1. Introduction
In the European Union (EU), 1.7 million persons younger than 75 years old died in 2013. Nearly 34% of those deaths could have been avoided if the patients had been provided with better healthcare [
1]. Similarly, in 2010, the World Health Organization (WHO) accredited 63% of global deaths to non-communicable diseases that are largely preventable [
2]. The published statistics clearly showcase that there is significant room for improvement in the field of personal healthcare in the EU and worldwide. Around half of avoidable deaths are related to heart attacks and strokes. Blood pressure (BP) increases, among other factors, heighten the risk of cardiovascular diseases, strokes, renal failure [
3], and arterial stiffness [
4]. Thus, hypertension thresholds need to be established for appropriate and timely treatments [
5]. A typical biomedical signal processing system encompasses the biological system of interest, the sensors used to capture the activity of the biomedical system, and the methodology developed to analyze the signals and extract the desired information from the activity under scrutiny. In our study, the biological system being analyzed is the heart system, whose electrical activity is illustrated through ECG signals. There are two phases for managing blood flow: a diastole phase known as the filling phase and a systole phase known as the pumping phase. Blood pressure is defined as the force of the blood pushing against the walls of the arteries as the heart pumps blood [
6] and is measured in millimeters of mercury (mmHg). A normal heart rate is considered to be 70 beats per minute [
7]. The maximum pressure during one heart beat is the SBP and the minimum pressure in between two heart beats is the DBP. Recent technological advances have brought wearable bio-sensors (e.g., ECG sensors, sweating-rate sensors, respiration-rate body sensors, etc.) into everyday life. Wearable bio-sensors provide an opportunity for real-time monitoring of human vital signs, thus enabling the possibility for preventive, timely notification and real-time diagnosis [
8]. Unlike commonly-used BP sensors, which demand a specific measurement procedure, modern wearable bio-sensors monitor vital signals on line and all day long, presenting no additional burden other than wearing the device. Many of the devices, even the low-cost ones, achieve reasonable results in real-life circumstances. Some of the systems developed for the purpose of non-invasive BP monitoring are: the Superficial Temporal Artery Tonometry-based device [
9], the PPG optical sensor [
10], ARTSENS (ARTerial Stiffness Evaluation for Non-invasive Screening) for brachial arterial pressure [
11], an electronic system based on the oscillometric method [
12], a BP estimation device based on the principle of volume compensation [
13], a Modulated Magnetic Signature of Blood mechanism [
14], and portable equipment that includes a cuff-based BP sensing system [
15]. However, all of these devices exist as standalone devices that are specialized only for BP measurements, and exclude the other vital signs. Additionally, some of them do not achieve the desired laboratory results when used in real life, in particular for specific groups of users.
Regarding the research on BP estimation, most studies use a combination of electrocardiogram (ECG) and photoplethysmogram (PPG) sensors [
16,
17,
18], making the problem even more complex and equipment necessary. The common techniques used for obtaining measurements on BP mainly rely on Pulse Wave Velocity (PWV), Pulse Arrival Time (PAT), and Pulse Transit Time (PTT) [
19,
20,
21,
22], all of which require an accurate PPG measurement, which is not simple to get unobtrusively yet, and no clear proof on the PPG measurement’s relation to the BP has been provided [
23,
24,
25]. Considering the methods that use ECG sensors, Chan et al. [
26], and Ahmad et al. [
27] have presented studies on this relationship; however, both methods used an additional sensor besides the ECG sensor, i.e., the PPG sensor. The ECG-BP relationship has previously been discussed and tested in few studies [
28,
29]; however, the results confirm no strong relationship between hypertension occurrence and morphological changes in ECG. For this reason, we approach the problem from a different perspective in that our approach does not rely on ECG morphological changes.
