Label-Free Aptasensor for Lysozyme Detection Using Electrochemical Impedance Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Equipment
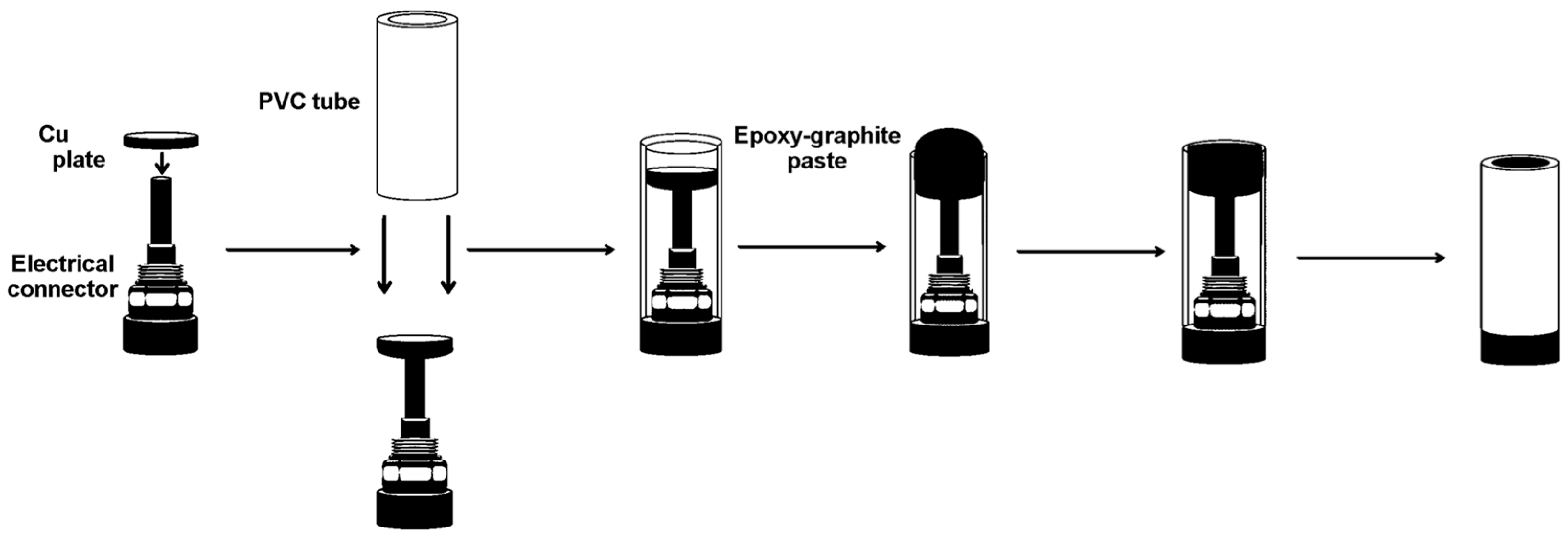
