UAVs and Machine Learning Revolutionising Invasive Grass and Vegetation Surveys in Remote Arid Lands
Abstract
:1. Introduction
2. Materials and Methods
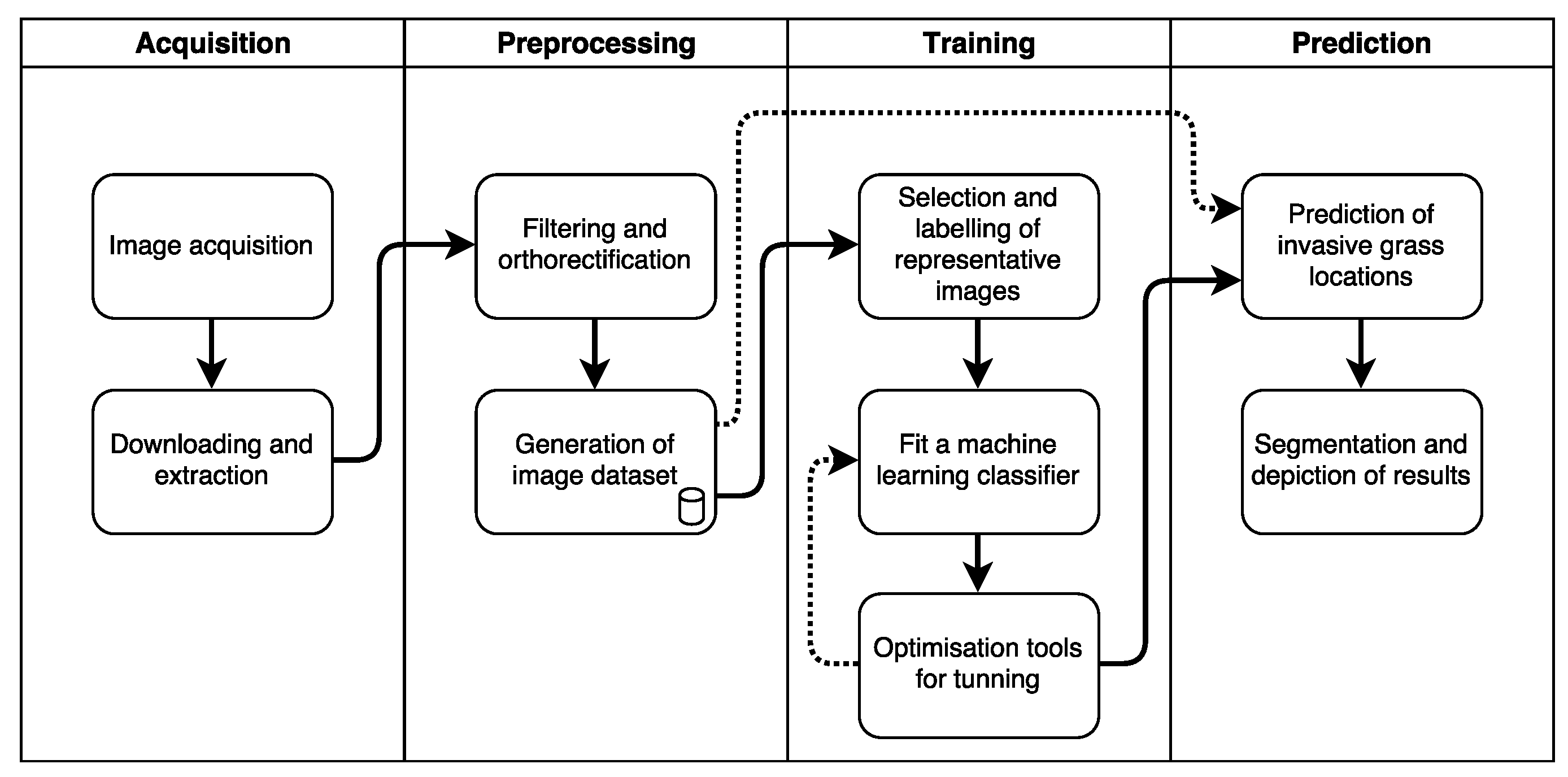
2.1. Process Pipeline
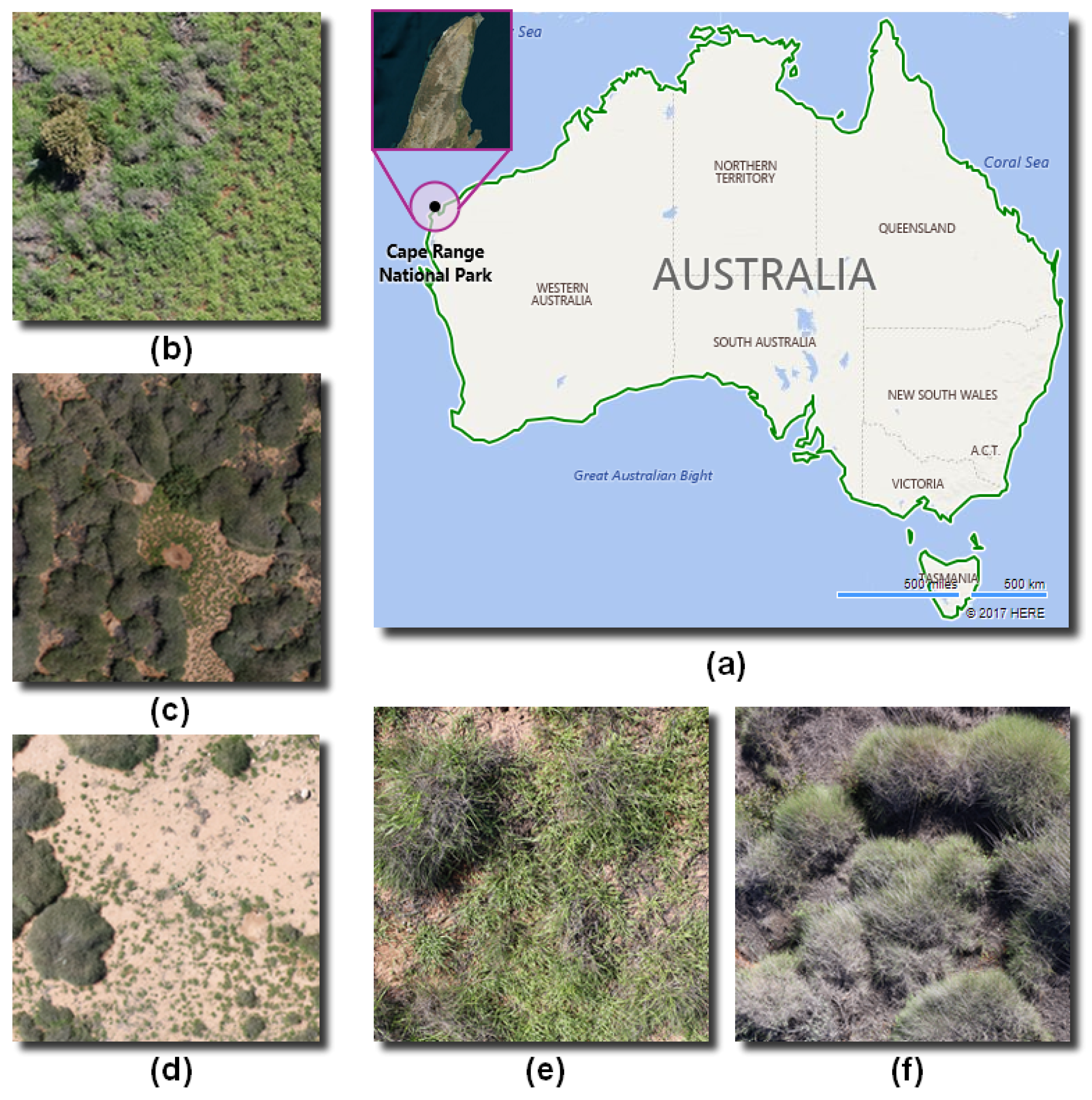
2.2. Site
2.3. Image Sensors
2.4. The UAV and Sample Acquisition
2.5. Software
2.6. Data Labelling
2.7. Classification Algorithm
| Algorithm 1 Detection and segmentation of invasive grasses using high-resolution RGB images. | ||
| Required: orthorectified image set I. Representative samples set G. Sample masks set H | ||
| Training | ||
| 1: | for do | ▹ total images in G (labelled data) |
| 2: | Load and images | |
| 3: | Convert colour space of into HSV | |
| 4: | Insert each colour channel into a feature array D | |
| 5: | Use 2D filters on and insert their outputs into D | |
| 6: | From and , filter only the pixels with assigned labelling on D | |
| 7: | end for | |
| 8: | Split D into training data and testing data | |
| 9: | Create a XGBoost classifier X and fit it using | |
| 10: | Use K-fold cross validation with | ▹ number of folds = 10 |
| 11: | Perform grid search to tune X parameters | |
| Prediction | ||
| 12: | for do | ▹ total images in I |
| 13: | Load image | |
| 14: | Convert colour space of into HSV | |
| 15: | Scan every pixel and predict the object using X | |
| 16: | Convert the data into a 2D image | |
| 17: | Export into TIF format | |
| 18: | end for | |
| 19: | return | |
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| CMOS | Complementary metal–oxide–semiconductor |
| Dry Veg. | Dry vegetation |
| GEOBIA | Geographic Object-Based Image Analysis |
| GIMP | GNU Image Manipulation Program |
| GIS | Geographic information system |