In this paper, we present a method that uses the ECG signal as the only source of information for estimating the maximum value of an arterial pressure tracing (SBP), the minimum value of an arterial pressure tracing (DBP), and the MAP value calculated from the SBP and DBP reference values. This method has the potential to be applied on wearable sensors, enabling unobtrusive on line all day measuring of BP parameters. Since wearable sensors technology has been applied in various situations, from ambulatory and clinical situations [
30], to military environments [
31], this tool may be an appropriate solution that will decrease the need for various sensors to be attached to the human body. Considering urgent medicine, the four parameters HR, RR, BP, and SPO2 are the essential vital signs to establish the hemostability of an injured person [
32]. Up until this point, in real situations, BP can be measured only by a standalone BP device in the vehicles or hospitals. On the other hand, modern telemedicine has allowed for the development of systems that use on-field patch-like ECG bio-sensors attached to a patient’s chest. The method we are proposing can derive BP measurement from the ECG signal only.
The approach is based on a combination of complexity analysis and machine learning to detect bio-system complexity and then use AI-based methods to infer medical relations. The complexity analysis [
33] of the ECG signals is used to exclude the morphological features of the ECG signals. The hypothesis that complexity decrease in the case of abnormality was proven empirically for other medical conditions [
34,
35,
36]. Following this hypothesis, we perform a complexity analysis of 3129 ECG signals obtained from 51 subjects of different age groups in either healthy, unhealthy or traumatic conditions. For experimental purposes, the measurements were performed by using three types of commercial bio-sensors and specialized medical equipment for comparison. The acquisition of the reference BP values for the bio-sensor measurements was performed manually by using an intermittent cuff-based method, whereas, for the clinical measurements, a continuous arterial BP monitor was used. The reference BP measurements were used to categorize the ECG signals into three BP classes (Normal, 0; Prehypertension, 1; and Hypertension, 2), which consisted of the following groups [
37]: hypotension (HPTN) and normal (N) as Normal class, prehypertension (PHTN) as Prehypertension class, and stage 1 hypertension (S1HTN), stage 2 hypertension (S2HTN), isolated systolic hypertension (ISHTN), and hypertensive crisis (HTNC) as Hypertension class. Having extracted features by using complexity analysis, a stacking Machine-Learning (ML) solution was applied to classify the ECG signals into the appropriate BP category and, consequently, regression models were developed to predict the SBP, DBP and MAP values for the given ECG signal. The rest of the paper is organized as follows. The proposed method and the data used are described in
Section 2.2. The experimental results are presented in
Section 3, followed by a discussion in
Section 4, where we make a direct comparison between our method and related methods for BP estimation. In
Section 5, we present the conclusions of the study.
3. Results
The data were randomly split into three different non-overlapping datasets: 60% of the subjects were included in the training, 10% in the validation and 30% in the testing set. Four different models were built: a classification model that predicts the BP class, which is needed for three different regression models built to predict the real SBP, DBP, and MAP values.
3.1. Training Experiments
The preprocessing and the feature extraction phase produced a total of 3129 vectors containing seven features mapped into three BP classes: normal, prehypertension and hypertension, according to the
Table 2. Considering the output of the classification model to be a very important as part of the feature vectors upon which the regression models are built, we chose the classification model that performed the best among 100 randomly-chosen train-validation-test sets. The performance of the stacking ML solution used for the classification was evaluated through the kappa statistics that in addition to the accuracy of the classifier also takes into account the possibility of guessing the class by chance as described in Equation (
10).
After the evaluation of the validation sets in the particular iteration, the number of candidate models was reduced to eight models that obtained kappa statistics of over 0.40. The accuracy, the kappa statistics from the classification, and the errors in mmHg obtained when used in the regression models for each of the validation sets are presented in
Table 5.
The choice of appropriate model depends on the performance of the regression models created for each of the eight candidate training sets according to the Kappa statistics, taking into account the errors and the correlation between the real and predicted BP values in the validation set.