2.3. Preparation of Working Electrodes
2.4. Experimental Procedure
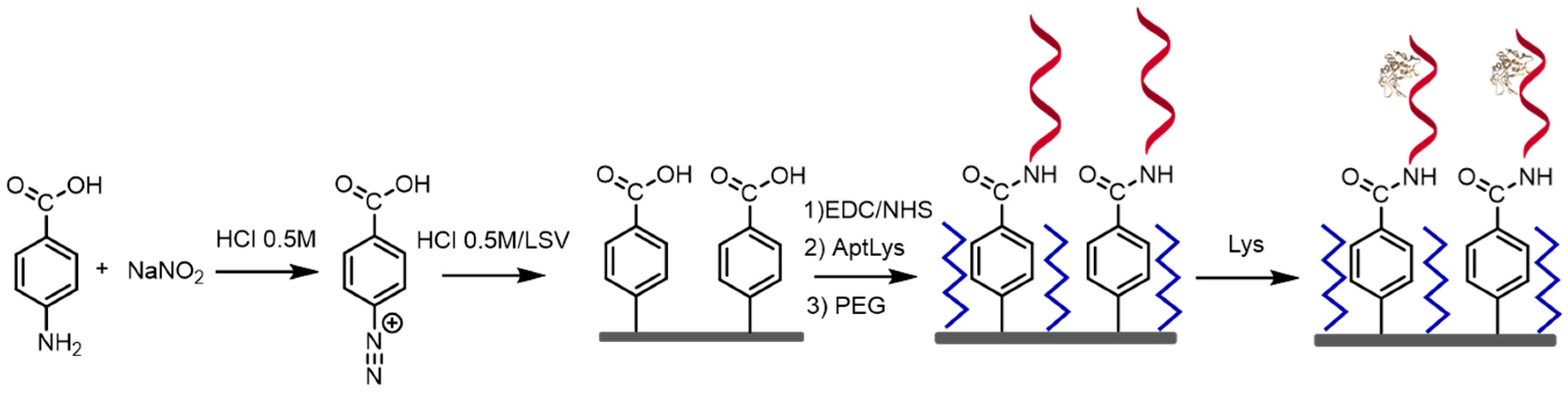
2.4.1. Aptamer Preconditioning
2.4.2. Aptasensor Immobilization onto the Electrode Surface
