehe, G.; Thies, B.; Reudenbach, C.; Bendix, J. Retrieval of grassland plant coverage on the Tibetan Plateau based on a multi-scale, multi-sensor and multi-method approach. Remote Sens. Environ. 2015, 164, 197–207. [Google Scholar] [CrossRef]
- Feng, T.; Brandt, M.; Liu, Y.Y.; Verger, A.; Tagesson, T.; Diouf, A.A.; Rasmussen, K.; Mbow, C.; Wang, Y.; Fensholt, R. Remote sensing of vegetation dynamics in drylands: Evaluating vegetation optical depth (VOD) using AVHRR NDVI and in situ green biomass data over West African Sahel. Remote Sens. Environ. 2016, 177, 265–276. [Google Scholar] [Green Version]
- Zhao, K.; Valle, D.; Popescu, S.; Zhang, X.; Mallick, B. Hyperspectral remote sensing of plant biochemistry using Bayesian model averaging with variable and band selection. Remote Sens. Environ. 2013, 132, 102–119. [Google Scholar] [CrossRef]
- Shao, T.T.; Song, K.S.; Jia, D.U.; Yang, G.; Zeng, L.H.; Lei, X.C.; Wu, Y.Q. Hyperspectral Remote Sensing Modeling of FPAR for Corn Based on Partial Least Squares (PLS) Regression Analysis. Geogr. Geo-Inf. Sci. 2012, 3, 9. [Google Scholar]
- Gil, A.; Yu, Q.; Lobo, A.; Lourenço, P.; Silva, L.; Calado, H. Assessing the effectiveness of high resolution satellite imagery for vegetation mapping in small islands protected areas. J. Coast. Res. 2011, 64, 1663–1667. [Google Scholar]
- White, J.C.; Wulder, M.A.; Grills, D. Detecting and mapping mountain pine beetle red-attack damage with SPOT5 10-m multispectral imagery. BC J. Ecosyst. Manag. 2006, 7, 105–118. [Google Scholar]
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
- Song, K.; Lu, D.; Li, L.; Li, S.; Wang, Z.; Du, J. Remote sensing of chlorophyll- a concentration for drinking water source using genetic algorithms (GA)-partial least square (PLS) modeling. Ecol. Inf. 2012, 10, 25–36. [Google Scholar] [CrossRef]
- Yin, X.J.; Zhao, S.F. Remote Sensing Estimation of Pigment Contents in Diseased Leaves Using PLS Algorithm. Remote Sens. Inf. 2012, 27, 50–56. [Google Scholar]
- Song, S. PLS Wavelength Selection by Hyperspectral Remote Sensing in Rice. Geomat. Inf. Sci. Wuhan Univ. 2010, 35, 219–223. [Google Scholar]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Investigation of fungal development in maize kernels using NIR hyperspectral imaging and multivariate data analysis. J. Cereal Sci. 2012, 55, 272–278. [Google Scholar] [CrossRef]
- Laurin, G.V.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Frate, F.D.; Guerriero, L.; Pirotti, F.; Valentini, R. Above ground biomass estimation in an African tropical forest with lidar and hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Peerbhay, K.Y.; Mutanga, O.; Ismail, R. Commercial tree species discrimination using airborne AISA Eagle hyperspectral imagery and partial least squares discriminant analysis (PLS-DA) in KwaZulu–Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2013, 79, 19–28. [Google Scholar] [CrossRef]
- Wilson, N.; Greenberg, J.; Jumpasut, A.; Collison, A. In-Orbit Radiometric Calibration of the Planet Dove Constellation. In Proceedings of the Conference on Characterization and Radiometric Calibration for Remote Sensing (CALCON), Logan, UT, USA, 21–24 August 2017. [Google Scholar]
- Traganos, D.; Cerra, D.; Reinartz, P. Cubesat-Derived Detection of Seagrasses Using Planet Imagery Following Unmixing-Based Denoising: Is Small the Next Big? ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, 42, 283–287. [Google Scholar] [CrossRef]