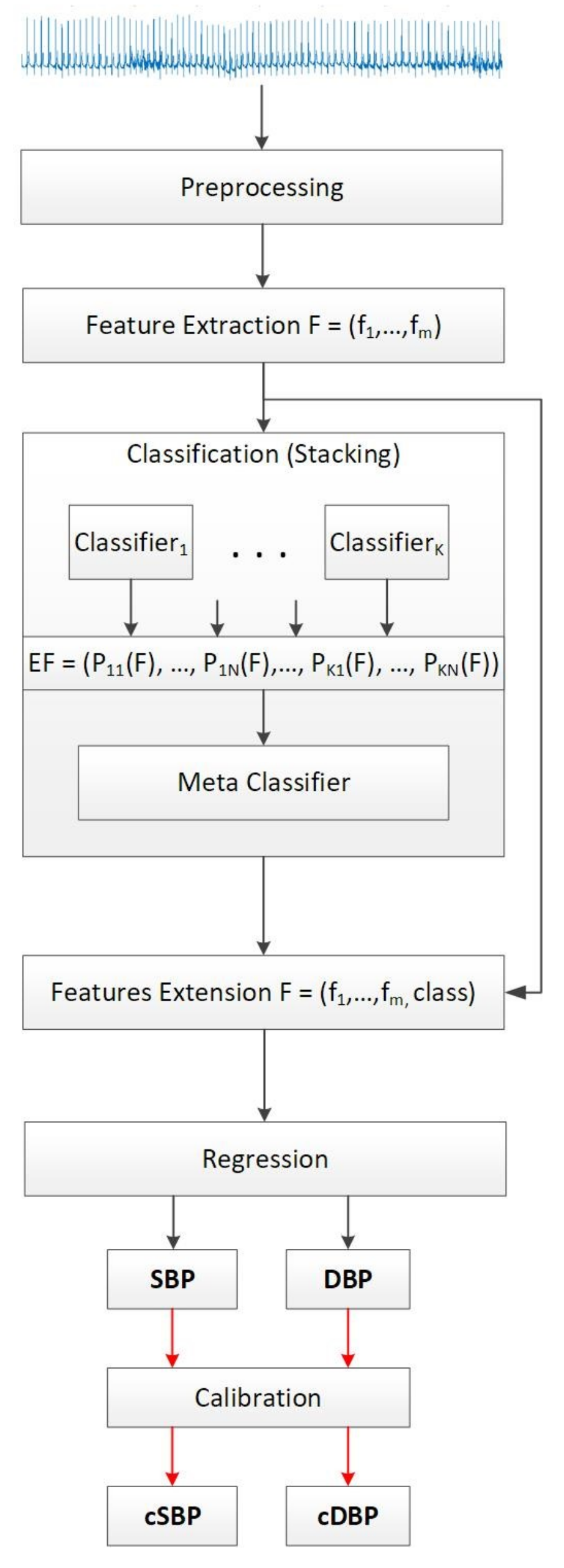
Three distinct models for predicting the SBP, DBP, and MAP were created for each of the candidate training sets that showed greatest accuracy when evaluated with the validation set. The original BP classes in the training sets were used to extend the initial feature vectors and prepare the data for regression, as depicted in
Figure 1. The regression models were evaluated by using the Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE). MAE is the average error obtained from the absolute differences between the real,
, and the predicted values
, for
, where
n is the number of instances per subject. MAE weights all the differences equally and is calculated as:
To obtain a higher weight for the large errors, which is important for the BP problem, the differences between the real absolute and the predicted values are first squared, then averaged, and afterwards a square root of the average is taken. The RMSE is calculated according to the following equation:
Those results are presented in
Table 5 for each SBP, DBP and MAP. The last performance metric that we take into account is the correlation of the real and the predicted BP values in the validation set. Those results are presented in the last column of
Table 5.
Given the results, the model with the highest kappa statistic of 0.78 has been chosen as most suitable with achieved accuracy of 85.71%, and errors of 7.86 mmHg, 6.00 mmHg and 11.19 mmHg for the SBP, DBP, and MAP, correspondingly, producing an average correlation of 0.77 between the actual and the predicted values.
3.2. Testing Experiments
Table 6 presents the MAE and RMSE evaluation for SBP, DBP, and MAP for each of the 15 subjects (denoted from 1 to 15 in Column 1) included in the testing set. The number of instances per test subject ranges from 1 to 436 (Column 2). The total number of tested instances is 786, producing an overall MAE ± SD (RMSE) in mmHg of 8.64 ± 10.74 (10.97) for the SBP case, 18.20 ± 8.45 (19.34) for the DBP case, and 13.52 ± 8.06 (15.07) for the MAP case. Considering the result for each subject separately, the worst results we saw were from subjects 9, 10 and 12. For those subjects, only one instance is available and we are not able to check the predictions for other instances from the same subject, thus we cannot be assured that the particular measurement obtained is reliable.