2.4.3. Blocking Step
2.4.4. Label Free Detection of Lysozyme
2.4.5. Regeneration of Aptasensor
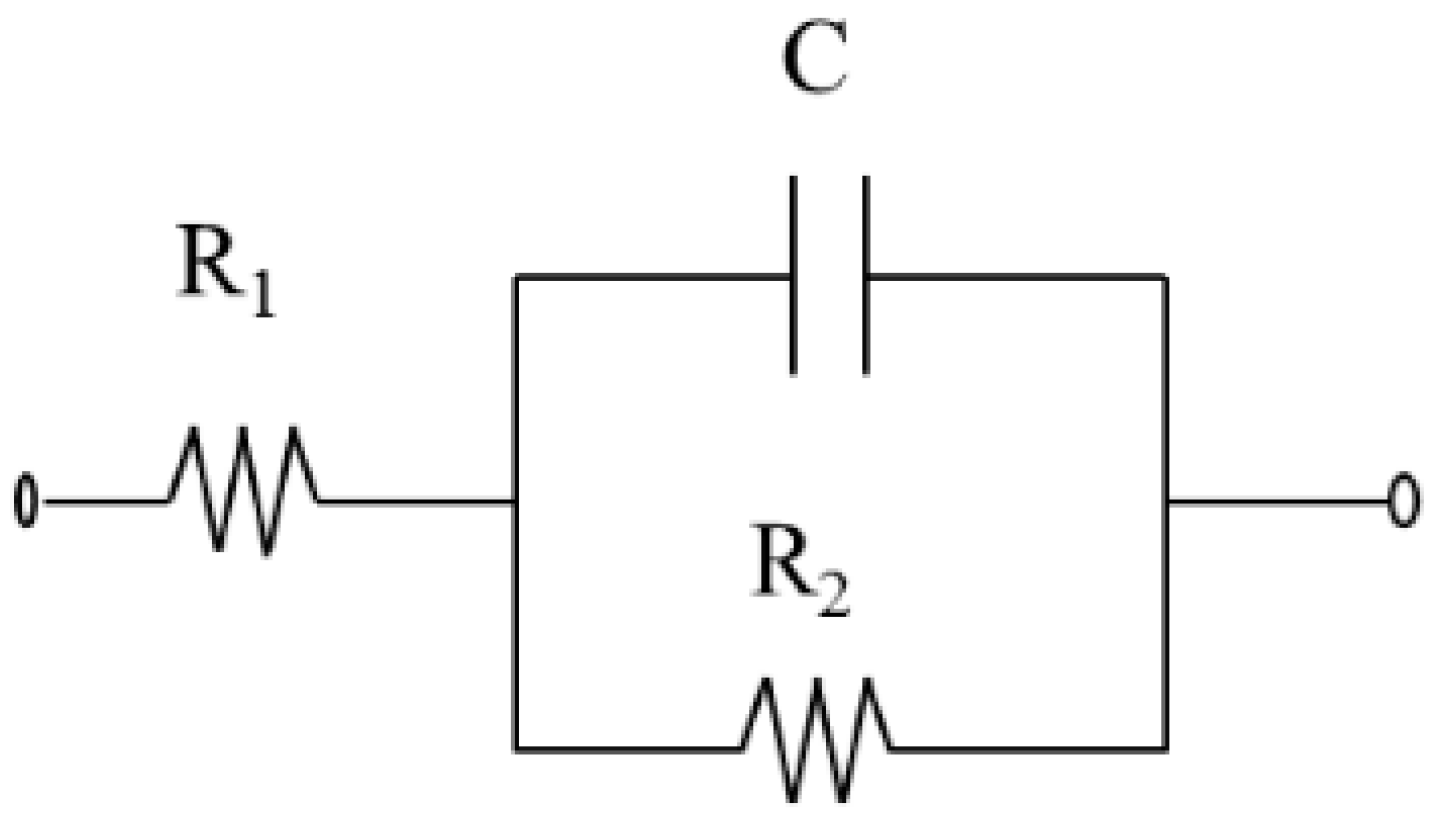
