Feature Extraction of Electronic Nose Signals Using QPSO-Based Multiple KFDA Signal Processing
Abstract
:1. Introduction
2. Methodology
2.1. Review of Kernel Fisher Discriminant Analysis
2.2. Properties of Mercer’s Kernels
- (1)
- ;
- (2)
- .
2.3. QPSO-Based Weighted Kernel Fisher Discriminant Analysis Model
3. Description of Experimental Data
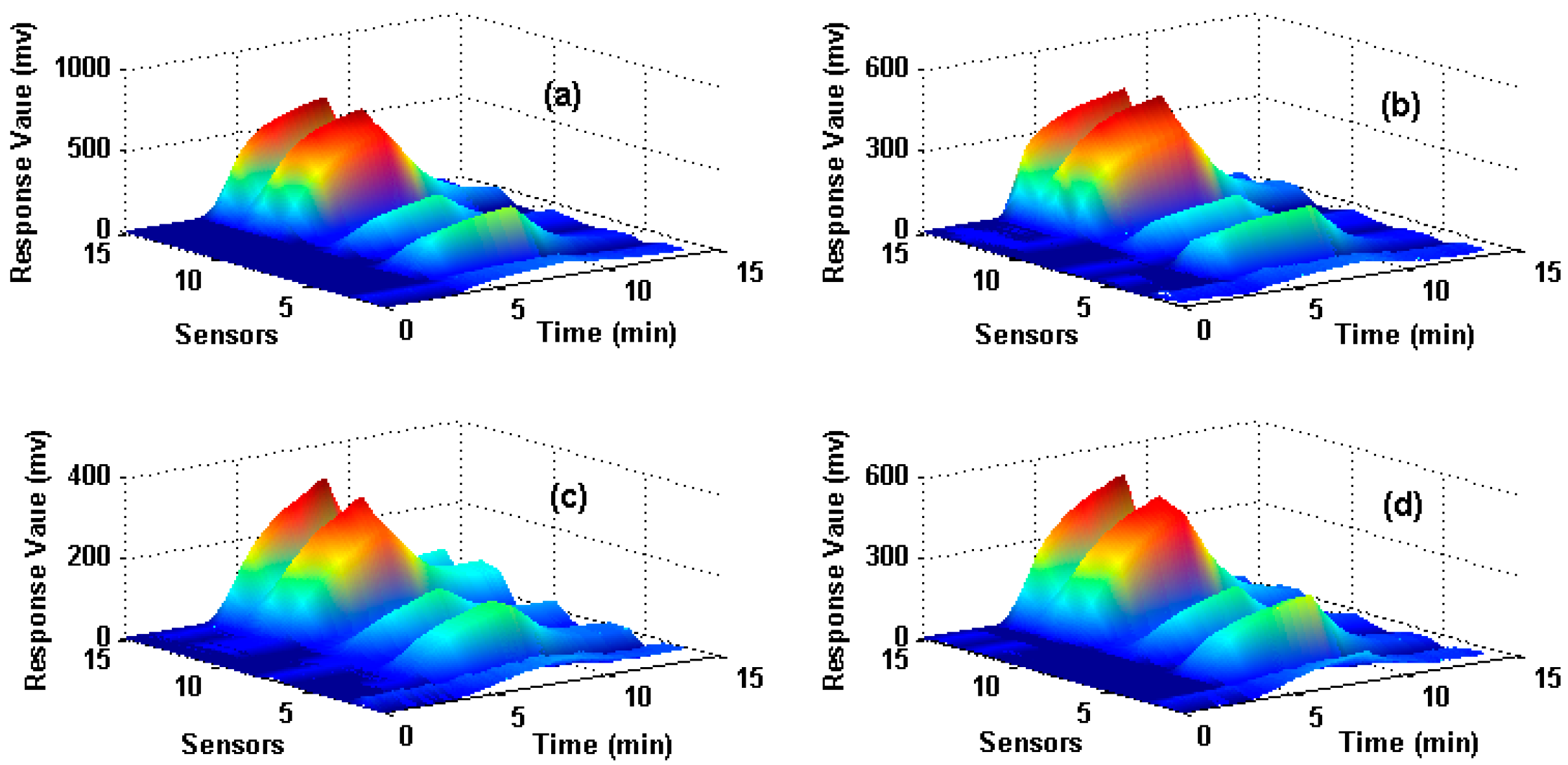
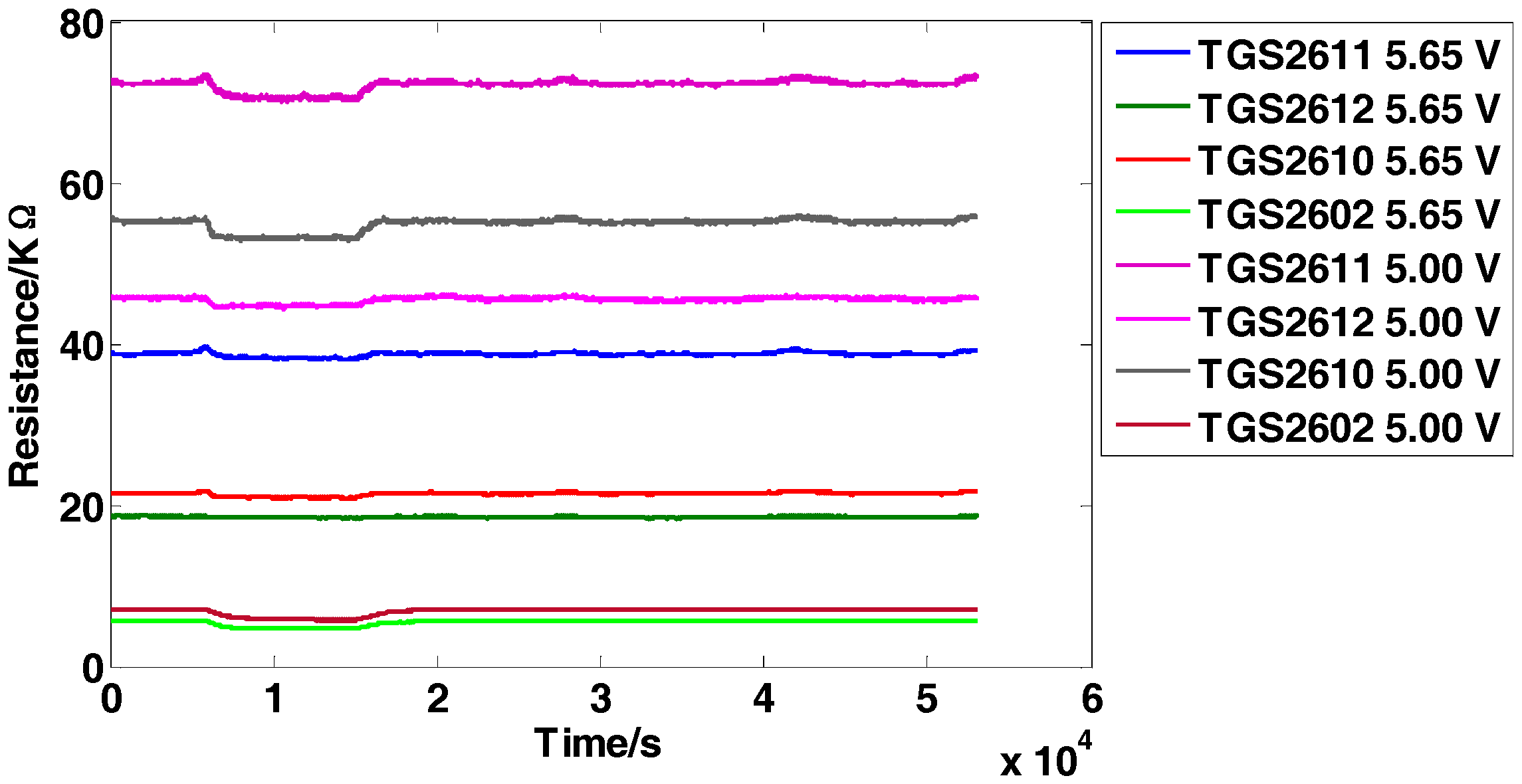
3.1. Dataset I
3.2. Dataset II
4. Results and Discussion
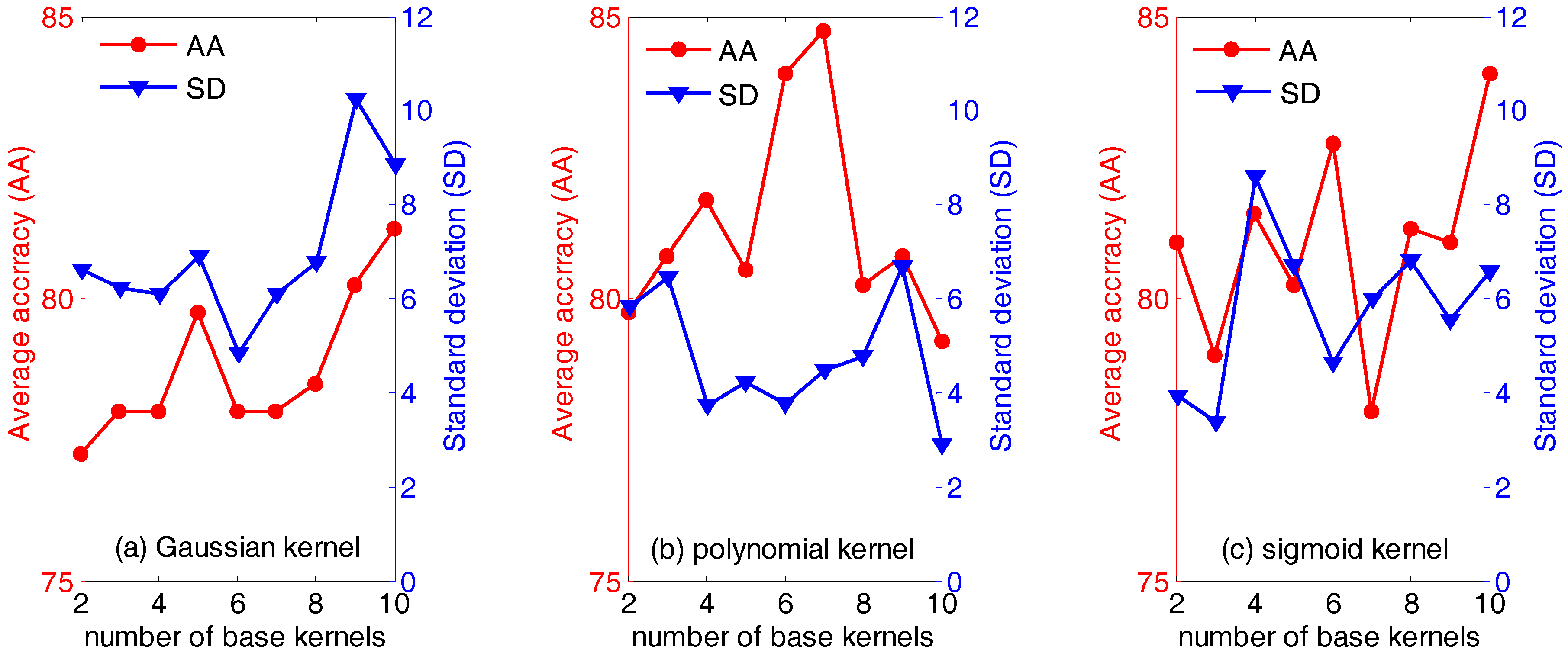
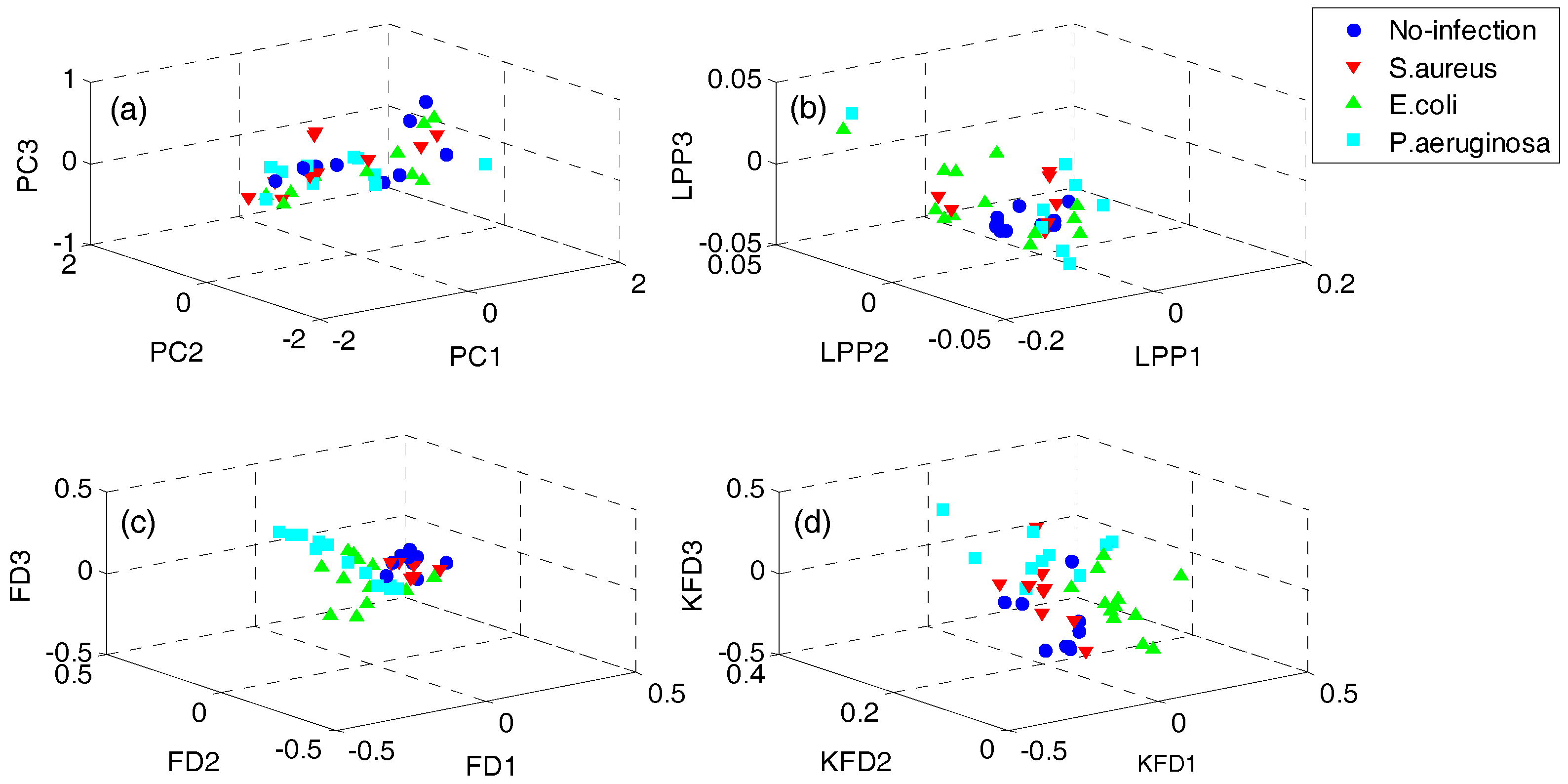
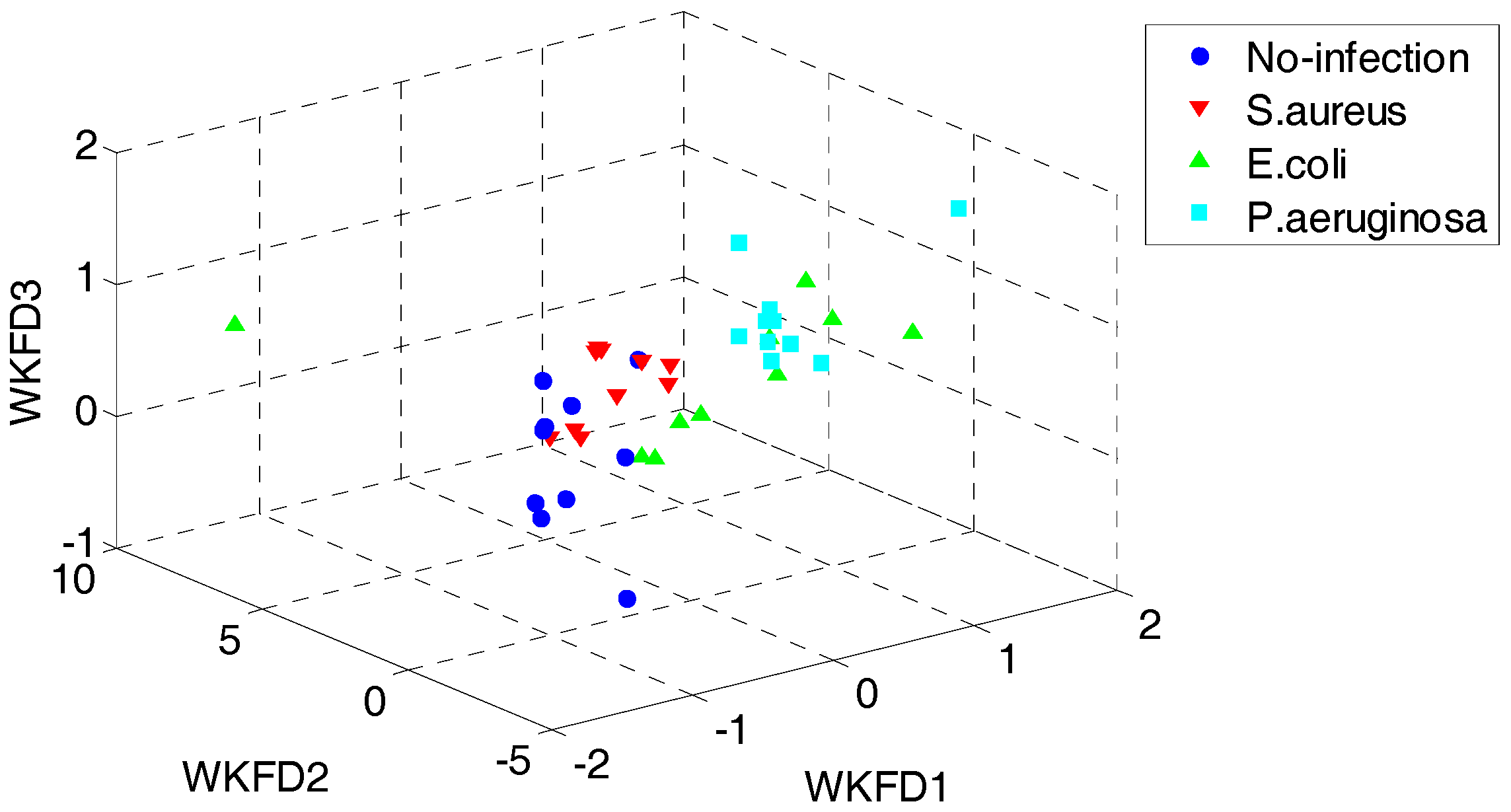
4.1. Results of Dataset I
4.2. Results of Dataset II
4.3. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lin, Y.J.; Guo, H.R.; Chang, Y.H.; Kao, M.T.; Wang, H.H.; Hong, R.I. Application of the Electronic Nose for Uremia Diagnosis. Sens. Actuators B Chem. 2001, 76, 177–180. [Google Scholar] [CrossRef]
- Adiguzel, Y.; Kulah, H. Breath Sensors for Lung Cancer Diagnosis. Biosens. Bioelectron. 