The absolute blood pressure values for the SBP and DBP prediction are depicted in
Figure 2. The actual BP values are marked with a black line. The green line represents the predicted BP values. All BP values are given in mmHg. It can be concluded that the models tend to predict higher BP values than the actual values, especially when predicting the DBP; however, the prediction line still follows the dynamic of the actual BP values.
To estimate the sensitivity of the results, we used the validation set to investigate its influence in the choice of the hyperparameters. We fed the classifier and the regression models one subject from the validation set in each iteration, obtaining the best hyperparameters for the particular setting. However, no significant changes in the parameters were noticed and the following results in
Table 7 were obtained when testing the models with the designated testing set.
3.3. Calibration Experiments
For solving the problem of miss-predicting the BP values, a calibration method based on the probability distributions of the MAE values obtained from the validation set in the training phase has been proposed. Each SBP, DBP, and MAP error highlighted a test for probability distribution that produced the best fit probability distributions. Their parameters are as follows. The SBP validation set errors showed to fit best in a Generalized Pareto distribution with ; the DBP validation set errors showed to fit best in a Logistic distribution with , and for MAP the validation set errors showed to fit best in Generalized Pareto distribution with .
Having the distributions parameters, for every subject in the testing set, an error from the particular distribution is generated and subtracted from the predicted value. Hereupon, as soon as a new subject is available, a random error from the given probability distribution is generated and is subtracted from the predicted value.
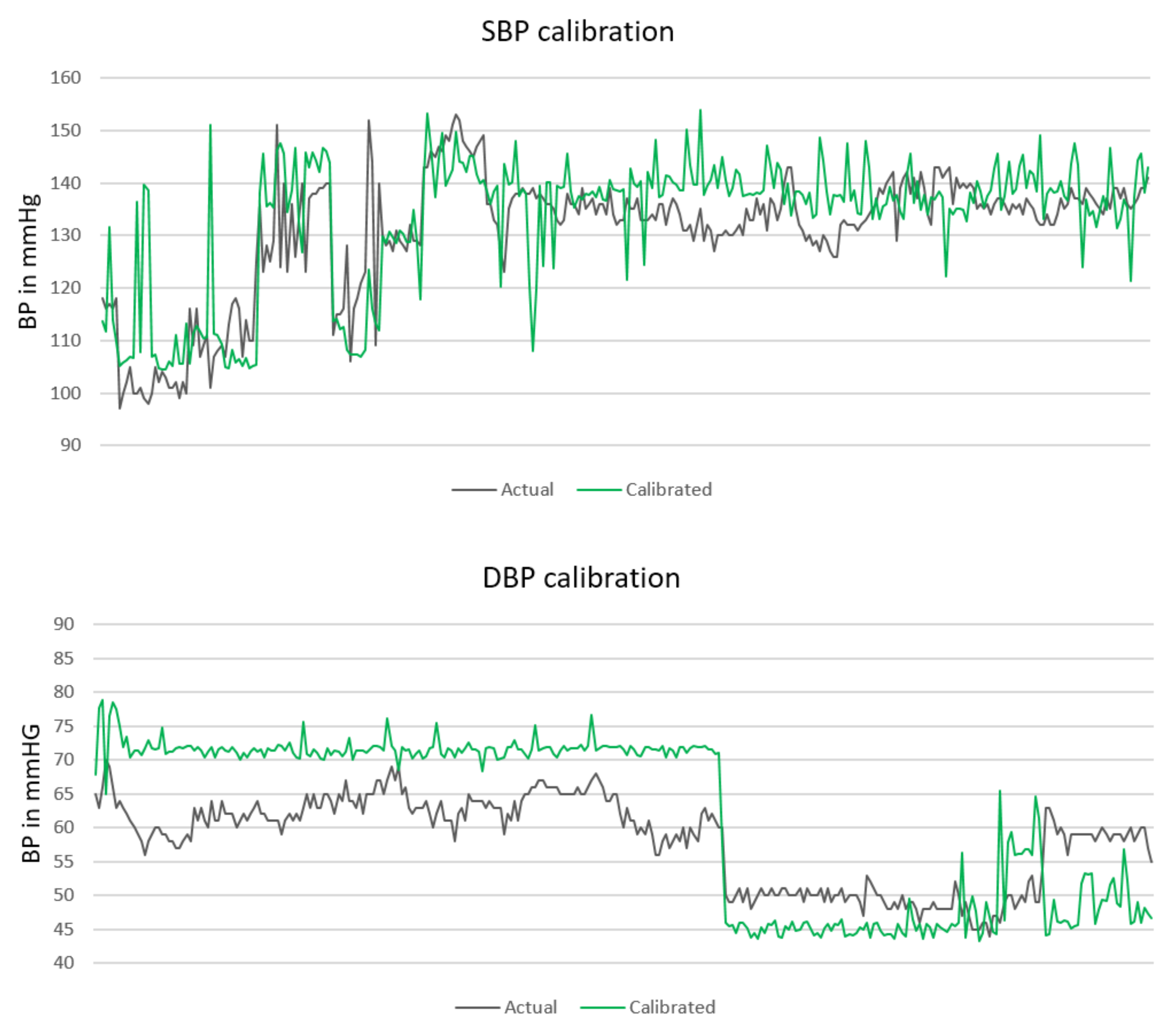