2.5. Impedimetric Measurements and Data Processing
3. Results and Discussions
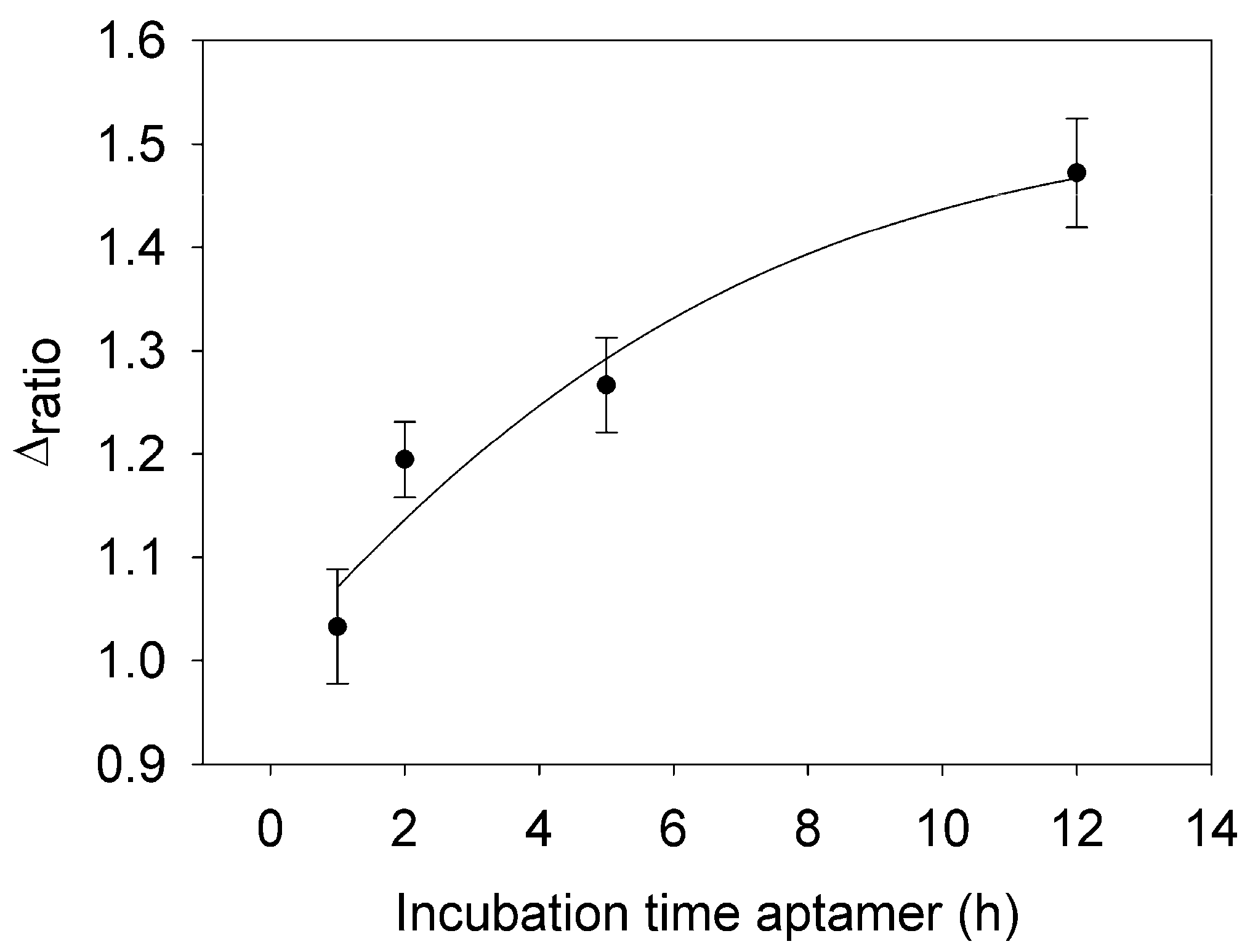
3.1. Incubation Time of Aptamer
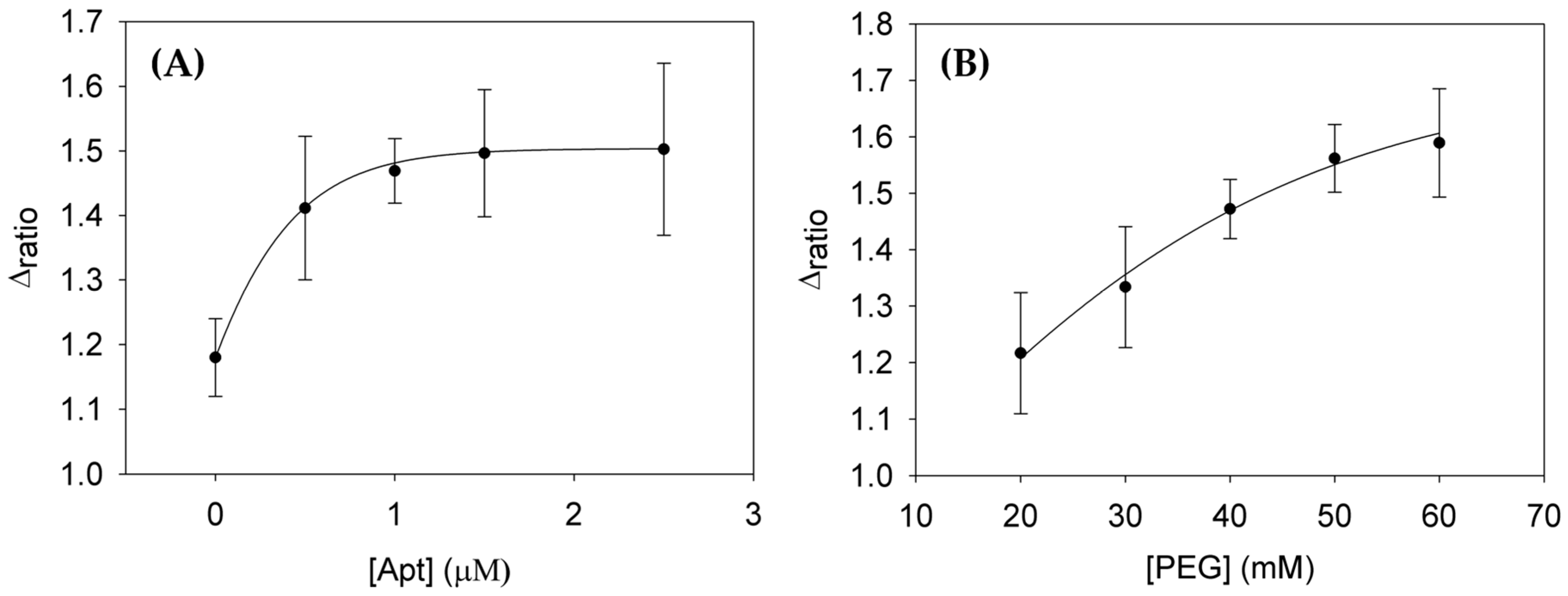