2015, 65, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak, T.; Wojnowski, W.; Dymerski, T.; Gębicki, J.; Namieśnik, J. Electronic noses in classification and quality control of edible oils: A review. Food Chem. 2018, 246, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wan, P.; Jia, M.; Hu, J.; Guan, Y.; Feng, L. A novel electronic nose based on porous In2O3 microtubes sensor array for the discrimination of VOCs. Biosens. Bioelectron. 2015, 64, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Fonollosa, J.; Fernández, L.; Gutiérrez-Gálvez, A.; Huerta, R.; Marco, S. Calibration transfer and drift counteraction in chemical sensor arrays using Direct Standardization. Sens. Actuators B Chem. 2016, 236, 1044–1053. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis and Factor Analysis. In Principal Component Analysis; Springer Series in Statistics; Springer: New York, NY, USA, 1986. [Google Scholar]
- Fukunaga, K. Introduction to Statistical Pattern Recognition, 2nd ed.; Academic Press: Boston, MA, USA, 1990. [Google Scholar]
- Baudat, G.; Anouar, F. Generalized Discriminant Analysis Using a Kernel Approach. Neural Comput. 2000, 12, 2385–2404. [Google Scholar] [CrossRef] [PubMed]
- Muller, K.R.; Mika, S.; Ratsch, G.; Tsuda, K.; Scholkopf, B. An Introduction to Kernel-Based Learning Algorithms. IEEE Trans. Neural Netw. 2001, 12, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Lanckriet, G.; Cristianini, N.; Bartlett, P.; Ghaoui, L.E. Learning the Kernel Matrix with Semi-definite Programming. J. Mach. Learn. Res. 2004, 5, 27–72. [Google Scholar]
- Gönen, M.; Alpaydın, E. Multiple Kernel Learning Algorithms. J. Mach. Learn. Res. 2011, 12, 2211–2268. [Google Scholar]
- Jia, P.; Tian, F.; He, Q.; Fan, S.; Liu, J.; Yang, S.X. Feature Extraction of Wound Infection Data for Electronic Nose Based on a Novel Weighted KPCA. Sens. Actuators B Chem. 2014, 201, 555–566. [Google Scholar] [CrossRef]
- Jian, Y.L.; Huang, D.Y.; Yan, J.; Lu, K.; Huang, Y.; Wen, T.L.; Zeng, T.Y.; Zhong, S.J.; Xie, Q.L. A Novel Extreme Learning Machine Classification Model for e-Nose Application Based on the Multiple Kernel Approach. Sensors 2017, 17, 1434. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Uda, S.; Kin, T.; Asai, K. Minimizing the Cross Validation Error to Mix Kernel Matrices of Heterogeneous Biological Data. Neural Process. Lett. 2004, 19, 63–72. [Google Scholar] [CrossRef]