Figure 3 presents the calibrated prediction (green line) for the same BP values presented in
Figure 2.
The overall MAE and RMSE after the calibration significantly decreases, especially in the DBP case. The new MAE ± SD (RMSE) in mmHg are 7.72 ± 10.22 (10.50) for SBP, 9.45 ± 10.03 (11.07) for DBP, and 8.13 ± 8.84 (10.26) for MAP. The calibration results for each distinct subject are presented in
Table 8. Several subjects for which we encountered problems for prediction without calibration, remained problematic in the calibration phase as well. However, a significant improvement was obtained for subjects 14 and 15 (presented in bold in
Table 8), which contain most of the instances, 436 and 258, correspondingly. For those subjects, the total error (MAE SBP + MAE DBP + MAE MAP) in
Table 8 decreased by 40% in the 14th subject and by 50% in the 15th subject when compared to the total error (MAE SBP + MAE DBP + MAE MAP) in
Table 6 for the same subjects.
3.4. Feature Analysis
As previously mentioned, we used a band-pass Butterworth filter for keeping the information that is carried between 0.3 Hz and 50 Hz. Choosing the lowest threshold of 0.3 Hz was both experimentally proven and supported by the literature. Considering the fact that valid ECG information is provided even in the range [0.05, 0.5] Hz [
76]; to completely remove the baseline, the cut-off frequency must be set higher than the lowest frequency in the ECG [
54]. This is necessary in order to prevent some of the baseline to pass as part of the ECG. Therefore, we performed an analysis of how baseline removal at different frequencies affects the performance of the created models. The results are presented in
Table 9. Starting from a cut-off frequency of 0.01 up to the threshold of 0.5, where both the baseline and ECG information exist, it can be perceived that the frequency of 0.3 is the point where the models achieve the highest accuracy, since after this threshold the accuracy starts to decrease again.