| GPS | Global positioning system |
| GSD | Ground sampling distance |
| HSV | Hue, saturation, value colour model |
| KML | Keyhole Markup Language |
| MDPI | Multidisciplinary Digital Publishing Institute |
| RAM | Random-access memory |
| RGB | Red, green, blue colour model |
| TIF | Tagged Image File |
| UAV | Unmanned Aerial Vehicles |
| WA | Western Australia |
| XGBoost | eXtreme Gradient Boosting |
References
- Godfree, R.; Firn, J.; Johnson, S.; Knerr, N.; Stol, J.; Doerr, V. Why non-native grasses pose a critical emerging threat to biodiversity conservation, habitat connectivity and agricultural production in multifunctional rural landscapes. Landsc. Ecol. 2017, 32, 1219–1242. [Google Scholar] [CrossRef]
- Schlesinger, C.; White, S.; Muldoon, S. Spatial pattern and severity of fire in areas with and without buffel grass (Cenchrus ciliaris) and effects on native vegetation in central Australia. Austral Ecol. 2013, 38, 831–840. [Google Scholar] [CrossRef]
- Fensham, R.J.; Wang, J.; Kilgour, C. The relative impacts of grazing, fire and invasion by buffel grass (Cenchrus ciliaris) on the floristic composition of a rangeland savanna ecosystem. Rangel. J. 2015, 37, 227. [Google Scholar] [CrossRef]
- Grice, A.C. The impacts of invasive plant species on the biodiversity of Australian rangelands. Rangel. J. 2006, 28, 27. [Google Scholar] [CrossRef]
- Marshall, V.; Lewis, M.; Ostendorf, B. Buffel grass (Cenchrus ciliaris) as an invader and threat to biodiversity in arid environments: A review. J. Arid Environ. 2012, 78, 1–12. [Google Scholar] [CrossRef]
- Bonney, S.; Andersen, A.; Schlesinger, C. Biodiversity impacts of an invasive grass: Ant community responses to Cenchrus ciliaris in arid Australia. Biol. Invasions 2017, 19, 57–72. [Google Scholar] [CrossRef]