- Fung, G.; Dundar, M.; Bi, J.; Rao, B. A Fast Iterative Algorithm for Fisher Discriminant Using Heterogeneous Kernels. In Proceedings of the 21st International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004; p. 40. [Google Scholar]
- Kim, S.J.; Magnani, A.; Boyd, S. Optimal Kernel Selection in Kernel Fisher Discriminant Analysis. In Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, PA, USA, 25–29 June 2006; pp. 465–472. [Google Scholar]
- Ye, J.; Ji, S.; Chen, J. Multi-Class Discriminant Kernel Learning via Convex Programming. J. Mach. Learn. Res. 2008, 9, 719–758. [Google Scholar]
- Yan, F.; Mikolajczyk, K.; Barnard, M.; Cai, H.; Kittler, J. Lp norm multiple kernel Fisher discriminant analysis for object and image categorization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010; pp. 3626–3632. [Google Scholar]
- Yan, F.; Kittler, J.; Mikolajczyk, K.; Tahir, A. Non-Sparse Multiple Kernel Fisher Discriminant Analysis. J. Mach. Learn. Res. 2012, 13, 607–642. [Google Scholar]
- Liu, X.Z.; Feng, G.C. Multiple Kernel Learning in Fisher Discriminant Analysis for Face Recognition. Int. J. Adv. Robot. Syst. 2013, 10, 142. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, X. Multiple Kernel Local Fisher Discriminant Analysis for Face Recognition. Signal Process. 2013, 93, 1496–1509. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Xie, H.; Liu, D. Multiple Data-Dependent Kernel Fisher Discriminant Analysis for Face Recognition. Math. Probl. Eng. 2014. [Google Scholar] [CrossRef] [PubMed]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Sun, J.; Feng, B.; Xu, W. Particle Swarm Optimization with Particles Having Quantum Behavior. In Proceedings of the IEEE Conference on Evolutionary Computation, Portland, OR, USA, 19–23 June 2004; pp. 325–331. [Google Scholar]
- Yao, F.; Dong, Z.Y.; Meng, K.; Xu, Z.; Iu, H.C.; Wong, K.P. Quantum-Inspired Particle Swarm Optimization for Power System Operations Considering Wind Power Uncertainty and Carbon Tax in Australia. IEEE Trans. Ind. Inform. 2012, 8, 880–888. [Google Scholar] [CrossRef]
- Peng, C.; Yan, J.; Duan, S.; Wang, L.; Jia, P.; Zhang, S. Enhancing Electronic Nose Performance Based on a Novel QPSO-KELM Model. Sensors 2016, 16, 520. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Tian, F.; Yan, J.; He, Q.; Shen, Y.; Pan, L. A background elimination method based on wavelet transform in wound infection detection by electronic nose. Sens. Actuators B Chem. 2011, 157, 395–400. [Google Scholar] [CrossRef]