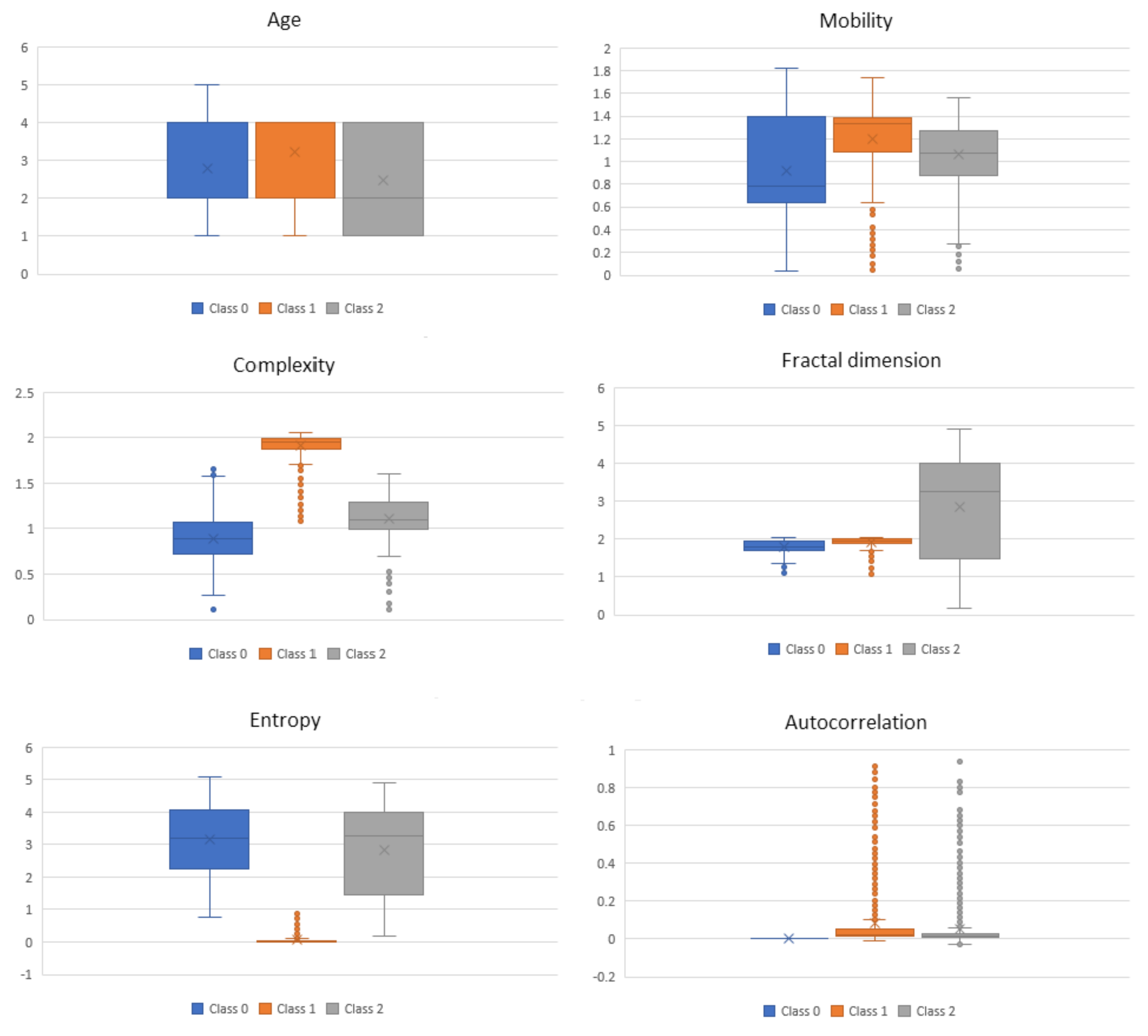
For the complexity features obtained from the ECG signals at 0.3–50 Hz, in
Figure 4, we present box-and-whisker plots to illustrate the shape of the distributions, the mean value, and the variability of each complexity feature with respect to the three BP classes as described in
Table 2. This was done for the purpose of analyzing if a certain feature is distinguishable between the three classes. It can be seen that, for some of the features, e.g., Mobility, Complexity and Entropy, just the mean value itself has a discriminatory power for the three classes. In addition to the mean value, the variability of the feature values also includes some additional information. However, in some cases, the mentioned variability of the feature values may indicate noise in the data.
4. Discussion
Our BP estimation system, based on ECG sensor inputs, enabled reliable monitoring of various BP parameters on data obtained from 51 different subjects and four different ECG sensors. The intention of the proposed method is to reveal new insight into the relation between ECG and BP. The relations are represented by the ML models that we determined from the data. We are not aware of any previous study that described this type of relations using this particular choice of features. The datasets we published are to be freely available for scientific purposes [
77].
