Biological Oscillators in Nanonetworks—Opportunities and Challenges
Abstract
:1. Introduction
1.1. Motivation
- Need for a biologically compatible time keeper: Quartz crystal oscillators provide and maintain the time information in almost every electronic device [14]. To stay true to the definition of molecular communication, however, it would be meaningful to integrate oscillators that are made with biological components (e.g., molecules) and are driven by biochemical processes (e.g., gene translation and transcription) or, in other words, oscillators that are biocompatible [15]. Fortunately, to begin with, nature has an abundance of such oscillators. The sleep-wake cycle driven by the circadian oscillator [16], cell division controlled by mitotic oscillators [17,18], and the periodical break-down of glucose to sugar that is maintained by glycolytic oscillators [19] are few examples of biological oscillators in nature. In this study, we refer to them as natural oscillators. For many decades, biologists, physicists, and mathematicians have extensively studied natural oscillators, mainly to understand the underlying principles of the oscillators, which, as we will see in Section 2.1, are of a very complex nature, even though we outline only the main principles.
- Avenues for developing a simpler system: While the understanding of the complex mechanism that drives natural oscillators is a challenge, to engineer such mechanisms was another challenge until the birth of a field called synthetic biology [20]. Ironically, the successful realization of the first in-vivo, artificially-realized oscillator, namely, the repressilator [21] represented the beginning of synthetic biology. The repressilator laid the path for other novel designs to follow [22,23]. In this study, we refer to them as synthetic oscillators. Synthetic biology offers several advantages to nanonetworks. Firstly, it has led to the development of oscillators that involve much simpler mechanisms than their natural counterparts [24] and such oscillators could be embedded within a nanomachine. Secondly, such an engineering feat is a benefit to nanonetwork applications that are targeted towards living tissues, where biocompatible components that can be biologically engineered are preferred. Although these systems are still far from perfection, recent studies have shown that they can indeed be improved [25,26,27,28].
- Investigations from a communication systems engineer perspective: Nearing two decades since the inception of nanonetworks, few studies on oscillators in the literature have surfaced from the nanonetwork research community [29,30,31]. Taking cues from nature, these studies have presented oscillatory systems that will be suitable, in particular, for molecular nanomachines and, in general, for a nanonetwork. The first two oscillator systems were designed to allow a nanonetwork to achieve synchronization by converging the period of oscillations [29,30], while the third system was designed to align the clock times and extend the purpose of the oscillator beyond synchronization, to provide timing information for scheduling channel access and decoding the signals or for coordinating other communication modules in a nanomachine [31]. We will present, for the first time, qualitative comparisons between them.
- Lack of a consolidated study: To date, a consolidated literature that brings biological oscillators under one single study is lacking. Motivated by the gaps in the literature regarding biological oscillators, more specifically to nanonetworks, we provide a comprehensive review of biological oscillators from the earliest to the latest developments. Additionally, unlike other recent surveys [32], we study each oscillator using parameters that are significant in the eye of a communication systems engineer.
1.2. Main Contributions
- Consolidating the biological oscillators into a single work, which, to the best of our knowledge, no work has ever done, making this survey the first one.