- He, X.; Niyogi, P. Locality preserving projections. Adv. Neural Inf. Process. Syst. 2003, 16, 186–197. [Google Scholar]
| Class | Predicted as * | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gaussian Kernels (n = 10) | Polynomial Kernels (n = 7) | Sigmoid Kernels (n = 10) | |||||||||||||
| N | 1 | 2 | 3 | 4 | N | 1 | 2 | 3 | 4 | N | 1 | 2 | 3 | 4 | |
| 1 | 11 | 10 | 1 | 0 | 0 | 10 | 9 | 1 | 0 | 0 | 9 | 9 | 0 | 0 | 0 |
| 2 | 10 | 1 | 9 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 8 | 1 | 7 | 0 | 0 |
| 3 | 8 | 0 | 0 | 7 | 1 | 10 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 9 | 1 |
| 4 | 11 | 0 | 0 | 0 | 11 | 10 | 0 | 0 | 1 | 9 | 13 | 0 | 0 | 1 | 12 |
| Accuracy | 92.5% | 95% | 92.5% | ||||||||||||
| Test Batch | Accuracy Rate (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Batch 2 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 |
| Batch 3 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 | 99.375 |
| Batch 4 | 100 | 100 | 100 | 98.75 | 100 | 98.75 | 100 | 97.5 | 97.5 |