The proposed BP estimation system introduced several novelties which led to a performance close to that of a certified medical device. The first novelty was feature extraction using complexity analysis. Based on the hypothesis that a normal and healthy biomedical system is of high complexity and once an abnormality occurs its complexity drops, these features extracted from the ECG signals seem to contain valuable information regarding BP. The complexity analysis excludes the morphological features of the ECG signals and highlights the entropy of the system, which enables better learning and, consequently, predictions. This was confirmed both by the performance of the overall ML system and later on by the feature analysis, where it can be clearly seen that the distributions of the features change with respect to the different BP classes (
Figure 4).
Another novelty that distinguishes our system from the typical “flat” ML approaches for BP estimation is the introduction of the stacking scheme. The stack of several classifiers allows for the meta-learner to receive multiple views over the relations, structures, and patterns in the data, which leads to good performance. The error (MAE) on an unseen testing set is 8.64 for the SBP, 18.20 for the DBP, and 13.52 for the MAP prediction. If a calibration based on validation set errors probability distribution is provided, the MAE significantly decreases to 7.72 for SBP, 9.45 for DBP and 8.13 for MAP. The summary of the results is provided in
Table 10. The goal is to achieve results as close as possible to results obtained by what is considered a certified medical device for BP estimation (±5 mmHg, and SD within 8 mmHg according to BHS and AAMI standards [
78]).
Considering the time performance of the method, the sensing time needed to acquire ECG signals from the sensor is 30 s; the average time needed for the complexity analysis of the signal is 0.1272 s; the average time needed for the methodology to build the model for prediction is 1.0547 s; and the average time needed for performing a prediction is 0.0001 s. Therefore, once the prediction model is built, the predictions can be considered real-time calculations. From related work, we identified only two other studies in which ECG was used for BP estimation. However, both methods used an additional sensor besides the ECG sensor (i.e., PPG sensor) and achieved errors of ±5.93 (SBP), ±4.76 (DBP), and ±4.23 (MAP) when considering 10 participants [
27]; and 7.49 ± 8.8 (SBP) and 4.07 ± 5.6 (DBP) [
26]. In contrast to these approaches, our system uses only one sensor and is trained and evaluated on data from 51 participants and four different sensors.
4.1. Limitations and Future Work
The method could achieve results close to those achieved by medical device by using probability distributions based calibration. If it is desired to measure BP in other than sedentary conditions, the method should be enriched by an activity recognition module (e.g., by using acceleration sensors [
79]). Moreover, a context-based BP estimation may be developed in the future [
80]. The method was based and evaluated on all suitable data that we found to be available for this kind of research together with our own developed database with 51 different subjects and four different ECG sensors. However, for robust testing, a larger study with a few hundred diverse participants will be considered.
4.2. Comparison with Prior Work
Considering the published results, the achieved mean error for the systolic BP (SBP) and diastolic BP (DBP) estimation is 5.1 ± 4.3 mmHg, and 4.6 ± 4.3 mmHg, respectively, in a case study that encompasses 78 PPG records and matching SBP and DBP values [
81]; an error of ±4.76 mmHg for DBP, ±4.23 mmHg for the mean arterial pressure (MAP), and ±5.93 mmHg for SBP is achieved in a pilot study of 150 recordings from 10 subjects [
27]; 9 ± 5.6 mmHg for SBP and 1.8 ± 1.3 mmHg for DBP is obtained from a method that uses Ballistocardiography (BCG) and PPG signals [
82]; 0.8 ± 7 mmHg for SBP and 0.9 ± 6 mmHg for DBP by using PPG signals [
83]; accuracy results of 7.487 ± 8.824 mmHg (mean ± SD) for SBP and 4.076 ± 5.617 mmHg (mean ± SD) for DBP from a PTT-based study [
26]; and a similar PTT-based method tested on 300 datasets from six subjects provides a SD of 6.492 mmHg [
84].
Table 11 presents a comparison of the results reported in this paper with the results reported in the literature. All results present the MAE ± SD. Our study uses least amount of sensors (one), analyzed data from 51 subjects with the widest age range (16–83), used complexity analysis with a stack of ML, and achieved comparable results to the rest of the studies.