- Classification of the biological oscillators based on whether they are found in nature or not.
- Reviewing the natural oscillators and their underlying mechanisms with sufficient detail, bearing in mind that not all researchers working in nanonetworks have biology backgrounds.
- Reviewing the synthetic oscillators and their design principles and properties, supported with simple and accurate visuals of the system’s schematics, bearing in mind that not all researchers working in nanonetworks have synthetic biology backgrounds.
- Reviewing the recent works on oscillatory systems proposed by the nanonetwork research community.
- Comparative analysis of the oscillators.
- Identification of open research issues for both the physical and communication aspects of the oscillators.
2. Biological Oscillators
2.1. Natural Oscillators
2.1.1. Glycolytic Oscillators
2.1.2. Cyclic Adenosine Monophosphate (cAMP) Oscillator
2.1.3. Circadian Oscillator
2.1.4. Calcium Oscillator
2.1.5. Mitotic Oscillators
2.2. Synthetic Oscillators
2.2.1. Goodwin Oscillator
2.2.2. Repressilator
2.2.3. Atkinson Oscillator
2.2.4. Hasty Oscillator
2.2.5. Metabolator
2.2.6. Dual Feedback Oscillator
2.2.7. Fussenegger Oscillator
2.2.8. miRNA-Regulated Oscillator
2.2.9. Displacillator
2.3. Oscillators Specific to the Nanonetwork
2.3.1. Moore Oscillator
2.3.2. Akgül Oscillator
2.3.3. Shitiri Oscillator
3. Open Research Issues
3.1. Noise
3.2. Design
3.3. Sustainability
3.4. Adoption and Implementation to Nanonetworks
3.4.1 Interfacing
3.4.2. Matched Oscillators
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Akyildiz, I.F.; Brunetti, F.; Blázquez, C. Nanonetworks: A new communication paradigm. Comput. Netw. 2008, 52, 2260–2279. [Google Scholar] [CrossRef]
- Freitas, R.A., Jr. Nanomedicine. Volume 1: Basic Capabilities, 1st ed.; Landes Bioscience: Georgetown, TX, USA, 1999. [Google Scholar]
- Hiyama, S.; Moritani, Y.; Suda, T.; Egashira, R.; Enomoto, A.; Moore, M.; Nakano, T. Molecular Communication. NSTI-Nanotech 2005, 3, 391–394. [Google Scholar]
- Guo, W.; Mias, C.; Farsad, N.; Wu, J.L. Molecular Versus Electromagnetic Wave Propagation Loss in Macro-Scale Environments. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2015, 1, 18–25. [Google Scholar] [CrossRef]
- Akyildiz, I.F.; Jornet, J.M.; Han, C. Terahertz band: Next frontier for wireless communications. Phys. Commun. 2014, 12, 16–32. [Google Scholar] [CrossRef]
- Berridge, M.J. Module 10: Neuronal Signalling. Cell Signal. Biol. 2014, 6, csb0001010. [Google Scholar] [CrossRef]
- Berridge, M.J. Module 9: Cell Cycle and Proliferation. Cell Signal. Biol. 2014, 6, csb0001009. [Google Scholar] [CrossRef]
- Berridge, M.J. Module 2: Cell Signalling Pathways. Cell Signal. Biol. 2014, 6, csb0001002. [Google Scholar] [CrossRef]
- Suda, T.; Moore, M.; Nakano, T.; Egashira, R.; Enomoto, A. Exploratory Research on Molecular Communication between Nanomachines. Late Break. Pap. 2005, 25, 29. [Google Scholar]
- Llatser, I.; Cabellos-Aparicio, A.; Alarcon, E. Networking challenges and principles in diffusion-based molecular communication. IEEE Wirel. Commun. 2012, 19, 36–41. [Google Scholar] [CrossRef]
- Nakano, T.; Suda, T.; Okaie, Y.; Moore, M.J.; Vasilakos, A.V. Molecular Communication Among Biological Nanomachines: A Layered Architecture and Research Issues. IEEE Trans. Nanobiosci. 2014, 13, 169–197. [Google Scholar] [CrossRef] [PubMed]
- DeMello, A.J. Control and detection of chemical reactions in microfluidic systems. Nature 2006, 442, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Eckford, A.W.; Haraguchi, T. Molecular Communication; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Horowitz, P.; Hill, W. The Art of Electronics, 3rd ed.; Cambridge University Press: New York, NY, USA, 2015. [Google Scholar]