- Jackson, J. Impacts and Management of Cenchrus ciliaris (buffel grass) as an Invasive Species in Northern Queensland. Ph.D. Thesis, James Cook University, Townsville, Australia, 2004. [Google Scholar]
- Jackson, J. Is there a relationship between herbaceous species richness and buffel grass (Cenchrus ciliaris)? Austral Ecol. 2005, 30, 505–517. [Google Scholar] [CrossRef]
- Martin, T.G.; Murphy, H.; Liedloff, A.; Thomas, C.; Chadès, I.; Cook, G.; Fensham, R.; McIvor, J.; van Klinken, R.D. Buffel grass and climate change: A framework for projecting invasive species distributions when data are scarce. Biol. Invasions 2015, 17, 3197–3210. [Google Scholar] [CrossRef]
- Miller, G.; Friedel, M.; Adam, P.; Chewings, V. Ecological impacts of buffel grass (Cenchrus ciliaris L.) invasion in central Australia—Does field evidence support a fire-invasion feedback? Rangel. J. 2010, 32, 353. [Google Scholar] [CrossRef]
- Smyth, A.; Friedel, M.; O’Malley, C. The influence of buffel grass (Cenchrus ciliaris) on biodiversity in an arid Australian landscape. Rangel. J. 2009, 31, 307. [Google Scholar] [CrossRef]
- Gonzalez, L.; Whitney, E.; Srinivas, K.; Periaux, J. Multidisciplinary aircraft design and optimisation using a robust evolutionary technique with variable fidelity models. In Proceedings of the 10th AIAA/ISSMO Multidisciplinary Analysis and Optimization Conference, Albany, NY, USA, 30 August–1 September 2004; pp. 3610–3624. [Google Scholar]
- Whitney, E.; Gonzalez, L.; Periaux, J.; Sefrioui, M.; Srinivas, K. A robust evolutionary technique for inverse aerodynamic design. In Proceedings of the 4th European Congress on Computational Methods in Applied Sciences and Engineering, Jyväskylä, Finland, 24–28 July 2004. [Google Scholar]