- Fukushi, T.; Shikata, E.; Kimura, I. Some morphological characters of rice dwarf virus. Virology 1962, 18, 192–205. [Google Scholar] [CrossRef]
- Phadikar, S. Classification of Rice Leaf Diseases Based on Morphological Changes. Int. J. Geogr. Inf. Sci. 2012, 2, 61–66. [Google Scholar]
- Phadikar, S.; Sil, J.; Das, A.K. Rice diseases classification using feature selection and rule generation techniques. Comput. Electron. Agric. 2013, 90, 76–85. [Google Scholar] [CrossRef]
- Rouse J.W., Jr; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with Erts. Nasa Spec. Publ. 1974, 351, 309–313. [Google Scholar]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- De Tomás, A.; Nieto, H.; Guzinski, R.; Mendiguren, G.; Sandholt, I.; Berline, P. In Multi-scale approach of the surface temperature/vegetation index triangle method for estimating evapotranspiration over heterogeneous landscapes. EGU Gen. Assem. 2012, 101, 131–138. [Google Scholar]
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of Vegetation Indices and a Modified Simple Ratio for Boreal Applications. Can. J. Remote Sens. 2014, 22, 229–242. [Google Scholar] [CrossRef]
- Peñuelas, J.; Inoue, Y. Reflectance Indices Indicative of Changes in Water and Pigment Contents of Peanut and Wheat Leaves. Photosynthetica 1999, 36, 355–360. [Google Scholar] [CrossRef]
- Husson, E.; Ecke, F.; Reese, H. Comparison of Manual Mapping and Automated Object-Based Image Analysis of Non-Submerged Aquatic Vegetation from Very-High-Resolution UAS Images. Remote Sens. 2016, 8, 724. [Google Scholar] [CrossRef]
- Satoh, K.; Shimizu, T.; Kondoh, H.; Hiraguri, A.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Relationship between Symptoms and Gene Expression Induced by the Infection of Three Strains of Rice dwarf virus. PLoS ONE 2011, 6, e18094. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, K.; Panja, B.N. Glume blight disease of high yielding boro (summer) rice from sub-Himalayan terai zone of West Bengal. Environ. Ecol. 2001, 19, 249–250. [Google Scholar]
- Suryadi, Y.; Susilowati, D.N.; Riana, E.; Mubarik, N.R. Management of rice blast disease (Pyricularia oryzae) using formulated bacterial consortium. Emir. J. Food Agric. 2012, 23, 349. [Google Scholar]
- Qin, Z.; Zhang, M.; Christensen, T.; Li, W. In Remote sensing analysis of rice disease stresses for farm pest management using wide-band airborne data. Geosci. Remote Sens. Symp. IGARSS 2003, 4, 2215–2217. [Google Scholar]
- Qin, Z.; Zhang, M. Detection of rice sheath blight for in-season disease management using multispectral remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2005, 7, 115–128. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Huang, J.F.; Shi, J.J.; Tao, R.X.; Wan, Z.; Zhang, L.L. Characterizing and estimating rice brown spot disease severity using stepwise regression, principal component regression and partial least-square regression. J. Zhejiang Univ.-Sci. B (Biomed. Biotechnol.) 2007, 8, 738–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]


| Parameters | Information |
|---|---|
| Sensor | PlanetScope |
| Acquisition date | 21 August 2017 and 30 October 2017 |
| Orbit altitude | 475 km |
| Spatial resolution (m) | 3 |
| Revisit time (days) | 1 |
| Wavelength range (nm) | |
| Band 1 | Blue: 455–515 |
| Band 2 | Green: 500–590 |
| Band 3 | Red: 590–670 |
| Band 4 | NIR: 780–860 |
| Signal-to-noise ratio (SNR) | 68.8 |
| Definition | Related Bands and Equations | Sensitive to | Reference |
|---|---|---|---|
| Normalized difference vegetation index, NDVI | (RNIR − RR)/(RNIR + RR) | Green biomass | [47] |