- Nakano, T.; Moore, M.J.; Wei, F.; Vasilakos, A.V.; Shuai, J. Molecular communication and networking: Opportunities and challenges. IEEE Trans. Nanobiosci. 2012, 11, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Winfree, A.T. Human body clocks and the timing of sleep. Nature 1982, 297, 23–27. [Google Scholar] [CrossRef]
- Murray, A.; Kirschner, M. Dominoes and clocks: The union of two views of the cell cycle. Science 1989, 246, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.W.; Kirschner, M.W. Cyclin synthesis drives the early embryonic cell cycle. Nature 1989, 339, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Hess, B. The glycolytic oscillator. J. Exp. Biol. 1979, 81, 7–14. [Google Scholar] [PubMed]
- Andrianantoandro, E.; Basu, S.; Karig, D.K.; Weiss, R. Synthetic biology: New engineering rules for an emerging discipline. Mol. Syst. Biol. 2006, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Stricker, J.; Cookson, S.; Bennett, M.R.; Mather, W.H.; Tsimring, L.S.; Hasty, J. A fast, robust and tunable synthetic gene oscillator. Nature 2008, 456, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Tigges, M.; Marquez-Lago, T.T.; Stelling, J.; Fussenegger, M. A tunable synthetic mammalian oscillator. Nature 2009, 457, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Novák, B.; Tyson, J.J. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 2008, 9, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Gérard, C.; Novák, B. microRNA as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a ceRNA Network. PLoS ONE 2013, 8, e83372. [Google Scholar] [CrossRef] [PubMed]
- Hussain, F.; Gupta, C.; Hirning, A.J.; Ott, W.; Matthews, K.S.; Josić, K.; Bennett, M.R. Engineered temperature compensation in a synthetic genetic clock. Proc. Natl. Acad. Sci. USA 2014, 111, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Reznik, E.; Kaper, T.J.; Segre, D. The dynamics of hybrid metabolic-genetic oscillators. Chaos Interdiscip. J. Nonlinear Sci. 2013, 23. [Google Scholar] [CrossRef] [PubMed]
- Ryback, B.M.; Odoni, D.I.; van Heck, R.G.; van Nuland, Y.; Hesselman, M.C.; dos Santos, V.A.M.; van Passel, M.W.; Hugenholtz, F. Design and analysis of a tunable synchronized oscillator. J. Biol. Eng. 2013, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Nakano, T. Oscillation and synchronization of molecular machines by the diffusion of inhibitory molecules. IEEE Trans. Nanotechnol. 2013, 12, 601–608. [Google Scholar] [CrossRef]
- Akgül, Ö.U.; Canberk, B. An interference-free and simultaneous molecular transmission model for multi-user nanonetworks. Nano Commun. Netw. 2014, 5, 83–96. [Google Scholar] [CrossRef]
- Shitiri, E.; Cho, H. A Biochemical Oscillator Using Excitatory Molecules for Nanonetworks. IEEE Trans. Nanobiosci. 2016, 15, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Purcell, O.; Savery, N.J.; Grierson, C.S.; di Bernardo, M. A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface 2010, 7, 1503–1524. [Google Scholar] [CrossRef] [PubMed]
- Goldbeter, A. Biochemical oscillations and cellular rhythms. Curr. Sci. 1996, 73, 933–939. [Google Scholar]
- Friesen, W.O.; Block, G.D. What is a biological oscillator? Am. J. Physiol. 1984, 246, R847–R853. [Google Scholar] [CrossRef] [PubMed]
- Betz, A.; Chance, B. Phase relationship of glycolytic intermediates in yeast cells with oscillatory metabolic control. Arch. Biochem. Biophys. 1965, 109, 585–594. [Google Scholar] [CrossRef]
- Chandra, F.A.; Buzi, G.; Doyle, J.C. Glycolytic Oscillations and Limits on Robust Efficiency. Science 2011, 333, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Duysens, L.N.; Amese, J. Fluorescence spectrophotometry of reduced phosphopyridine nucleotide in intact cells in the near-ultraviolet and visible region. Biochim. Biophys. Acta 1957, 24, 19–26. [Google Scholar] [CrossRef]
- Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry; W.H. Freeman and Company: San Francisco, CA, USA, 2008. [Google Scholar]