- Gonzalez, L.; Montes, G.; Puig, E.; Johnson, S.; Mengersen, K.; Gaston, K. Unmanned Aerial Vehicles (UAVs) and Artificial Intelligence Revolutionizing Wildlife Monitoring and Conservation. Sensors 2016, 16, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chahl, J. Unmanned Aerial Systems (UAS) Research Opportunities. Aerospace 2015, 2, 189–202. [Google Scholar] [CrossRef]
- Allison, R.S.; Johnston, J.M.; Craig, G.; Jennings, S. Airborne Optical and Thermal Remote Sensing for Wildfire Detection and Monitoring. Sensors 2016, 16, 1310. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.E.; Wood, T.A.; Gullino, M.L.; Ortu, G. Diagnostic Tools for Plant Biosecurity. In Practical Tools for Plant and Food Biosecurity: Results from a European Network of Excellence; Gullino, M.L., Stack, J.P., Fletcher, J., Mumford, J.D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 209–226. [Google Scholar]
- Olsson, A.D.; van Leeuwen, W.J.; Marsh, S.E. Feasibility of Invasive Grass Detection in a Desertscrub Community Using Hyperspectral Field Measurements and Landsat TM Imagery. Remote Sens. 2011, 3, 2283–2304. [Google Scholar] [CrossRef]
- Marshall, V.M.; Lewis, M.M.; Ostendorf, B. Detecting new Buffel grass infestations in Australian arid lands: Evaluation of methods using high-resolution multispectral imagery and aerial photography. Environ. Monit. Assess. 2014, 186, 1689–1703. [Google Scholar] [CrossRef] [PubMed]
- Alexandridis, T.; Tamouridou, A.A.; Pantazi, X.E.; Lagopodi, A.; Kashefi, J.; Ovakoglou, G.; Polychronos, V.; Moshou, D. Novelty Detection Classifiers in Weed Mapping: Silybum marianum Detection on UAV Multispectral Images. Sensors 2017, 17, 2007. [Google Scholar] [CrossRef] [PubMed]