| Batch 5 | 75 | 72.5 | 75 | 75 | 75 | 75 | 73.75 | 75 | 75 |
| Average | 93.44 | 92.81 | 93.44 | 93.13 | 93.44 | 93.13 | 93.13 | 92.81 | 92.81 |
| Test Batch | Accuracy Rate (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Batch 2 | 98.75 | 98.75 | 99.375 | 97.5 | 98.75 | 96.875 | 98.125 | 96.875 | 93.75 |
| Batch 3 | 100 | 100 | 100 | 100 | 100 | 100 | 99.375 | 100 | 100 |
| Batch 4 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Batch 5 | 57.5 | 62.5 | 52.5 | 52.5 | 50 | 50 | 52.5 | 48.75 | 50 |
| Average | 89.06 | 90.31 | 87.97 | 87.50 | 87.19 | 86.72 | 87.50 | 86.41 | 85.94 |
| Test Batch | Accuracy Rate (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Batch 2 | 98.75 | 96.875 | 98.75 | 96.25 | 98.75 | 91.25 | 91.875 | 93.125 | 96.25 |
| Batch 3 | 96.875 | 98.75 | 99.375 | 100 | 100 | 100 | 100 | 99.375 | 100 |
| Batch 4 | 100 | 100 | 100 | 100 | 98.75 | 100 | 100 | 100 | 93.75 |
| Batch 5 | 71.25 | 70 | 70 | 70 | 71.25 | 67.5 | 72.5 | 70 | 68.75 |
| Average | 91.72 | 91.41 | 92.03 | 91.56 | 92.19 | 89.69 | 91.09 | 90.63 | 89.69 |
| Test Batch | Accuracy Rate (%) | |||||
|---|---|---|---|---|---|---|