- Higgins, J. A chemical mechanism for oscillation of glycolytic intermediates in yeast cells. Proc. Natl. Acad. Sci. USA 1964, 51, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Sel’Kov, E.E. Self-Oscillations in Glycolysis. Eur. J. Biochem. 1968, 4, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Goldbeter, A.; Lefever, R. Dissipative structures for an allosteric model. Biophys. J. 1972, 12, 1302–1315. [Google Scholar] [CrossRef]
- Goldbeter, A. Modulation of the adenylate energy charge by sustained metabolic oscillations. FEBS Lett. 1974, 43, 327–330. [Google Scholar] [CrossRef]
- Hess, B.; Boiteux, A. Mechanism of Glycolytic Oscillation in Yeast, I Aerobic and anaerobic growth conditions for obtaining glycolytic oscillation. Hoppe Seylers Z. Physiol. Chem. 1968, 349, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, S.; Sakakibara, R.; Uyeda, K. Significance of phosphorylation of phosphofructokinase. J. Biol. Chem. 1983, 258, 13292–13298. [Google Scholar] [PubMed]
- Keener, J.S.J. Mathematical Physiology; Springer: New York, NY, USA, 1998. [Google Scholar]
- Fall, C.P. Computational Cell Biology; Springer: New York, NY, USA, 2004. [Google Scholar]
- Gustavsson, A.K.; van Niekerk, D.D.; Adiels, C.B.; Preez, F.B.D.; Goksör, M.; Snoep, J.L. Sustained glycolytic oscillations in individual isolated yeast cells. FEBS J. 2012, 279, 2837–2847. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, A.-K.; Adiels, C.B.; Mehlig, B.; Goksör, M. Entrainment of heterogeneous glycolytic oscillations in single cells. Sci. Rep. 2015, 5, 9404. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, G.; Suprunenko, Y.F.; Jenkins, K.; Stefanovska, A. Modelling chronotaxicity of cellular energy metabolism to facilitate the identification of altered metabolic states. Sci. Rep. 2016, 6, 29584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerisch, G.; Hess, B. Cyclic-AMP-controlled oscillations in suspended Dictyostelium cells: Their relation to morphogenetic cell interactions. Proc. Natl. Acad. Sci. USA 1974, 71, 2118–2122. [Google Scholar] [CrossRef] [PubMed]
- Gerisch, G. Cell Aggregation and Differentiation in Dictyostelium. In Current Topics in Developmental Biology; Elsevier: New York, NY, USA, 1968. [Google Scholar]
- Newell, P.C. Cellular communication during aggregation of Dictyostelium. J. Gen. Microbiol. 1978, 104, 1–13. [Google Scholar] [CrossRef]
- Loomis, W.F. Cell signaling during development of Dictyostelium. Dev. Biol. 2014, 391, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Konijn, T.M.; Raper, K.B. Cell aggregation in Dictyostelium discoideum. Dev. Biol. 1961, 3, 725–756. [Google Scholar] [CrossRef]
- Tyson, J.J.; Murray, J.D. Cyclic AMP waves during aggregation of Dictyostelium amoebae. Development 1989, 106, 421–426. [Google Scholar] [PubMed]
- Brian, P.I. Mathematical Modelling in Systems Biology: An Introduction; The MIT Press: Massachusetts, MA, USA, 2013. [Google Scholar]
- Gerisch, G.; Wick, U. Intracellular oscillations and release of cyclic AMP from Dictyostelium cells. Biochem. Biophys. Res. Commun. 1975, 65, 364–370. [Google Scholar] [CrossRef]
- Roos, W.; Nanjundiah, V.; Malchow, D.; Gerisch, G. Amplification of cyclic-AMP signals in aggregating cells of Dictyostelium discoideum. FEBS Lett. 1975, 53, 139–142. [Google Scholar] [CrossRef]
- Shaffer, B.M. Secretion of cyclic AMP induced by cyclic AMP in the cellular slime mould Dictyostelium discoideum. Nature 1975, 255, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Halloy, J.; Lauzeral, J.; Goldbeter, A. Modeling oscillations and waves of cAMP in dictyostelium discoideum cells. Biophys. Chem. 1998, 72, 9–19. [Google Scholar] [CrossRef]
- Devreotes, P.N.; Sherring, J.A. Kinetics and concentration dependence of reversible cAMP-induced modification of the surface cAMP receptor in Dictyostelium. J. Biol. Chem. 1985, 260, 6378–6384. [Google Scholar] [PubMed]
- Klein, P.; Theibert, A.; Fontana, D.; Devreotes, P.N. Identification and cyclic AMP-induced modification of the cyclic AMP receptor in Dictyostelium discoideum. J. Biol. Chem. 1985, 260, 1757–1764. [Google Scholar] [PubMed]