- Blaschke, T.; Hay, G.J.; Kelly, M.; Lang, S.; Hofmann, P.; Addink, E.; Queiroz Feitosa, R.; van der Meer, F.; van der Werff, H.; van Coillie, F.; Tiede, D. Geographic Object-Based Image Analysis—Towards a new paradigm. ISPRS J. Photogramm. Remote Sens. 2014, 87, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sánchez, J.; López-Granados, F.; Peña, J. An automatic object-based method for optimal thresholding in UAV images: Application for vegetation detection in herbaceous crops. Comput. Electron. Agric. 2015, 114, 43–52. [Google Scholar] [CrossRef]
- Ashourloo, D.; Aghighi, H.; Matkan, A.A.; Mobasheri, M.R.; Rad, A.M. An Investigation Into Machine Learning Regression Techniques for the Leaf Rust Disease Detection Using Hyperspectral Measurement. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 4344–4351. [Google Scholar] [CrossRef]
- Robinson, T.; Wardell-Johnson, G.; Pracilio, G.; Brown, C.; Corner, R.; van Klinken, R. Testing the discrimination and detection limits of WorldView-2 imagery on a challenging invasive plant target. Int. J. Appl. Earth Obs. Geoinform. 2016, 44, 23–30. [Google Scholar] [CrossRef]
- Lin, F.; Zhang, D.; Huang, Y.; Wang, X.; Chen, X. Detection of Corn and Weed Species by the Combination of Spectral, Shape and Textural Features. Sustainability 2017, 9, 1335. [Google Scholar] [CrossRef]
- Schmittmann, O.; Lammers, P. A True-Color Sensor and Suitable Evaluation Algorithm for Plant Recognition. Sensors 2017, 17, 1823. [Google Scholar] [CrossRef] [PubMed]
- Bureau of Meteorology. Learmonth, WA—July 2016—Daily Weather Observations; Bureau of Meteorology: Learmonth Airport (station 005007), Australia, 2016. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Bradski, G. The OpenCV library. Dr. Dobb’s J. Softw. Tools 2000, 25, 122–125. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]