| Soil-adjusted vegetation index, SAVI | (1 + L) × (RNIR − RR)/(RNIR + RR + L); L = 0.5 | Canopy structure | [48] |
| Triangular vegetation index, TVI | 0.5 × [120 × (RNIR − RG) − 200 × (RR − RG)] | Radiant absorption of chlorophyll | [49] |
| Re-normalize difference vegetation index, RDVI | (RNIR − RR)/(RNIR + RR)0.5 | Vegetation coverage | [50] |
| Modified Simple Ratio, MSR | (RNIR/RR)/(RNIR/RR)0.5 | Leaf area, Biomass | [51] |
| Structural Independent Pigment Index, SIPI | (RNIR − RB)/(RNIR − RR) | Pigments content | [52] |
| Means of Normalized Two-Stage VIs | Means on 30 October | |||||||
|---|---|---|---|---|---|---|---|---|
| Healthy Rice | Rice Dwarf | Rice Blast | Glume Blight | Healthy Rice | Rice Dwarf | Rice Blast | Glume Blight | |
| NDVI | ** | ** | ** | ** | ** | * | ** | * |
| SAVI | *** | * | *** | * | * | ** | ** | * |
| TVI | ** | * | *** | ** | ** | * | ** | * |
| RDVI | ** | ** | *** | ** | * | * | ** | * |
| MSR | ** | ** | *** | ** | * | * | ** | * |
| SIPI | *** | *** | ** | *** | * | * | ** | ** |
| Means of Normalized Two-Stage VIs | Means on 30 October | |||||||
|---|---|---|---|---|---|---|---|---|
| Healthy Rice | Rice Dwarf | Rice Blast | Glume Blight | Healthy Rice | Rice Dwarf | Rice Blast | Glume Blight | |
| NDVI | 0.032 ** | 0.014 ** | 0.058 | 0.065 | 0.078 * | 0.018 ** | 0.08 * | 0.102 |
| SAVI | 0.047 * | 0.046 * | 0.036 * | 0.079 | 0.032 * | 0.031 * | 0.166 | 0.052 * |
| TVI | 0.032 * | 0.02 ** | 0.04 * | 0.012 ** | 0.037 * | 0.104 | 0.142 | 0.051 * |
| RDVI | 0.01 * | 0.009 ** | 0.045 * | 0.066 | 0.048 * | 0.074 * | 0.125 | 0.091 |
| MSR | 0.057 | 0.095 | 0.027 ** | 0.091 | 0.073 | 0.125 | 0.077 | 0.054 * |
| SIPI | 0.039 * | 0.027 * | 0.087 | 0.036 * | 0.056 * | 0.124 | 0.145 | 0.09 |
| Predicted Class | Healthy Rice | Rice Dwarf | Rice Blast | Glume Blight | User’s Accuracy (%) | Overall Accuracy (%) | Kappa Coefficient |
|---|---|---|---|---|---|---|---|
| Normalized two-stage VIs | |||||||
| Healthy rice | 54 | 0 | 6 | 2 | 87.1 | 75.62 | 0.47 |
| Rice dwarf | 4 | 60 | 5 | 9 | 76.92 | ||
| Rice blast | 11 | 4 | 48 | 5 | 70.59 | ||
| Glume blight | 5 | 8 | 2 | 27 | 64.29 | ||
| Producer’s accuracy (%) | 72.97 | 83.33 | 78.69 | 62.79 | |||
| single-date VIs | |||||||
| Healthy rice | 47 | 3 | 8 | 4 | 75.81 | 61.67 | 0.27 |
| Rice dwarf | 8 | 48 | 5 | 17 | 61.54 | ||
| Rice blast | 16 | 6 | 39 | 7 | 57.35 | ||
| Glume blight | 7 | 11 | 4 | 20 | 47.62 | ||
| Producer’s accuracy (%) | 60.26 | 70.59 | 69.64 | 41.67 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Huang, W.; Ye, H.; Ruan, C.; Xing, N.; Geng, Y.; Dong, Y.; Peng, D. Partial Least Square Discriminant Analysis Based on Normalized Two-Stage Vegetation Indices for Mapping Damage from Rice Diseases Using PlanetScope Datasets. Sensors 2018, 18, 1901. https://doi.org/10.3390/s18061901
Shi Y, Huang W, Ye H, Ruan C, Xing N, Geng Y, Dong Y, Peng D. Partial Least Square Discriminant Analysis Based on Normalized Two-Stage Vegetation Indices for Mapping Damage from Rice Diseases Using PlanetScope Datasets. Sensors. 2018; 18(6):1901. https://doi.org/10.3390/s18061901
Chicago/Turabian StyleShi, Yue, Wenjiang Huang, Huichun Ye, Chao Ruan, Naichen Xing, Yun Geng, Yingying Dong, and Dailiang Peng. 2018. "Partial Least Square Discriminant Analysis Based on Normalized Two-Stage Vegetation Indices for Mapping Damage from Rice Diseases Using PlanetScope Datasets" Sensors 18, no. 6: 1901. https://doi.org/10.3390/s18061901