- Shaulsky, G.; Fuller, D.; Loomis, W.F. A cAMP-phosphodiesterase controls PKA-dependent differentiation. Development 1998, 125, 691–699. [Google Scholar] [PubMed]
- Laub, M.T.; Loomis, W.F. A Molecular Network That Produces Spontaneous Oscillations in Excitable Cells of Dictyostelium. Mol. Biol. Cell 1998, 9, 3521–3532. [Google Scholar] [CrossRef] [PubMed]
- Daan, S. A history of chronobiological concepts. Protein Rev. 2010, 12, 1–35. [Google Scholar]
- Konopka, R.J.; Benzer, S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1971, 68, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc. Natl. Acad. Sci. USA 1992, 89, 11711–11715. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.J.; Edery, I.; Rosbash, M. PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature 1993, 364, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S.; Kornhauser, J.M.; Koumenis, C.; Eskin, A. Molecular approaches to understanding circadian oscillations. Annu. Rev. Physiol. 1993, 55, 729–753. [Google Scholar] [CrossRef] [PubMed]
- Goldbeter, A. A Model for Circadian Oscillations in the Drosophila Period Protein (PER). Proc. R. Soc. B Biol. Sci. 1995, 261, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Edery, I.; Zwiebel, L.J.; Dembinska, M.E.; Rosbash, M. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 1994, 91, 2260–2264. [Google Scholar] [CrossRef] [PubMed]
- Curtin, K.D.; Huang, Z.J.; Rosbash, M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 1995, 14, 365–372. [Google Scholar] [CrossRef]
- Berridge, M.J. Calcium oscillations. J. Biol. Chem. 1990, 265, 9583–9586. [Google Scholar] [PubMed]
- Dupont, G.; Combettes, L.; Bird, G.S.; Putney, J. Calcium oscillations Cold. Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Cuthbertson, K.S.R.; Cobbold, P.H. Phorbol ester and sperm activate mouse oocytes by inducing sustained oscillations in cell Ca2+. Nature 1985, 313, 541–542. [Google Scholar] [CrossRef]
- Woods, N.M.; Cuthbertson, K.S.R.; Cobbold, P.H. Repetitive transient rises in cytoplasmic free calcium in hormone-stimulated hepatocytes. Nature 1986, 319, 600–602. [Google Scholar] [CrossRef] [PubMed]
- Woods, N.M.; Cuthbertson, K.S.R.; Cobbold, P.H. Agonist-induced oscillations in cytoplasmic free calcium concentration in single rat hepatocytes. Cell Calcium 1987, 8, 79–100. [Google Scholar] [CrossRef]
- De Young, G.W.; Keizer, J. A single pool IP3R based model for agonist stimulated oscillations in Ca2+. Concentration. Nature 1992, 89, 9895–9899. [Google Scholar]
- Li, Y.X.; Rinzel, J. Equations for InsP3 receptor-mediated [Ca2+]i oscillations derived from a detailed kinetic model: A Hodgkin-Huxley like formalism. J. Theor. Biol. 1994, 166, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Falcke, M. Reading the patterns in living cells—The physics of Ca2+ signaling. Adv. Phys. 2004, 53, 255–440. [Google Scholar] [CrossRef]
- Shuai, Z.; Bing, L.; Shaoqun, Z.; Shangbin, C. Simulation of spontaneous Ca2+ oscillations in astrocytes mediated by voltage-gated calcium channels. Biophys. J. 2009, 97, 2429–2437. [Google Scholar]
- Meyer, T.; Stryer, L. Molecular model for receptor-stimulated calcium spiking. Proc. Natl. Acad. Sci. USA 1988, 85, 5051–5055. [Google Scholar] [CrossRef] [PubMed]
- Kummer, U.; Krajnc, B.; Pahle, J.; Green, A.K.; Dixon, C.J.; Marhl, M. Transition from stochastic to deterministic behavior in calcium oscillations. Biophys. J. 2005, 89, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Höfer, T.; Venance, L.; Giaume, C. Control and plasticity of intercellular calcium waves in astrocytes: A modeling approach. J. Neurosci. 2002, 22, 4850–4859. [Google Scholar] [CrossRef] [PubMed]
- De Pittà, M.; Goldberg, M.; Volman, V.; Berry, H.; Ben-Jacob, E. Glutamate regulation of calcium and IP3 oscillating and pulsating dynamics in astrocytes. J. Biol. Phys. 2009, 35, 383–411. [Google Scholar] [CrossRef] [PubMed]