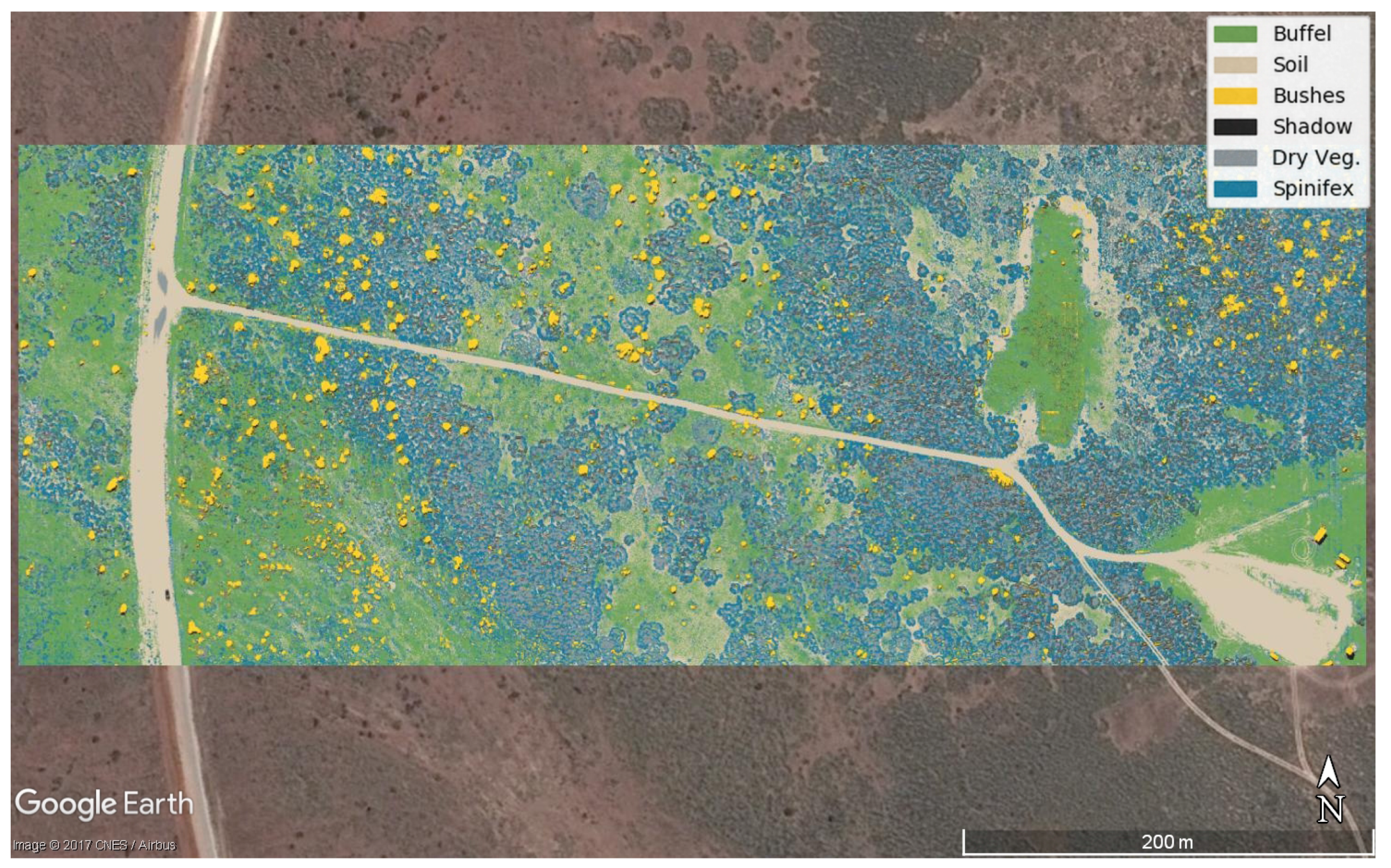
| Predicted | Buffel | Soil | Bushes | Shadow | Dry vegetation | Spinifex | |
|---|---|---|---|---|---|---|---|
| Labelled | Buffel | 25,256 | 17 | 156 | 0 | 4 | 362 |
| Soil | 15 | 25,196 | 1 | 0 | 1 | 0 | |
| Bushes | 632 | 1 | 3913 | 2 | 21 | 81 | |
| Shadow | 0 | 1 | 0 | 7729 | 0 | 0 | |
| Dry vegetation | 8 | 10 | 6 | 2 | 5734 | 159 | |
| Spinifex | 508 | 2 | 20 | 0 | 171 | 15,649 |
| Class | Precision (%) | Recall (%) | F-Score (%) | Support |
|---|---|---|---|---|
| Buffel | 95.60 | 97.91 | 96.75 | 25,795 |
| Soil | 99.88 | 99.93 | 99.90 | 25,213 |
| Bushes | 95.53 | 84.15 | 89.84 | 4650 |
| Shadow | 99.95 | 99.99 | 99.97 | 7730 |
| Dry vegetation | 96.68 | 96.87 | 96.78 | 5919 |
| Spinifex | 96.30 | 95.71 | 96.00 | 16,350 |
| Mean | 97.32 | 95.76 | 96.54 | ∑ = 85,657 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandino, J.; Gonzalez, F.; Mengersen, K.; Gaston, K.J. UAVs and Machine Learning Revolutionising Invasive Grass and Vegetation Surveys in Remote Arid Lands. Sensors 2018, 18, 605. https://doi.org/10.3390/s18020605
Sandino J, Gonzalez F, Mengersen K, Gaston KJ. UAVs and Machine Learning Revolutionising Invasive Grass and Vegetation Surveys in Remote Arid Lands. Sensors. 2018; 18(2):605. https://doi.org/10.3390/s18020605
Chicago/Turabian StyleSandino, Juan, Felipe Gonzalez, Kerrie Mengersen, and Kevin J. Gaston. 2018. "UAVs and Machine Learning Revolutionising Invasive Grass and Vegetation Surveys in Remote Arid Lands" Sensors 18, no. 2: 605. https://doi.org/10.3390/s18020605