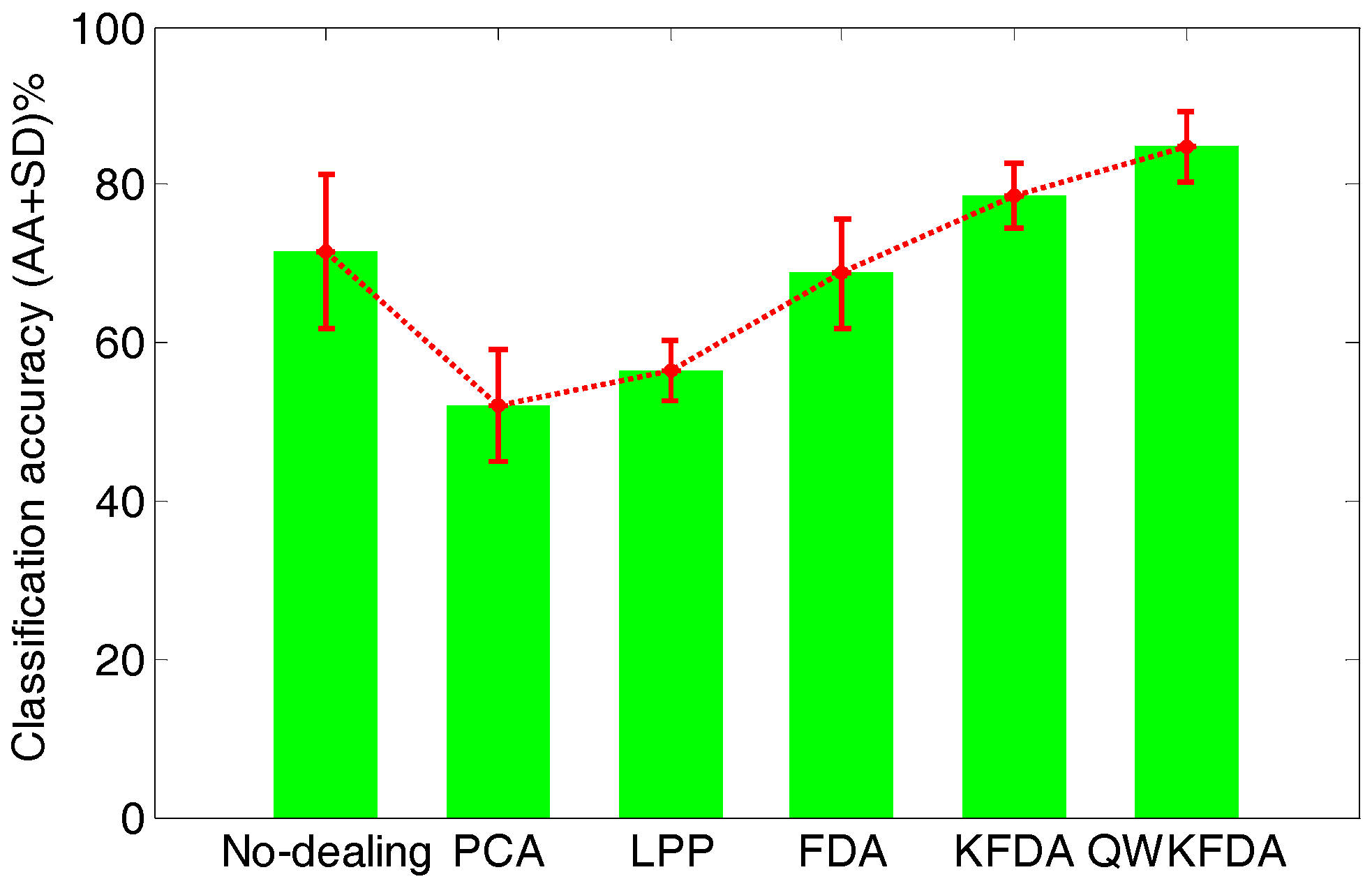
| No-Dealing | PCA | LPP | FDA | KFDA | QWKFDA | |
| Batch 2 | 99.375 | 95.625 | 90.625 | 96.875 | 93.75 | 99.375 |
| Batch 3 | 100 | 98.125 | 61.25 | 71.25 | 96.875 | 99.375 |
| Batch 4 | 97.5 | 98.75 | 93.75 | 91.25 | 100 | 100 |
| Batch 5 | 61.25 | 61.25 | 75 | 62.5 | 75 | 75 |
| Average | 89.53 | 88.44 | 80.16 | 80.47 | 91.41 | 93.44 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, T.; Yan, J.; Huang, D.; Lu, K.; Deng, C.; Zeng, T.; Yu, S.; He, Z. Feature Extraction of Electronic Nose Signals Using QPSO-Based Multiple KFDA Signal Processing. Sensors 2018, 18, 388. https://doi.org/10.3390/s18020388
Wen T, Yan J, Huang D, Lu K, Deng C, Zeng T, Yu S, He Z. Feature Extraction of Electronic Nose Signals Using QPSO-Based Multiple KFDA Signal Processing. Sensors. 2018; 18(2):388. https://doi.org/10.3390/s18020388
Chicago/Turabian StyleWen, Tailai, Jia Yan, Daoyu Huang, Kun Lu, Changjian Deng, Tanyue Zeng, Song Yu, and Zhiyi He. 2018. "Feature Extraction of Electronic Nose Signals Using QPSO-Based Multiple KFDA Signal Processing" Sensors 18, no. 2: 388. https://doi.org/10.3390/s18020388