- Paweletz, N. Walther Flemming: Pioneer of mitosis research. Nat. Rev. Mol. Cell Biol. 2001, 2, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Romond, P.-C.; Guilmot, J.-M.; Goldbeter, A. The mitotic oscillator: Temporal self-organization in a phosphorylation-dephosphorylation enzymatic cascade. Ber. Bunsenges. Phys. Chem. 1994, 98, 1152–1159. [Google Scholar] [CrossRef]
- Mitchison, T.J.; Salmon, E.D. Mitosis: A history of division. Nat. Cell Biol. 2001, 3, E17–E21. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, D.A. The nature of the cell cycle and the control of cell proliferation. Biosystems 1974, 5, 197–206. [Google Scholar] [CrossRef]
- Gilbert, D.A. The relationship between the transition probability and oscillator concepts of the cell cycle and the nature of the commitment to replication. Biosystems 1978, 10, 235–240. [Google Scholar] [CrossRef]
- Kauffman, S. Measuring a mitotic oscillator: The arc discontinuity. Bull. Math. Biol. 1974, 36, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.; Wille, J.J. The mitotic oscillator in Physarum polycephalum. J. Theor. Biol. 1975, 55, 47–93. [Google Scholar] [CrossRef]
- Goldbeter, A. A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase. Proc. Natl. Acad. Sci. USA 1991, 88, 9107–9111. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P. Universal control mechanism regulating onset of M-phase. Nature 1990, 344, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Draetta, G. Cell cycle control in eukaryotes: Molecular mechanisms of cdc2 activation. Trends Biochem. Sci. 1990, 15, 378–383. [Google Scholar] [CrossRef]
- Chen, B.-S.; Wang, Y.-C. Synthetic Gene Network; CRC Press: New York, NY, USA, 2014. [Google Scholar]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Butzin, N.C.; Mather, W.H. Synthetic Genetic Oscillators. In Reviews in Cell Biology and Molecular Medicine; Meyers, R.A., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006. [Google Scholar]
- Goodwin, B.C. Temporal Organization in Cells a Dyanamic Theroy of Cellular Control Processes; Academic Press: London, UK, 1963. [Google Scholar]
- Goodwin, B.C. Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 1965, 3, 425–438. [Google Scholar] [CrossRef]
- Griffith, J.S. Mathematics of Cellular Control Processes I. Negative Feedback to One Gene. J. Theor. Biol. 1968, 20, 202–208. [Google Scholar] [CrossRef]
- Aronson, B.; Johnson, K.; Loros, J.; Dunlap, J. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science 1994, 263, 1578–1584. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Hardin, P.E.; Rosbash, M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994, 13, 3590–3598. [Google Scholar] [PubMed]
- Smith, W.R. Qualitative mathematical models of endocrine systems. Am. J. Physiol. 1983, 245, R473–R477. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, M.R.; Savageau, M.A.; Myers, J.T.; Ninfa, A.J. Development of Genetic Circuitry Exhibiting Toggle Switch or Oscillatory Behavior in Escherichia coli. Cell 2003, 113, 597–607. [Google Scholar] [CrossRef]
- Hasty, J.; Isaacs, F.; Dolnik, M.; McMillen, D.; Collins, J.J. Designer gene networks: Towards fundamental cellular control. Chaos Interdiscip. J. Nonlinear Sci. 2001, 11, 207. [Google Scholar] [CrossRef] [PubMed]
- Fung, E.; Wong, W.W.; Suen, J.K.; Bulter, T.; Lee, S.G.; Liao, J.C. A synthetic gene-metabolic oscillator. Nature 2005, 435, 118. [Google Scholar] [CrossRef] [PubMed]
- Smolen, P.; Baxter, D.A.; Byrne, J.H. Frequency selectivity, multistability, and oscillations emerge from models of genetic regulatory systems. Am. Physiol. Soc. 1998, 274, C531–C541. [Google Scholar] [CrossRef]
- Hasty, J.; Dolnik, M.; Rottschäfer, V.; Collins, J.J. Synthetic gene network for entraining and amplifying cellular oscillations. Phys. Rev. Lett. 2002, 88, 148101. [Google Scholar] [CrossRef] [PubMed]
- Tigges, M.; Dénervaud, N.; Greber, D.; Stelling, J.; Fussenegger, M. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res. 2010, 38, 2702–2711. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, N.; Parkin, J.; Seelig, G.; Winfree, E.; Soloveichik, D. Enzyme-free nucleic acid dynamical systems. Science 2017, 358, eaal2052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009, 131, 17303–17314. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Nakano, T. Synchronization of Inhibitory Molecular Spike Oscillators. In Proceedings of the Bio-Inspired Models of Networks, Information, and Computing Systems, Lugano, Switzerland, 10–11 December 2012; pp. 183–195. [Google Scholar]
- Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Batzer, A.G. Stochastic gene expression in a single cell. Sci. STKE 2002, 297, 1183–1187. [Google Scholar] [CrossRef] [PubMed]
- Gonze, D.; Halloy, J.; Goldbeter, A. Robustness of circadian rhythms with respect to molecular noise. Proc. Natl. Acad. Sci. USA 2002, 99, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, D.T. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 1976, 22, 403–434. [Google Scholar] [CrossRef]
- Gillesple, D.T. Exact Stochastic Simulation of couple chemical reactions. J. Phys. Chem. 1977, 81, 2340–2361. [Google Scholar] [CrossRef]
- Goldbeter, A.; Gérard, C.; Gonze, D.; Leloup, J.C.; Dupont, G. Systems biology of cellular rhythms. FEBS Lett. 2012, 586, 2955–2965. [Google Scholar] [CrossRef] [PubMed]
- Gonze, D.; Halloy, J.; Gaspard, P. Biochemical clocks and molecular noise: Theoretical study of robustness factors. J. Chem. Phys. 2002, 116, 10997–11010. [Google Scholar] [CrossRef]
- Raser, J.M. Noise in Gene Expression: Origins, Consequences, and Control. Science 2005, 309, 2010–2013. [Google Scholar] [CrossRef] [PubMed]
- Becskel, A.; Serrano, L. Engineering stability in gene networks by autoregulation. Nature 2000, 405, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Purich, D.; Allison, R. Handbook of Biochemical Kinetics; Academic Press: New York, NY, USA, 2000. [Google Scholar]
- Harootunian, A.T.; Kao, J.P.Y.; Tsien, R.Y. Agonist-induced Calcium Oscillations in Depolarized Fibroblasts and Their Manipulation by Photoreleased Ins(l,4,5)P3, and Ca++, and Ca++ Buffer. Cold Spring Harb. Symp. Quant. Biol. 1988, 53, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Shitiri, E.; Cho, H.-S. Achieving in-phase synchronization in a diffusion-based nanonetwork with unknown propagation delay. In Proceedings of the 4th ACM International Conference on Nanoscale Computing and Communication, Washington, DC, USA, 27–29 September 2017; pp. 1–6. [Google Scholar]
- Lin, L.; Yang, C.; Ma, M. Offset and Skew Estimation for Clock Synchronization in Molecular Communication Systems. In Proceedings of the 9th EAI International Conference on Bio-inspired Information and Communications Technologies, New York, NY, USA, 3–5 December 2015. [Google Scholar]

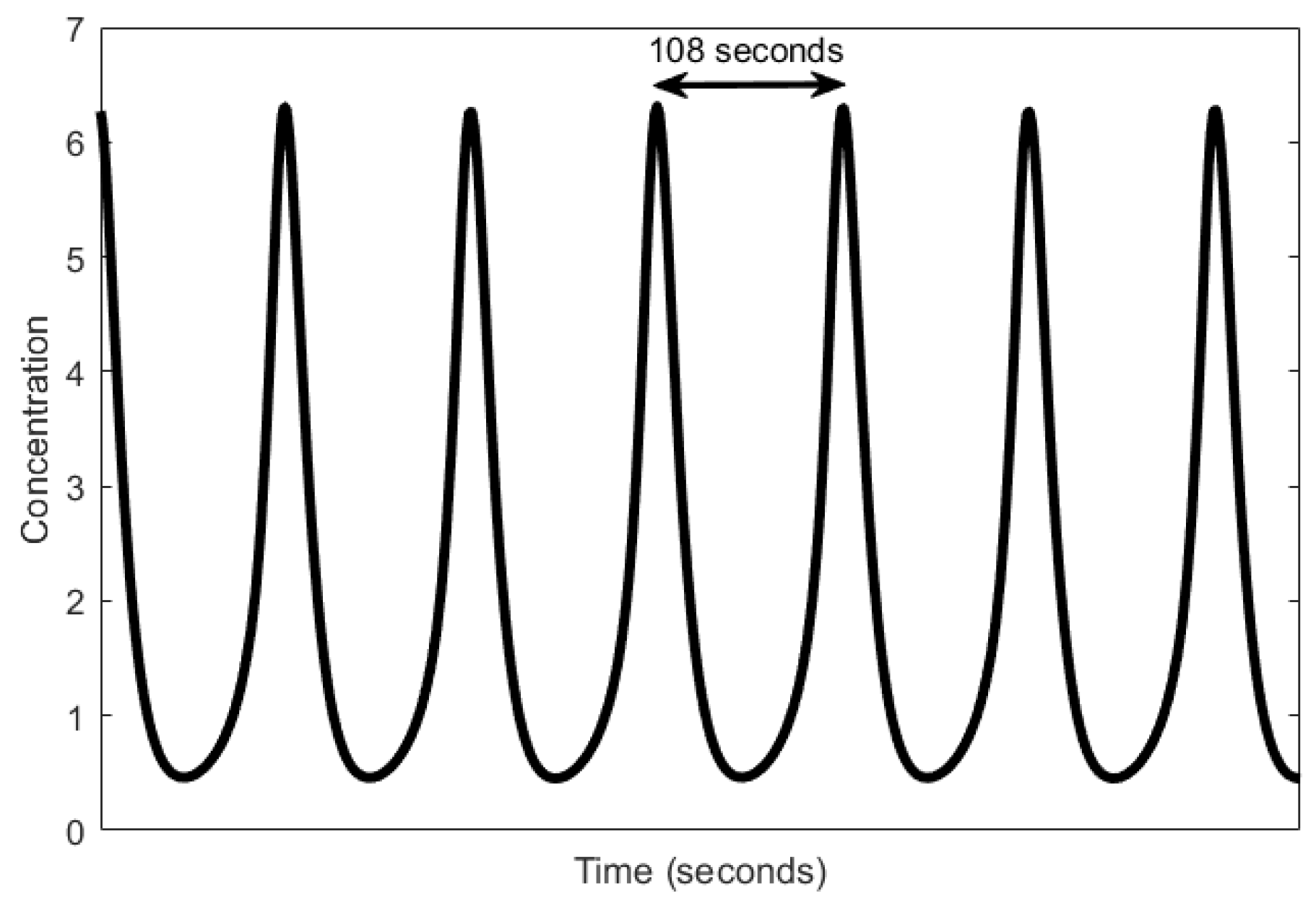
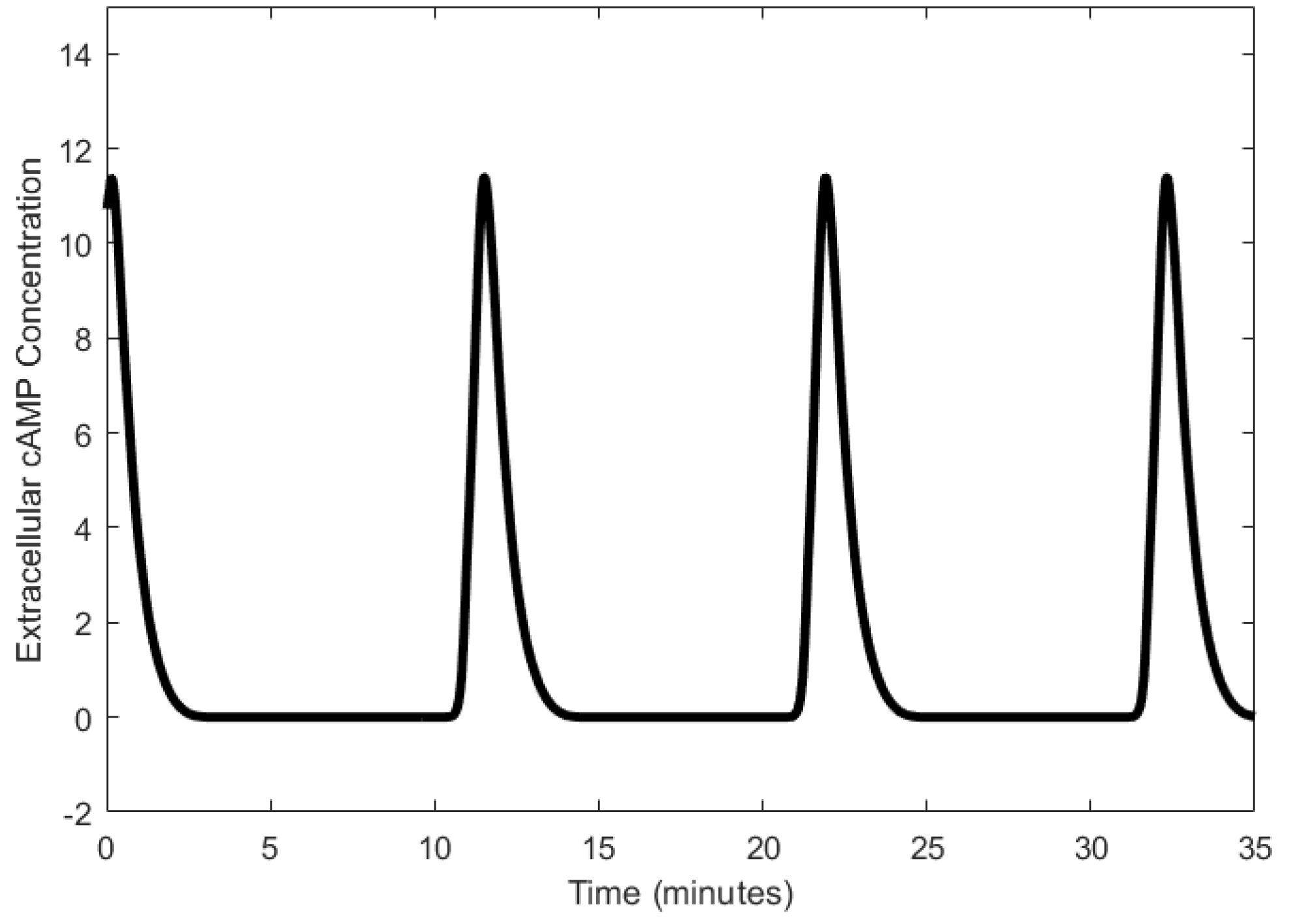

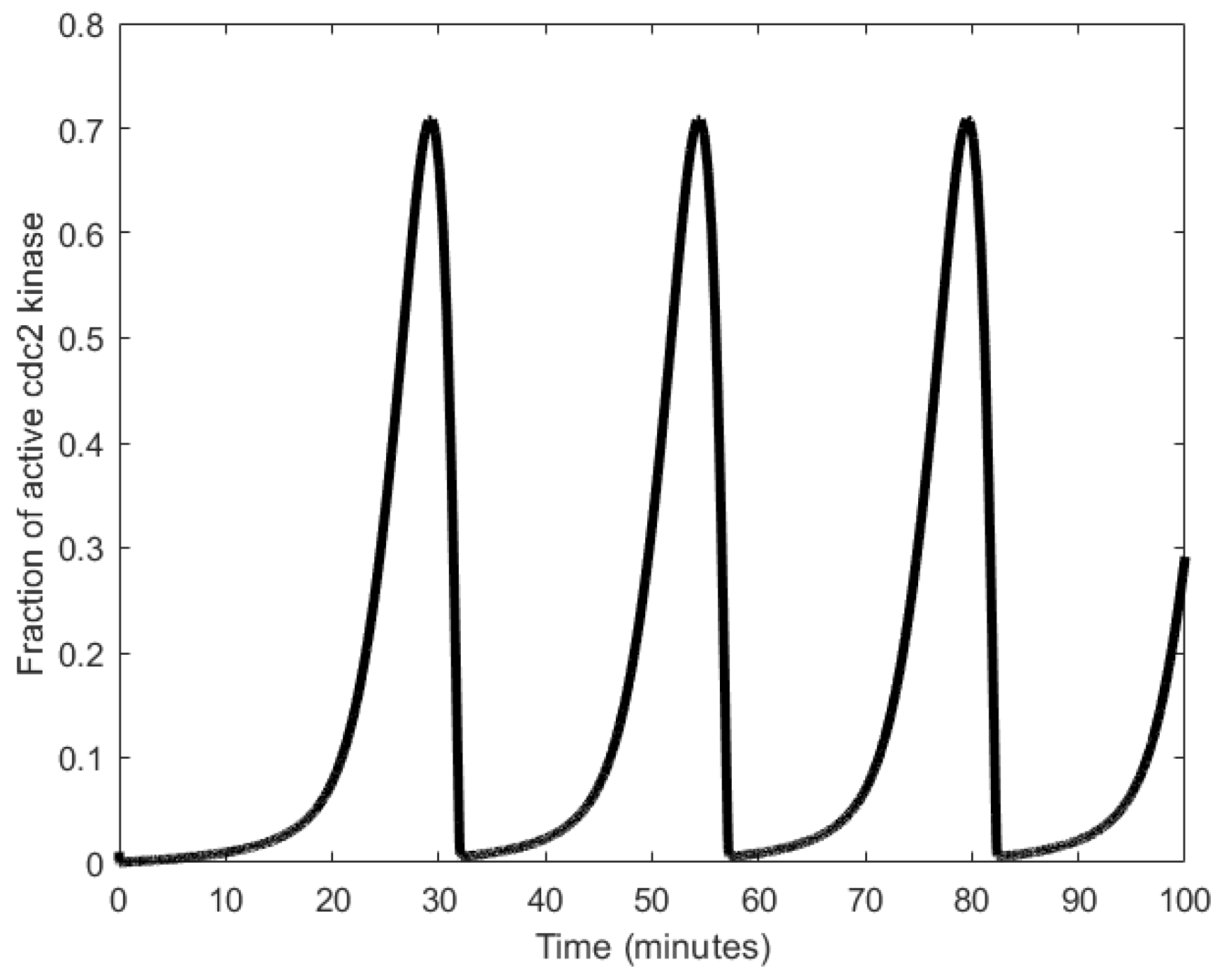

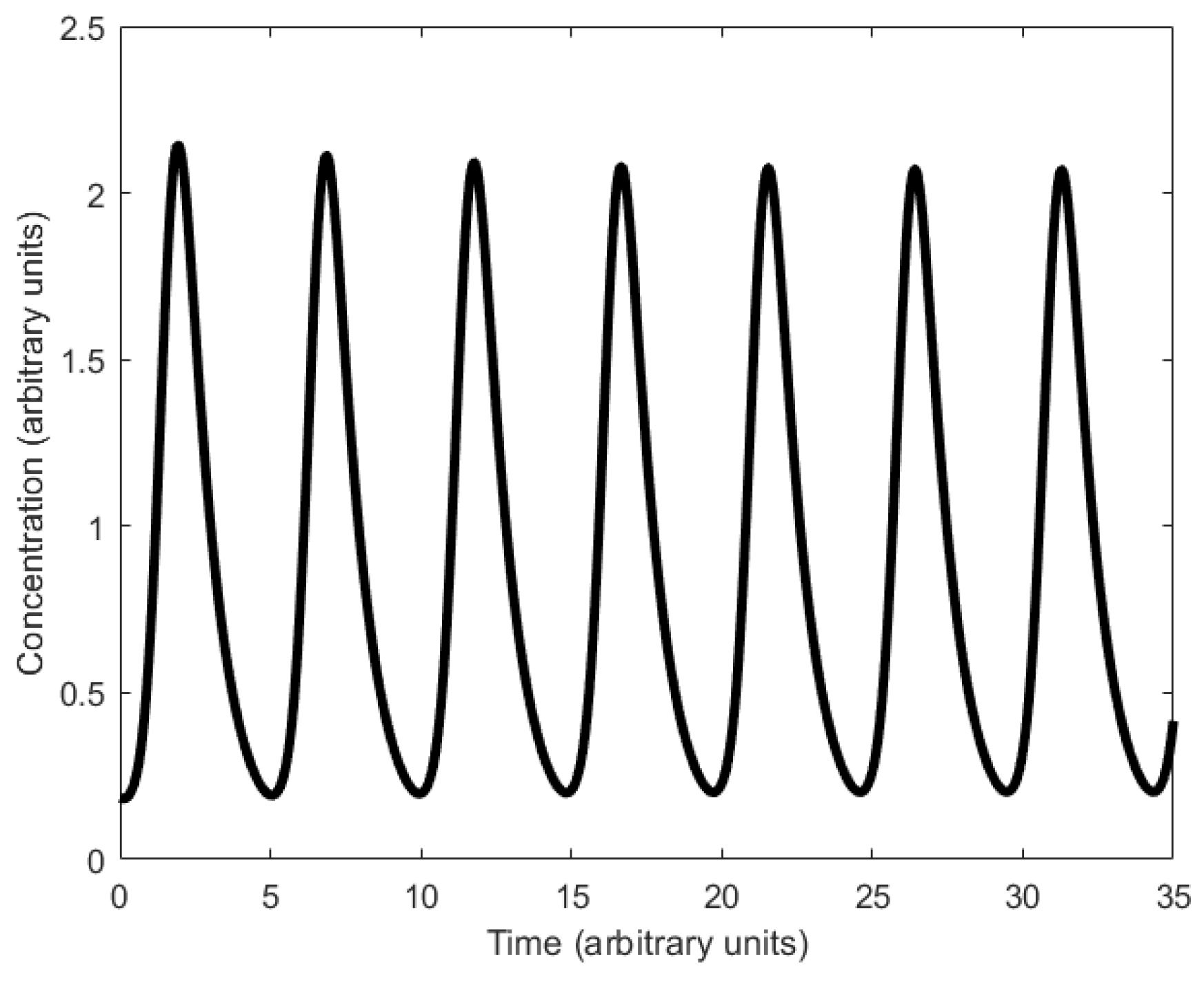
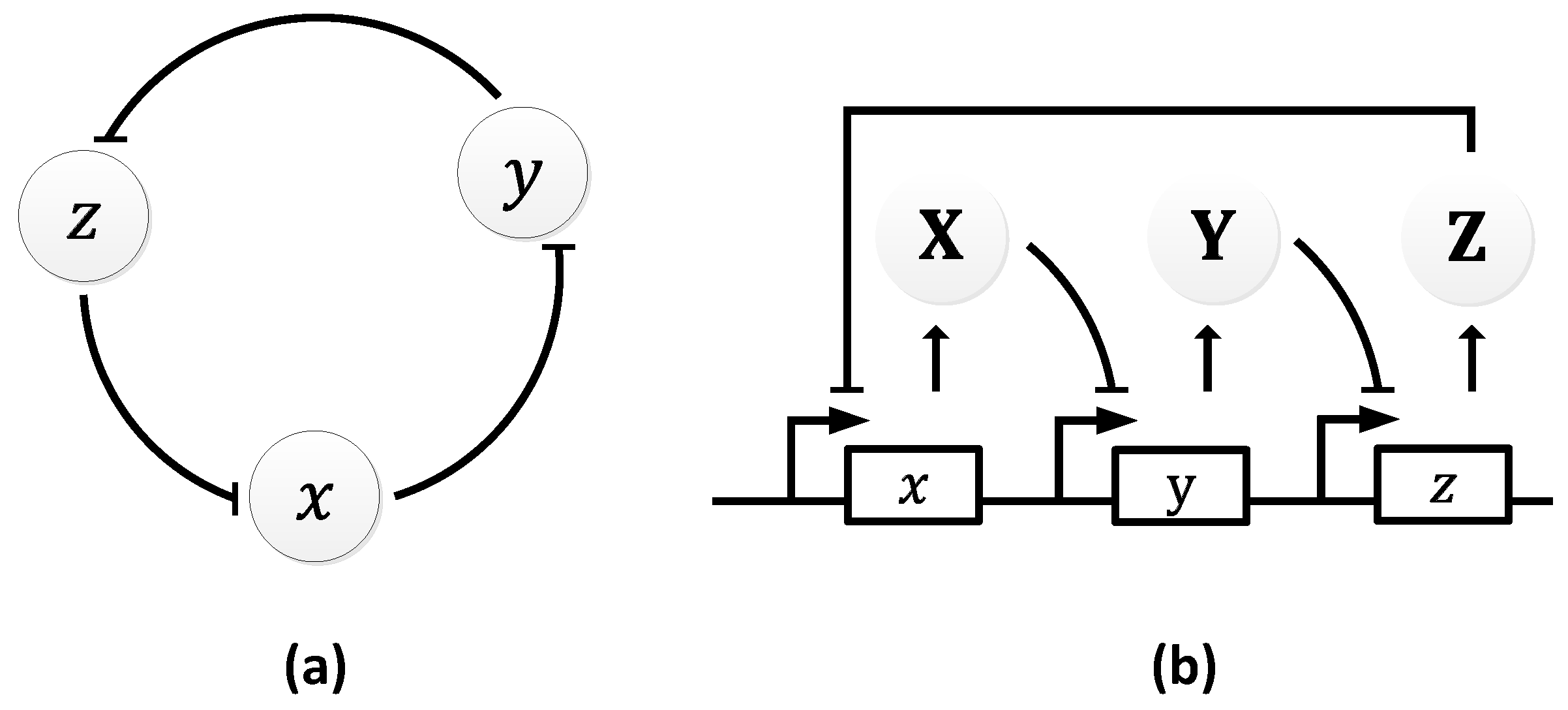

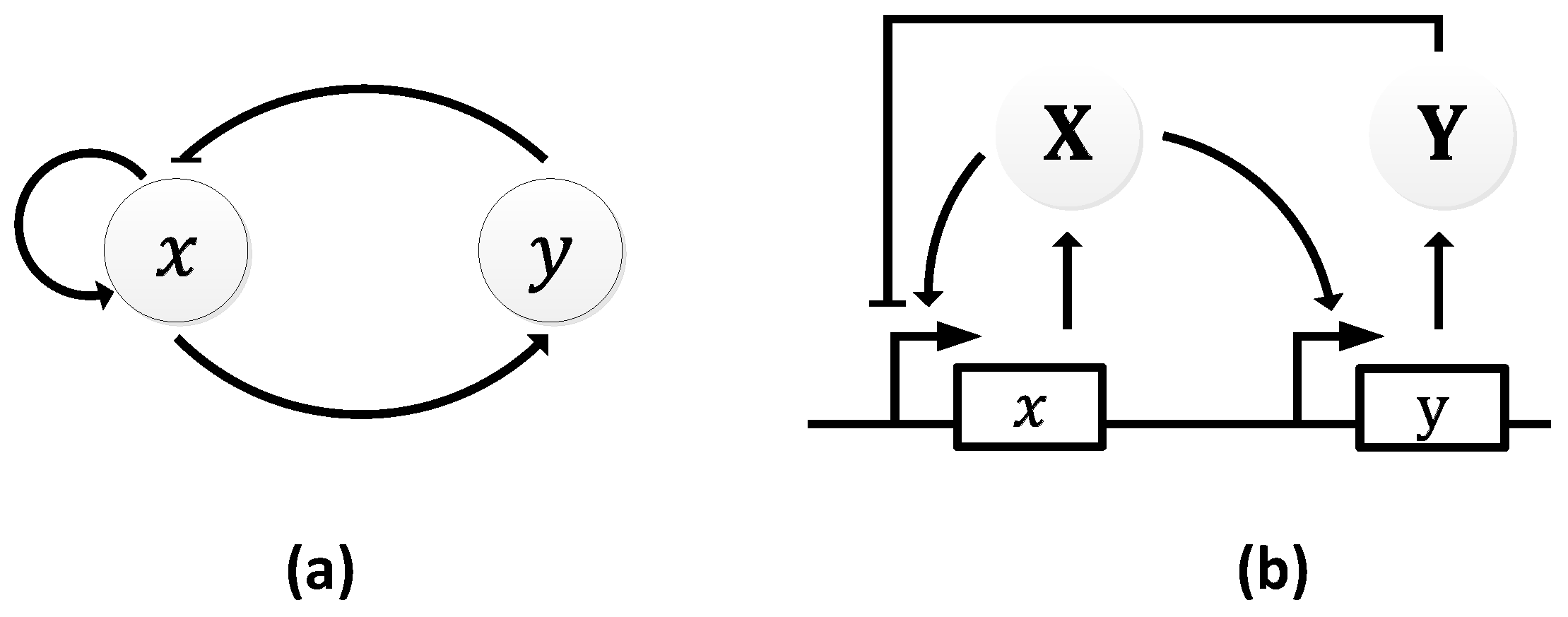



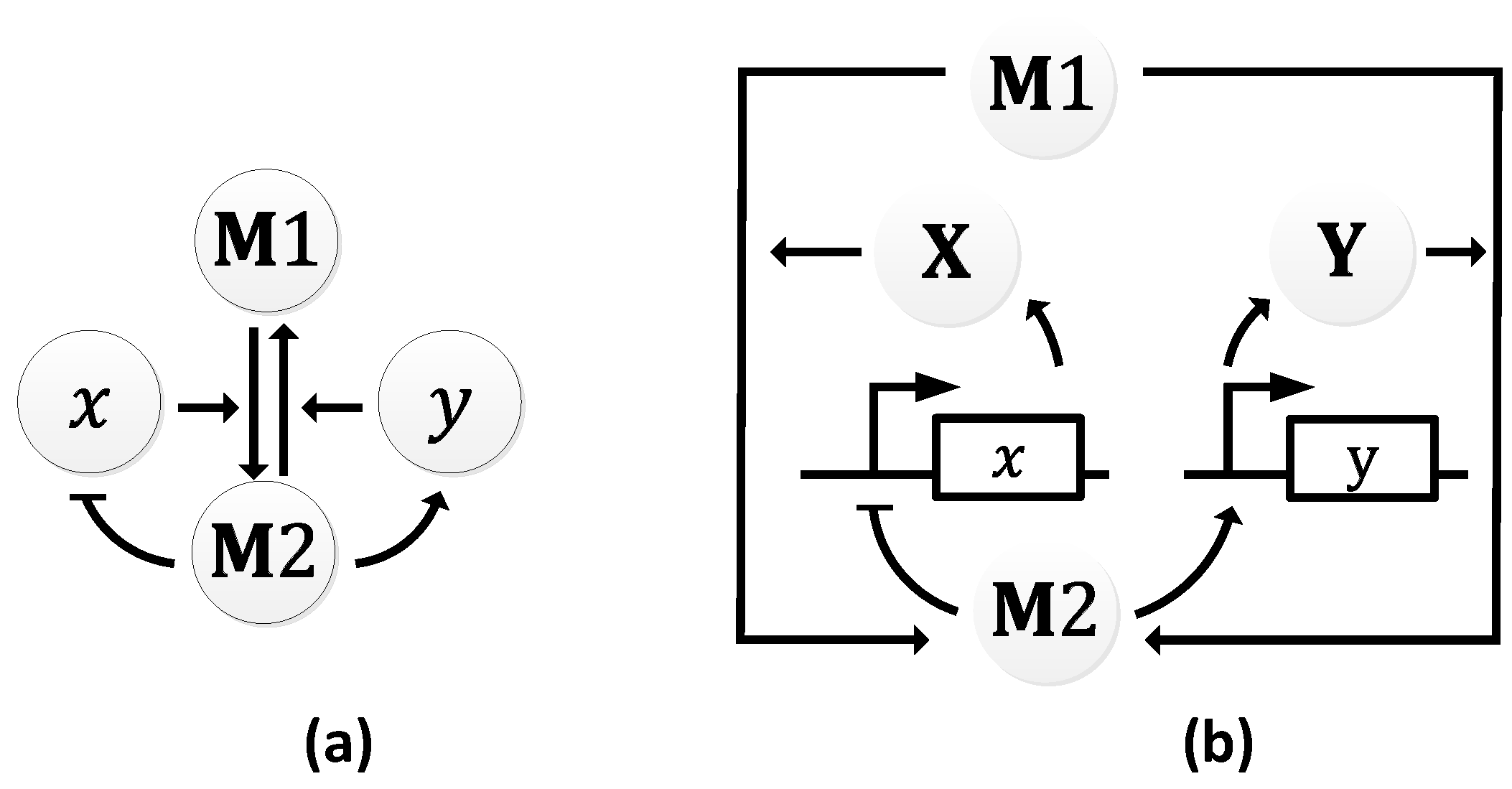

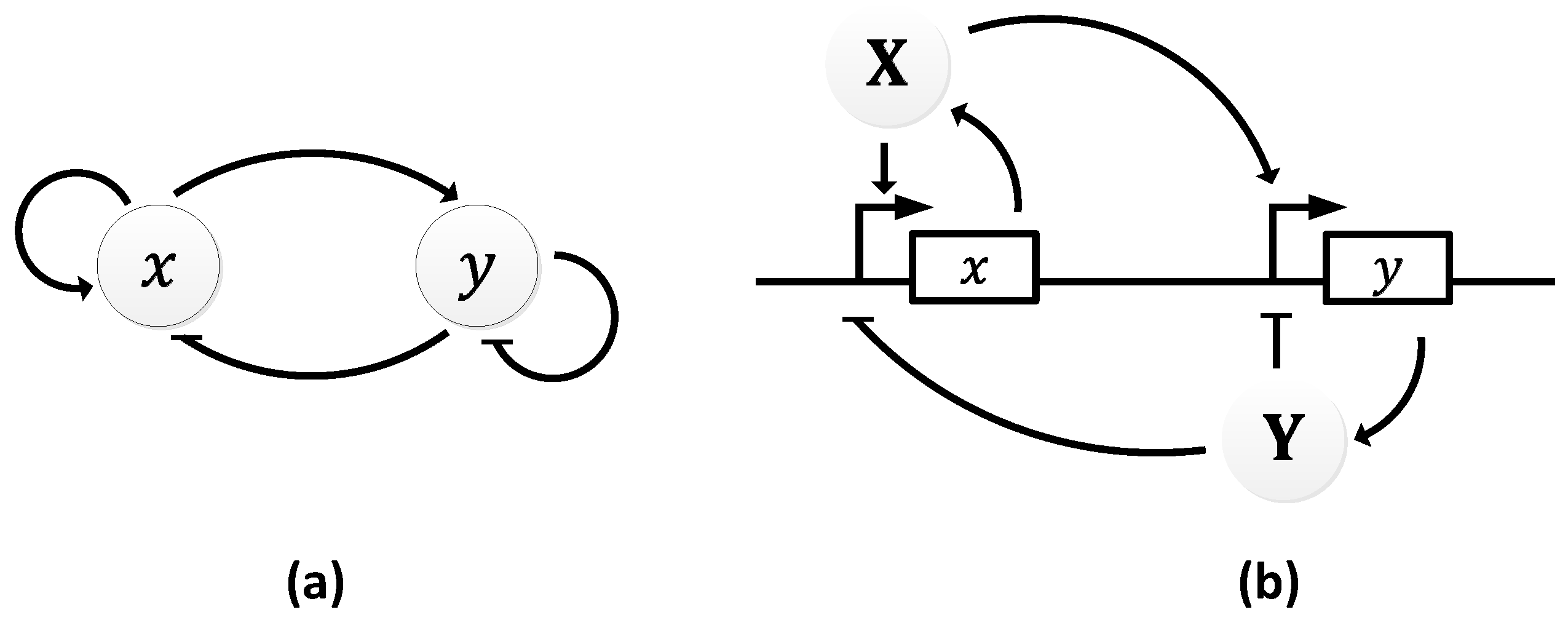
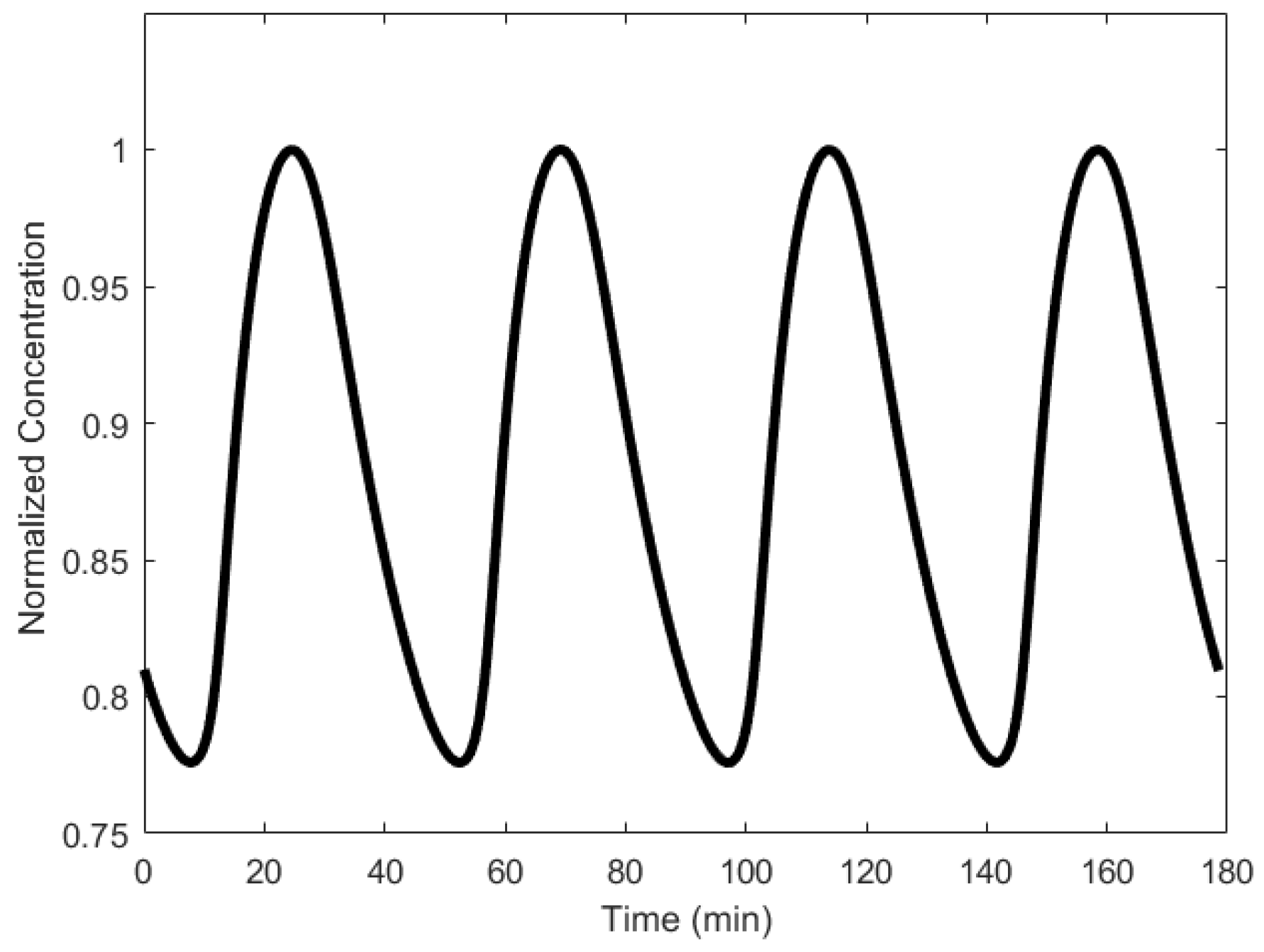
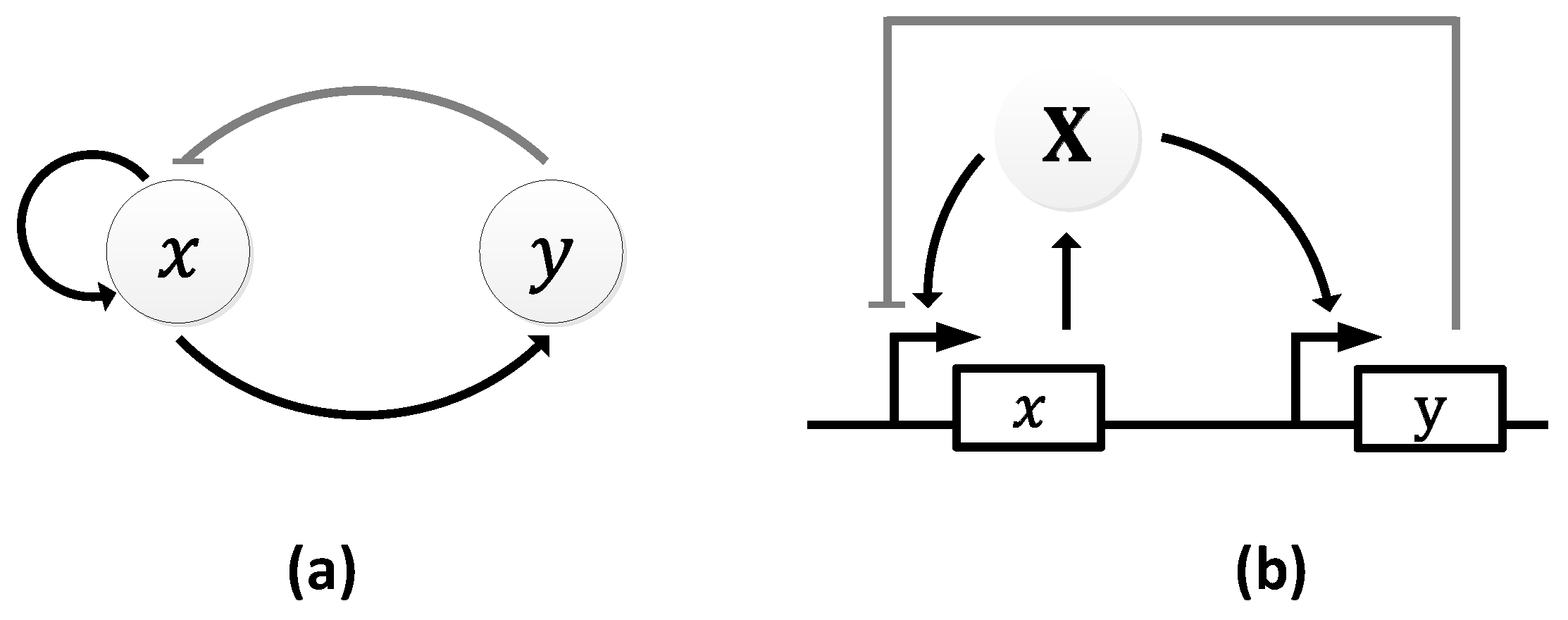
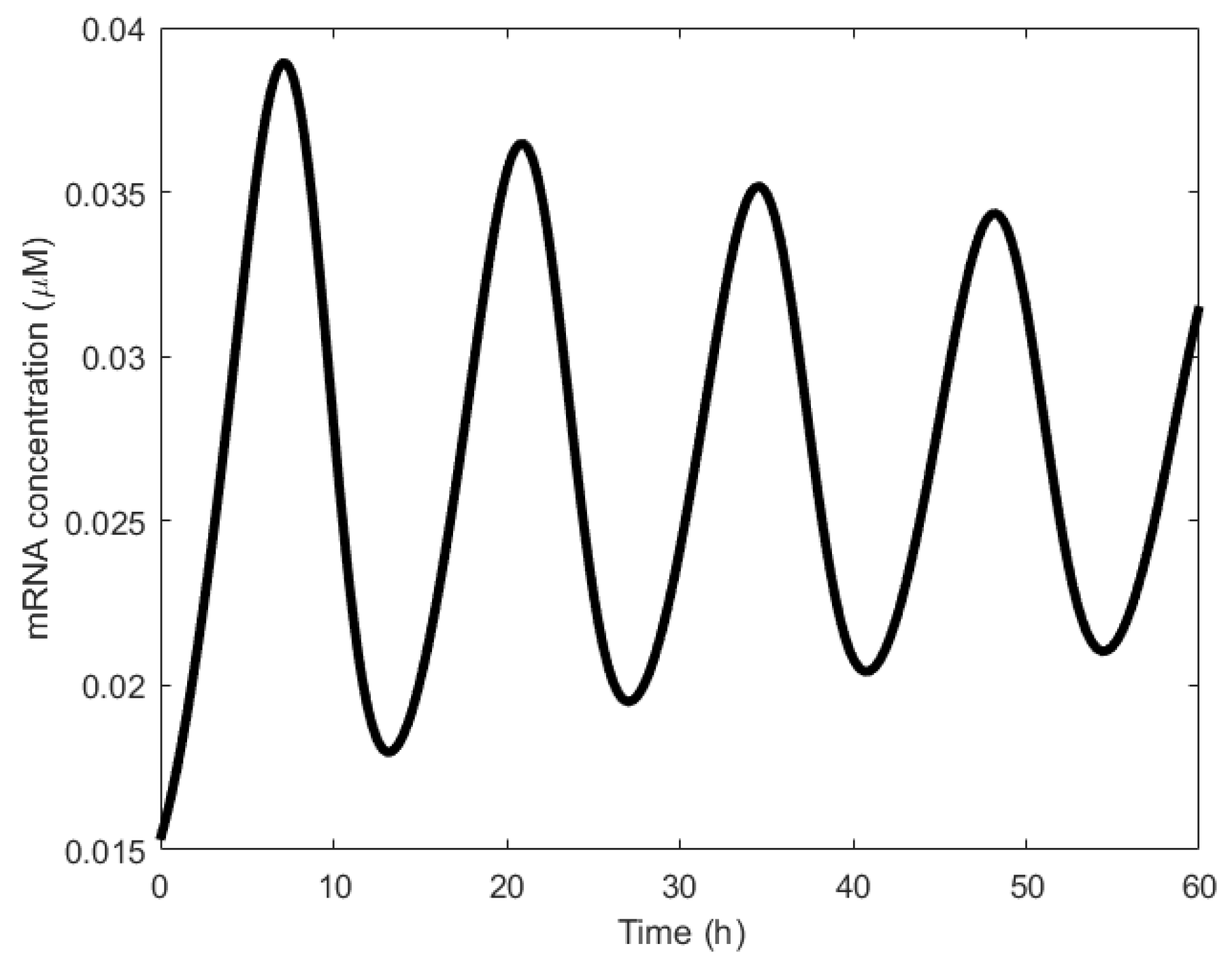


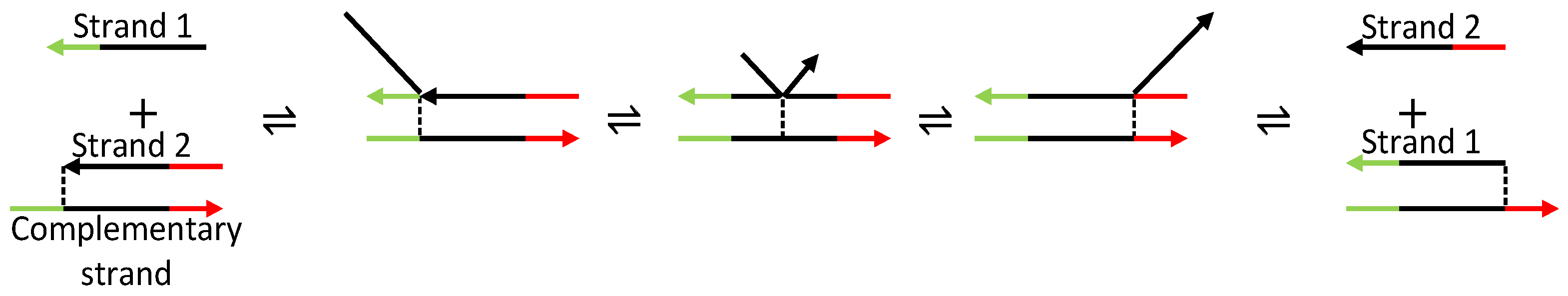
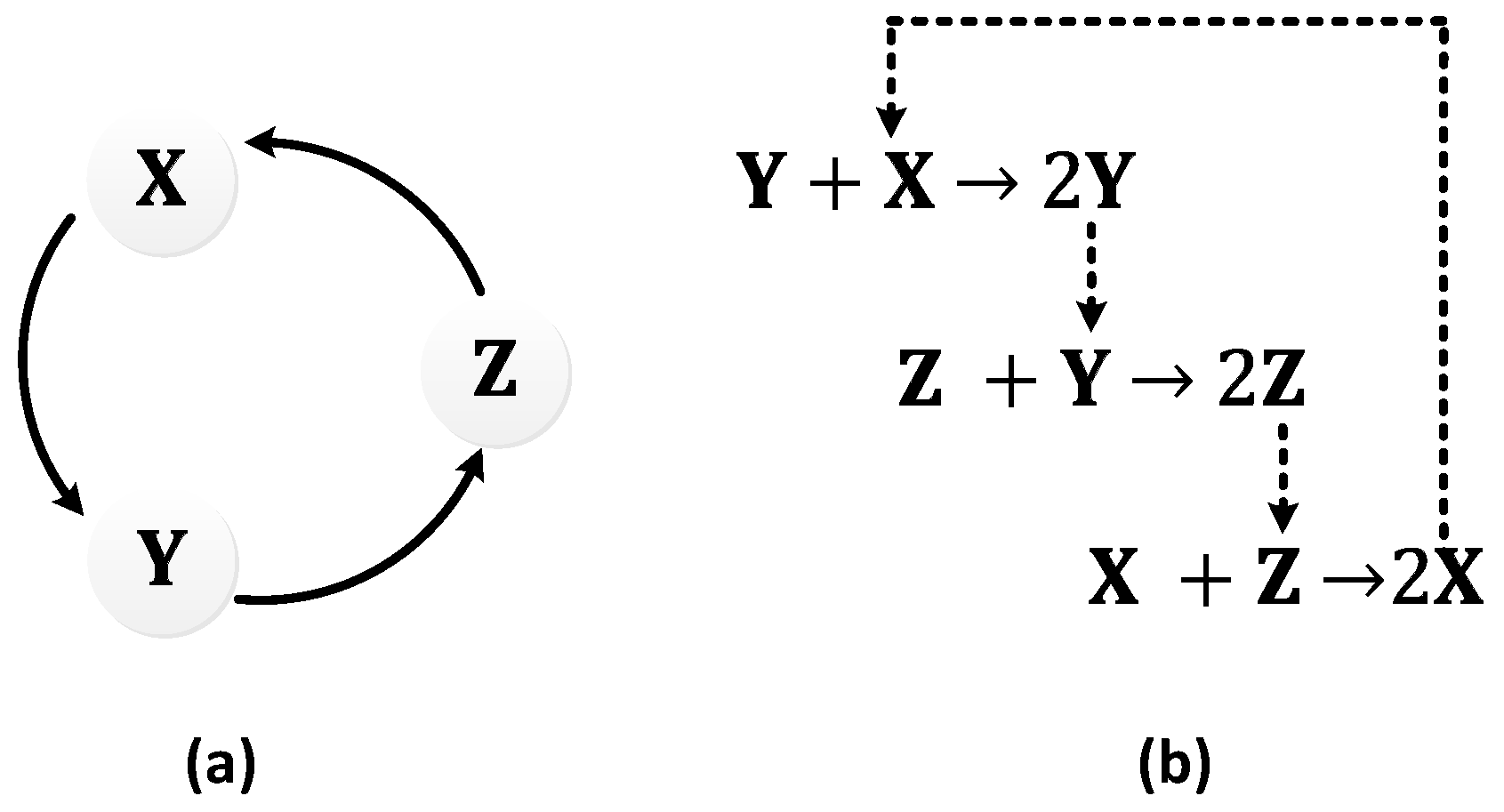
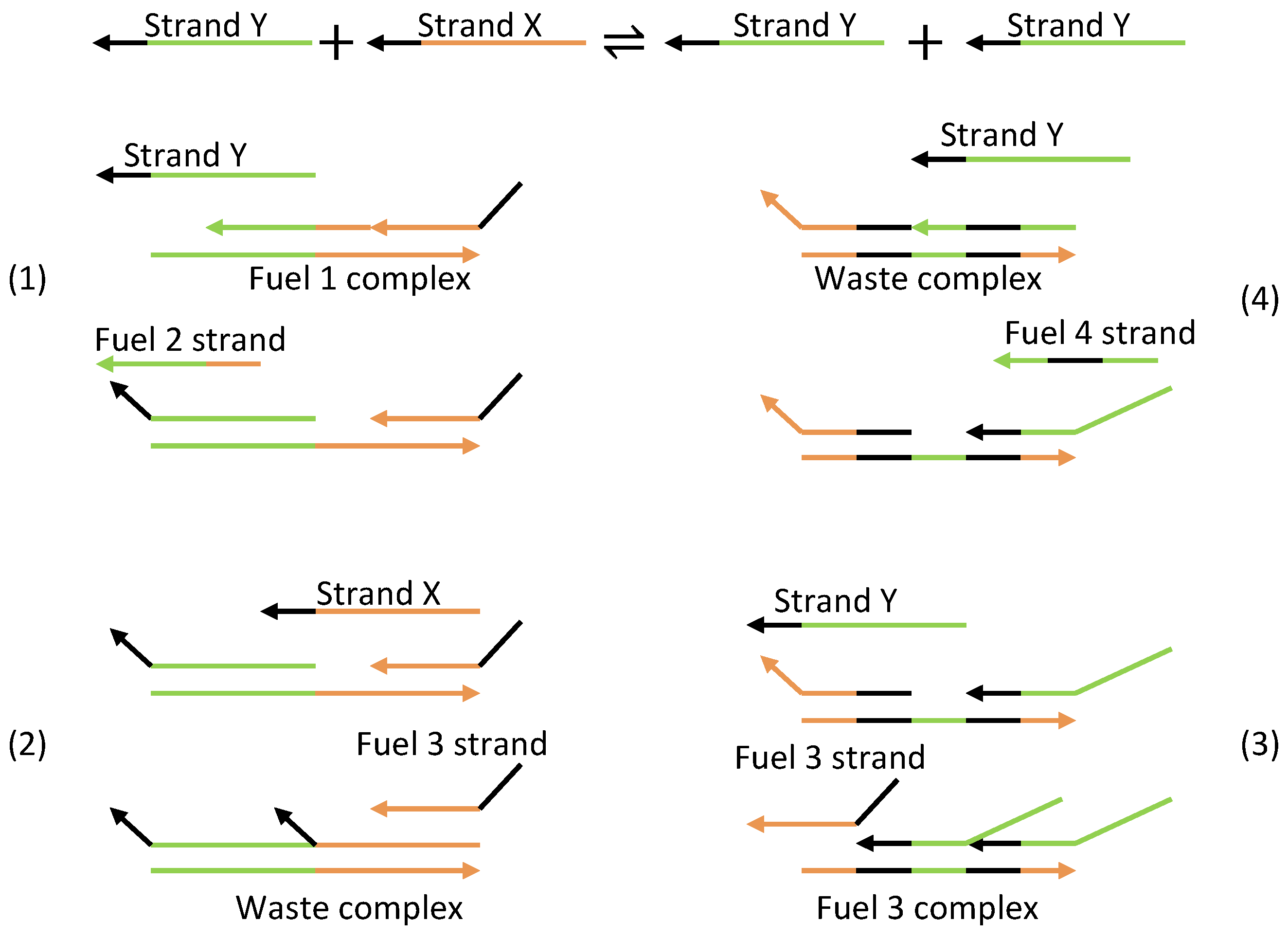
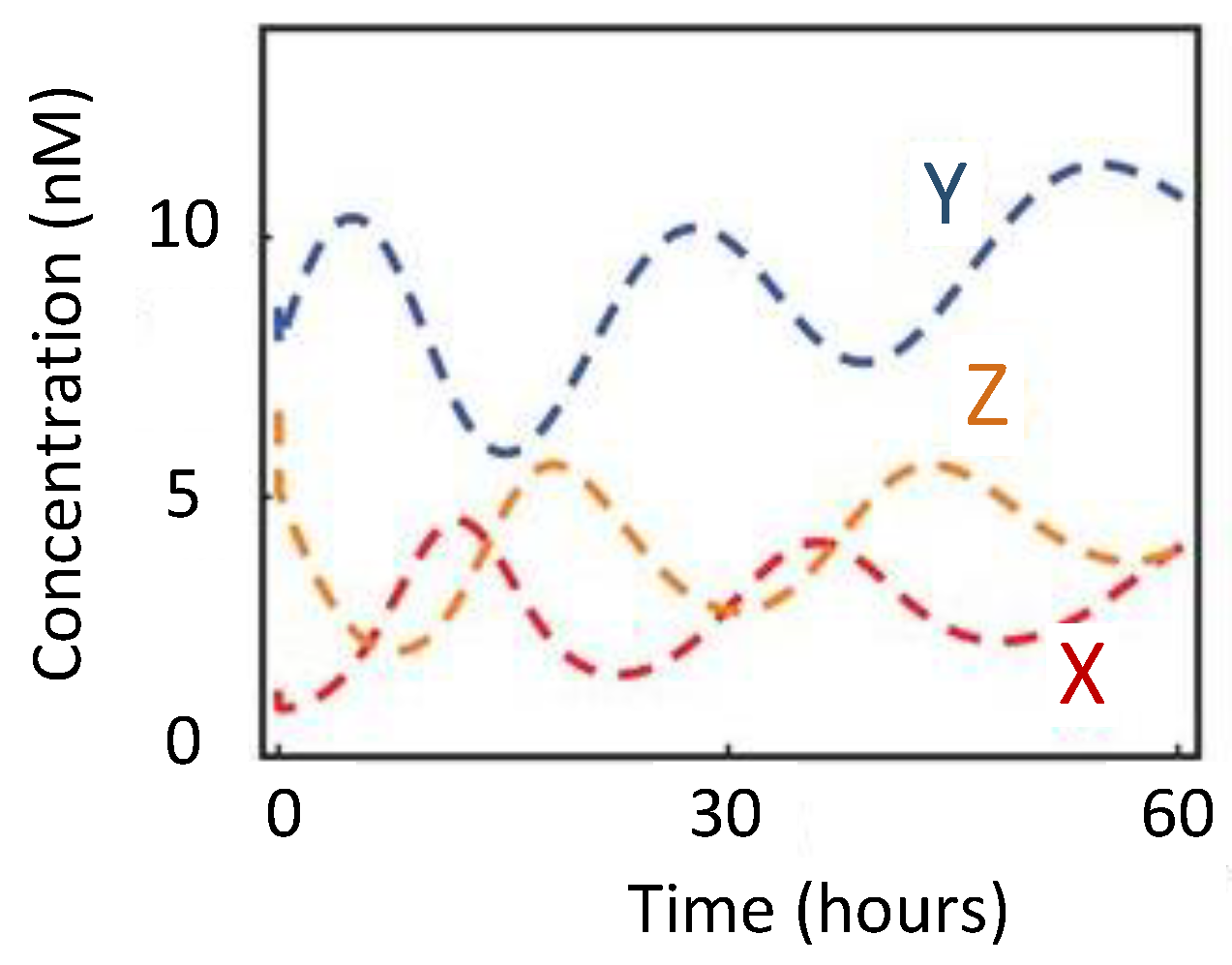
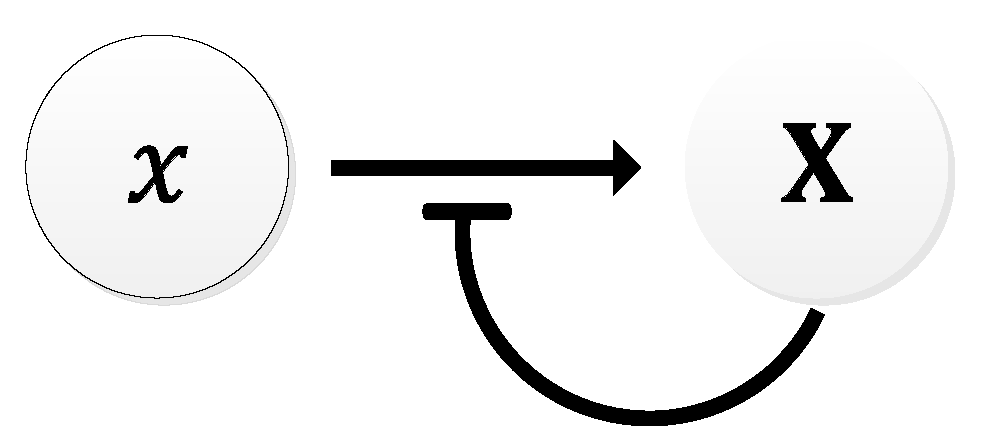
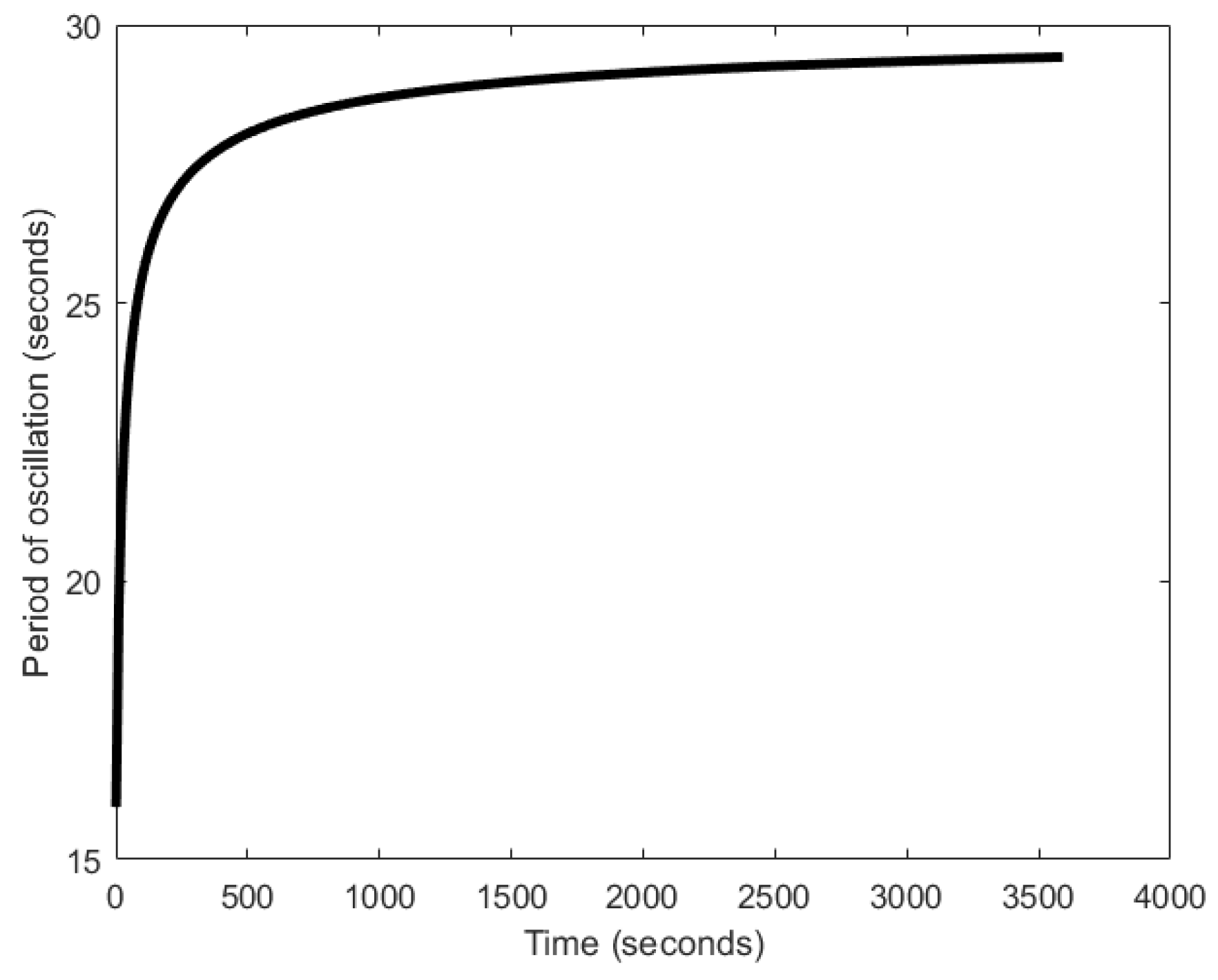

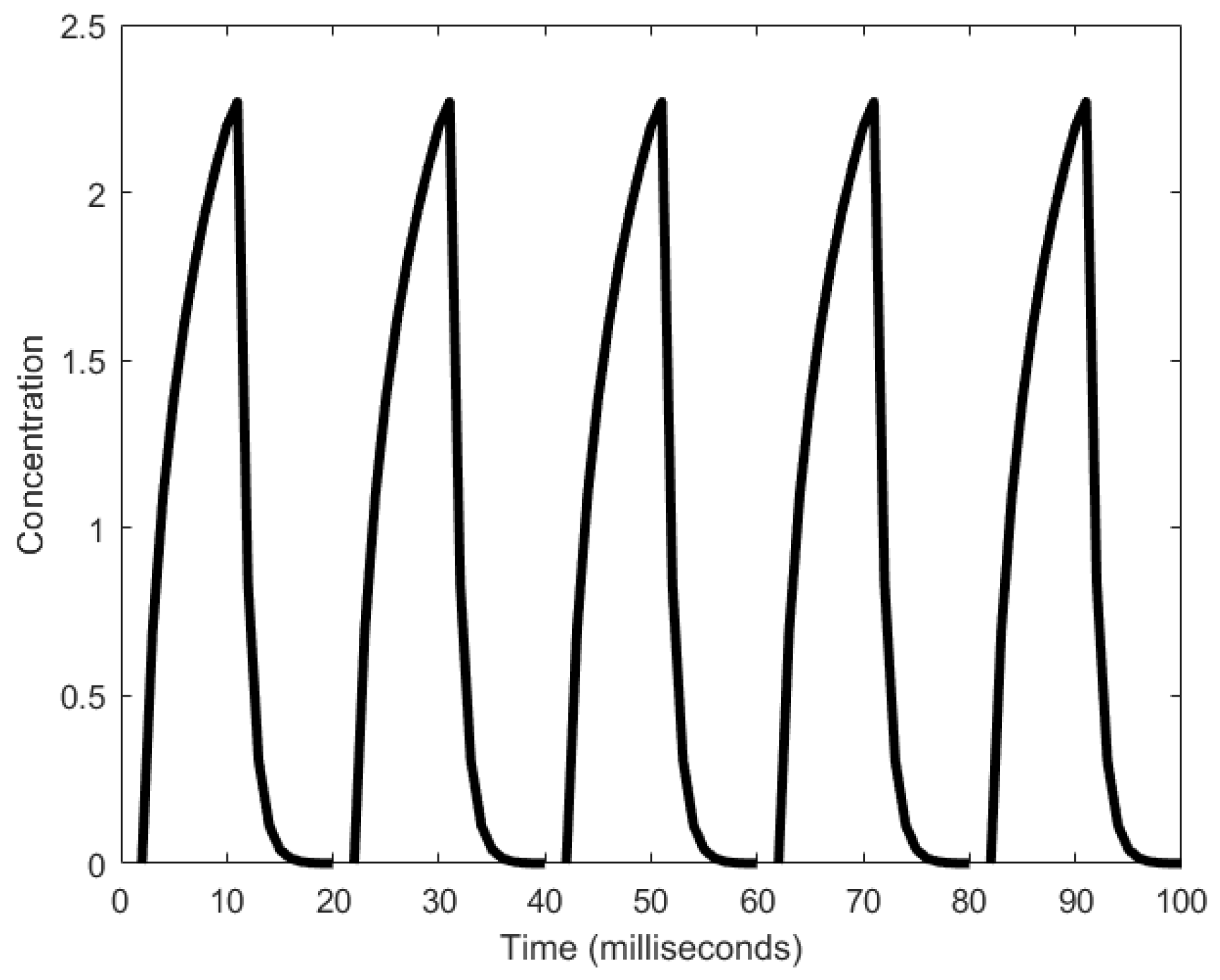
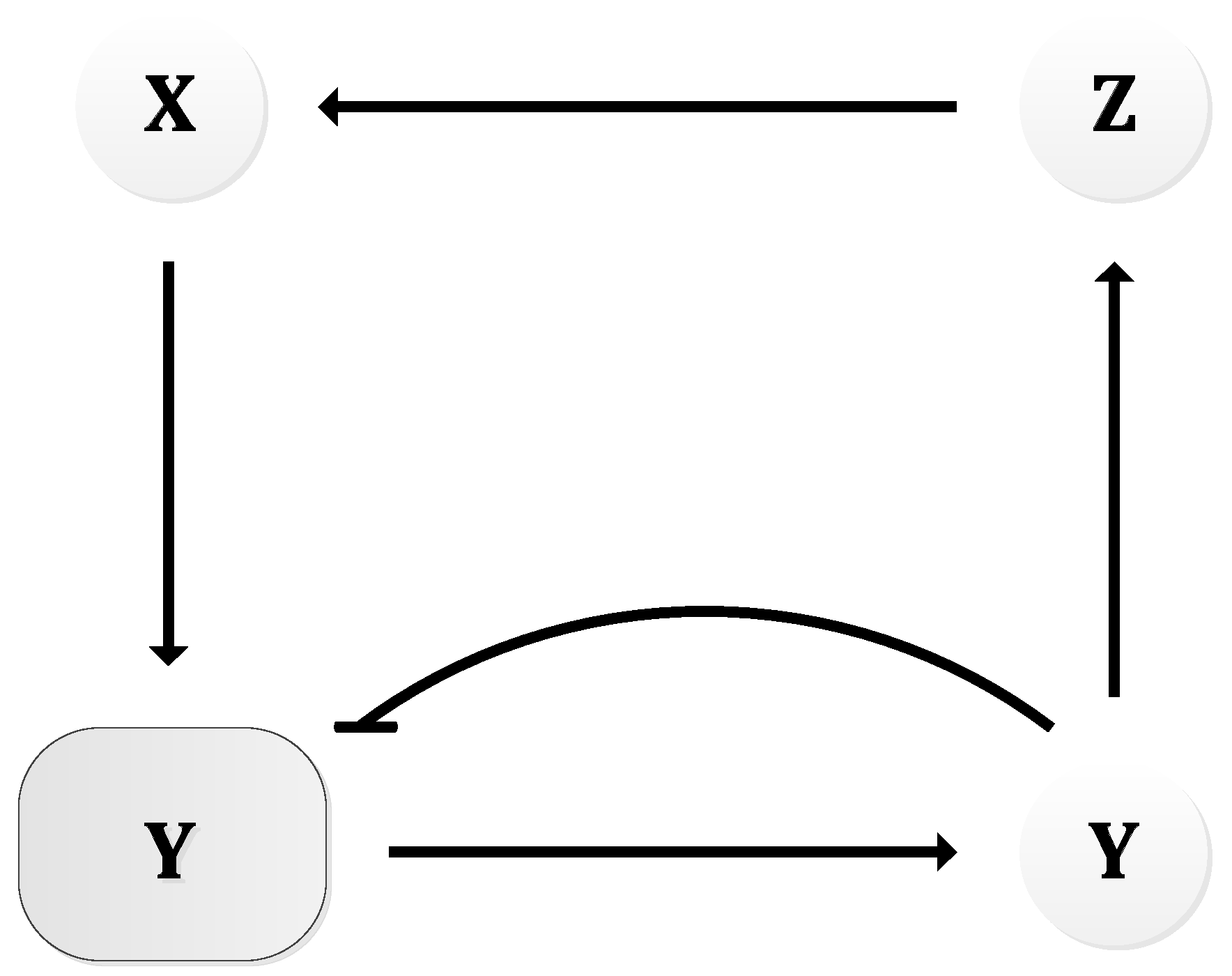

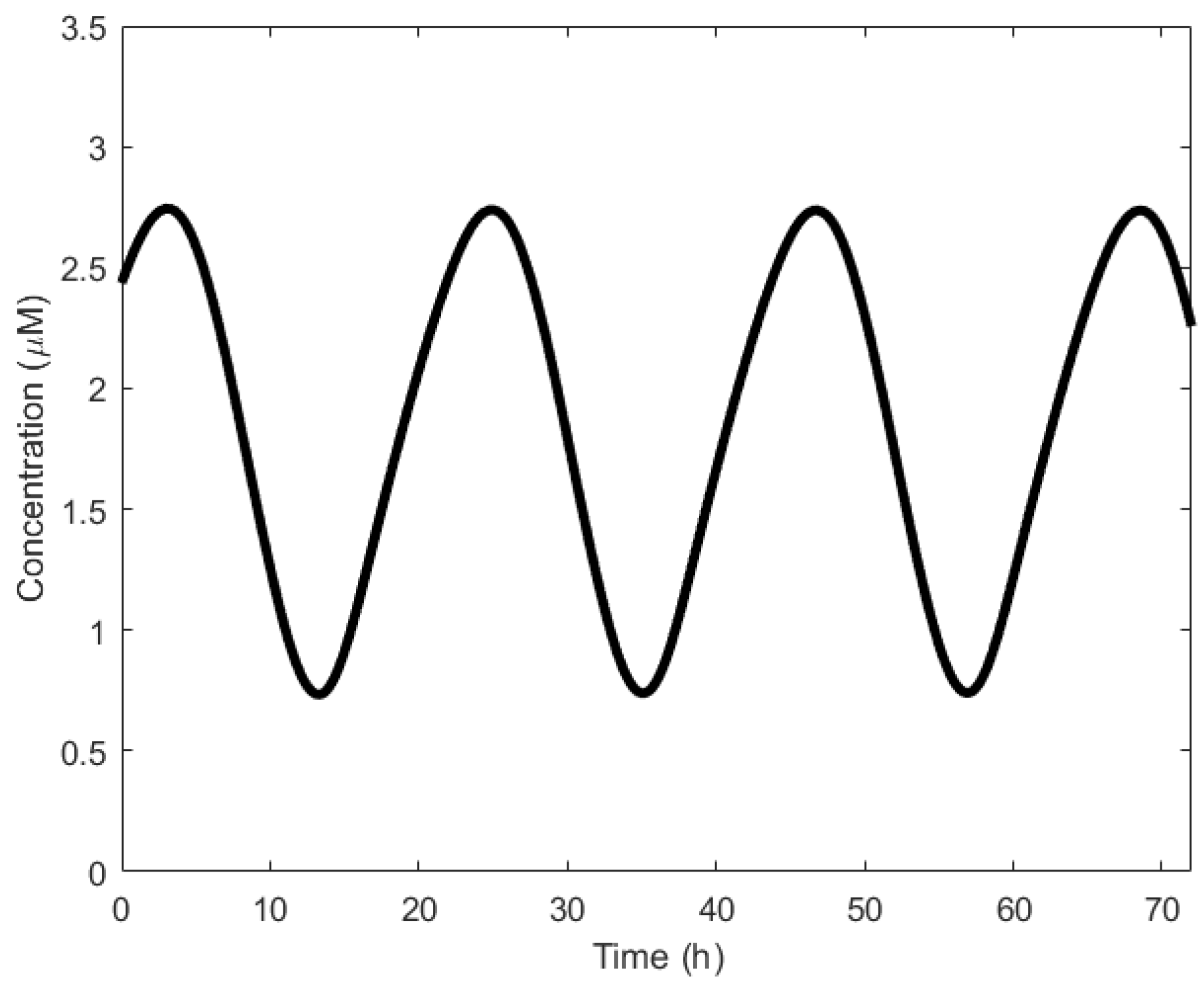
| Oscillator | Year of Discovery | Cell/Organism | Frequency | Process Regulating the Oscillations |
|---|---|---|---|---|
| Circadian oscillator | 1729 [65] | Unicellular and multicellular | 11.57 μHz | Transcription regulation |
| Mitotic oscillator | 1974 [92] | Eukaryotic cells | 11.57 μHz to 1.7 mHz | Enzyme regulation |
| Glycolytic oscillator | 1965 [35] | Yeast cells; heart cells; muscle extracts; Pancreatic beta cells; | 0.3–16.7 mHz | Enzyme regulation |
| cAMP oscillator | 1974 [50] | Dictyostelium discoideum | 1.7–3.33 mHz | Receptor-enzyme interactions |
| Calcium oscillator | 1985 [76] | Variety of cells | 16.7 mHz to 1 Hz | Transport between cells and their environment or between various intracellular compartments |
| Oscillator | Year | Frequency: Theoretical/Experimental | Oscillations Observed | Process Involved in the Oscillations |
|---|---|---|---|---|
| miRNA-regulated oscillator | 2013 [25] | 11.57 μHz/n.a | n.a. | Post-transcription regulation |
| Displacillator | 2017 [114] | 0.0139 mHz/0.0139 mHz | n.a. | Strand displacement |
| Fussenegger oscillator | 2009 [23] | 0.019 mHz/0.098 ± 0.23 mHz | n.a. | Post-transcription regulation |
| Atkinson oscillator | 2003 [106] | 0.028 mHz/0.02 mHz | n.a. | Transcription regulation |
| Repressilator | 2000 [21] | 0.11 mHz/0.1 ± 0.42 mHz | 40% | Transcription regulation |
| Dual feedback oscillator | 2008 [22] | 0.38 mHz/0.42 mHz | 99% | Transcription regulation |
| Hasty oscillator | 2001 [107] | (0.38–2.08) mHz/n.a. | n.a. | Transcription regulation |
| Metabolator | 2005 [108] | 0.42 mHz/1.67 ± 0.37 mHz | 60% | Metabolic and transcription regulation |
| Goodwin oscillator | 1963 [100] | 0.56 mHz/n.a. | n.a. | Transcription regulation |
| Oscillator | Year | Frequency: Theoretical/Experimental | Oscillations Observed | Process Involved in the Oscillations |
|---|---|---|---|---|
| Moore oscillator | 2013 [29] | 50 mHz–mHz/n.a. | n.a. | Auto-inhibition |
| Akgül oscillator | 2014 [30] | 100 mHz/n.a. | n.a. | Auto-inducer |
| Shitiri oscillator | 2016 [31] | 500 mHz/n.a. | n.a. | Molecule-receptor interactions |
| Oscillator | Feedback Loops | Model-to-Wet Lab Agreement | Robustness | Tunability | Oscillations |
|---|---|---|---|---|---|
| Goodwin oscillator | One self-(−) | Bad | Low | No | Sustained |
| Repressilator | Three (−) | Good | Medium | n.a. | Sustained |
| Atkinson oscillator | One self-(−) and one (−) | Excellent | n.a. | n.a. | Damped |
| Hasty oscillator | One self-(±), one (±), and one (−) | n.a. | n.a. | n.a. | Sustained |
| Metabolator | One (−) and one (+) | Good | Medium | n.a. | Sustained |
| Dual feedback oscillator | One (−), one self-(−), one (+), and one self-(+) | Good | High | Yes | Sustained |
| Fussenegger oscillator | One self-(−) and one (−) | Good after revision | n.a. | n.a. | Damped |
| miRNA-regulated oscillator | One self-(−) and one coupled (−) | n.a. | n.a. | n.a. | Sustained |
| Displacillator | Three (+) | Good | n.a. | Yes | Damped |
| Moore oscillator | Two self-(−) and two (−) | n.a. | n.a. | n.a. | Sustained |
| Akgül oscillator | One self-(+) | n.a. | n.a. | n.a. | Sustained |
| Shitiri oscillator | Two self-(−) and one (+) | n.a. | n.a. | Yes | Sustained |
| Oscillator | Advantages | Disadvantages | Type | Frequencies |
|---|---|---|---|---|
| Natural | Not embedded into a nanomachine Simpler to implement | Close proximity Additional interfacing | Broadcasting only | Low |
| Synthetic | Embedded into a nanomachine Allows more functionalities | Synchronization required | Peer-to-peer or broadcasting | Low–medium |
| Synthetic nanonetwork | Embedded into a nanomachine Allows more functionalities | Synchronization required Lacks laboratory validation | Peer-to-peer or broadcasting | Medium–high |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shitiri, E.; Vasilakos, A.V.; Cho, H.-S. Biological Oscillators in Nanonetworks—Opportunities and Challenges. Sensors 2018, 18, 1544. https://doi.org/10.3390/s18051544
Shitiri E, Vasilakos AV, Cho H-S. Biological Oscillators in Nanonetworks—Opportunities and Challenges. Sensors. 2018; 18(5):1544. https://doi.org/10.3390/s18051544
Chicago/Turabian StyleShitiri, Ethungshan, Athanasios V. Vasilakos, and Ho-Shin Cho. 2018. "Biological Oscillators in Nanonetworks—Opportunities and Challenges" Sensors 18, no. 5: 1544. https://doi.org/10.3390/s18051544