3.2. Optimization of Aptamer and PEG Blocking Agent Concentrations
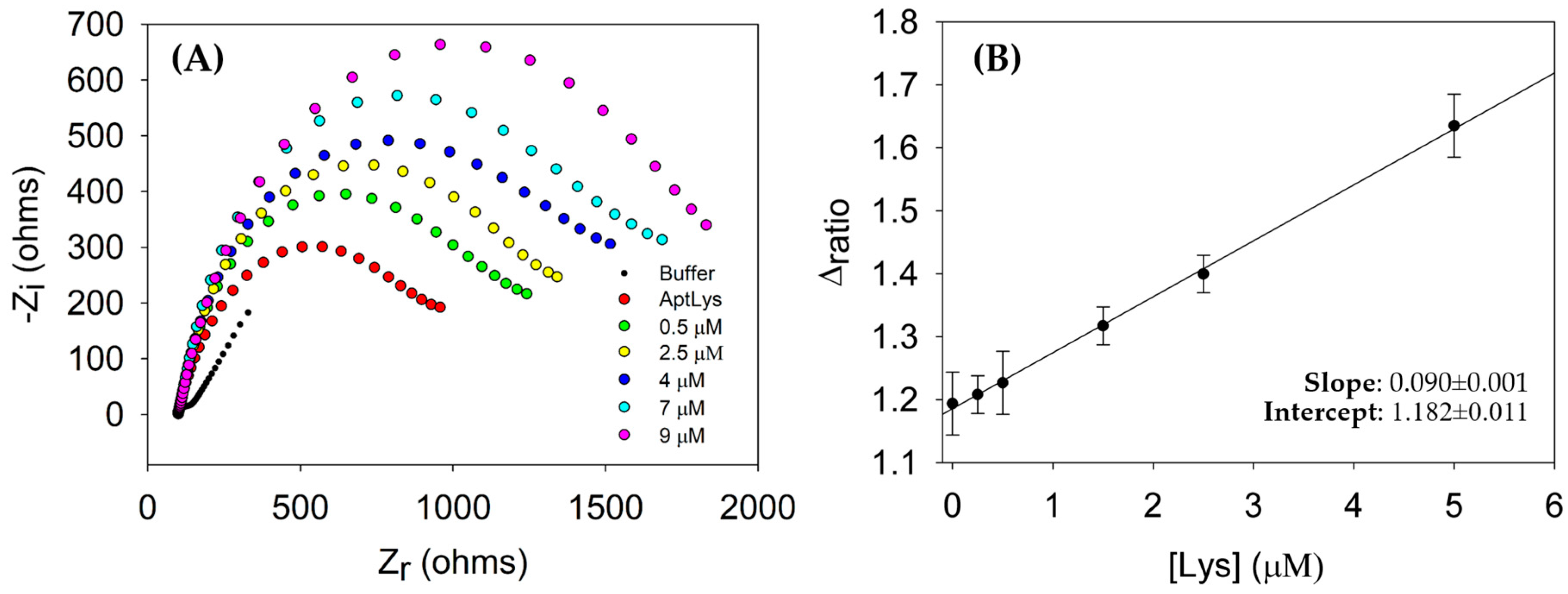
3.3. Analytical Performance of the Aptasensor for Lysozyme Detection
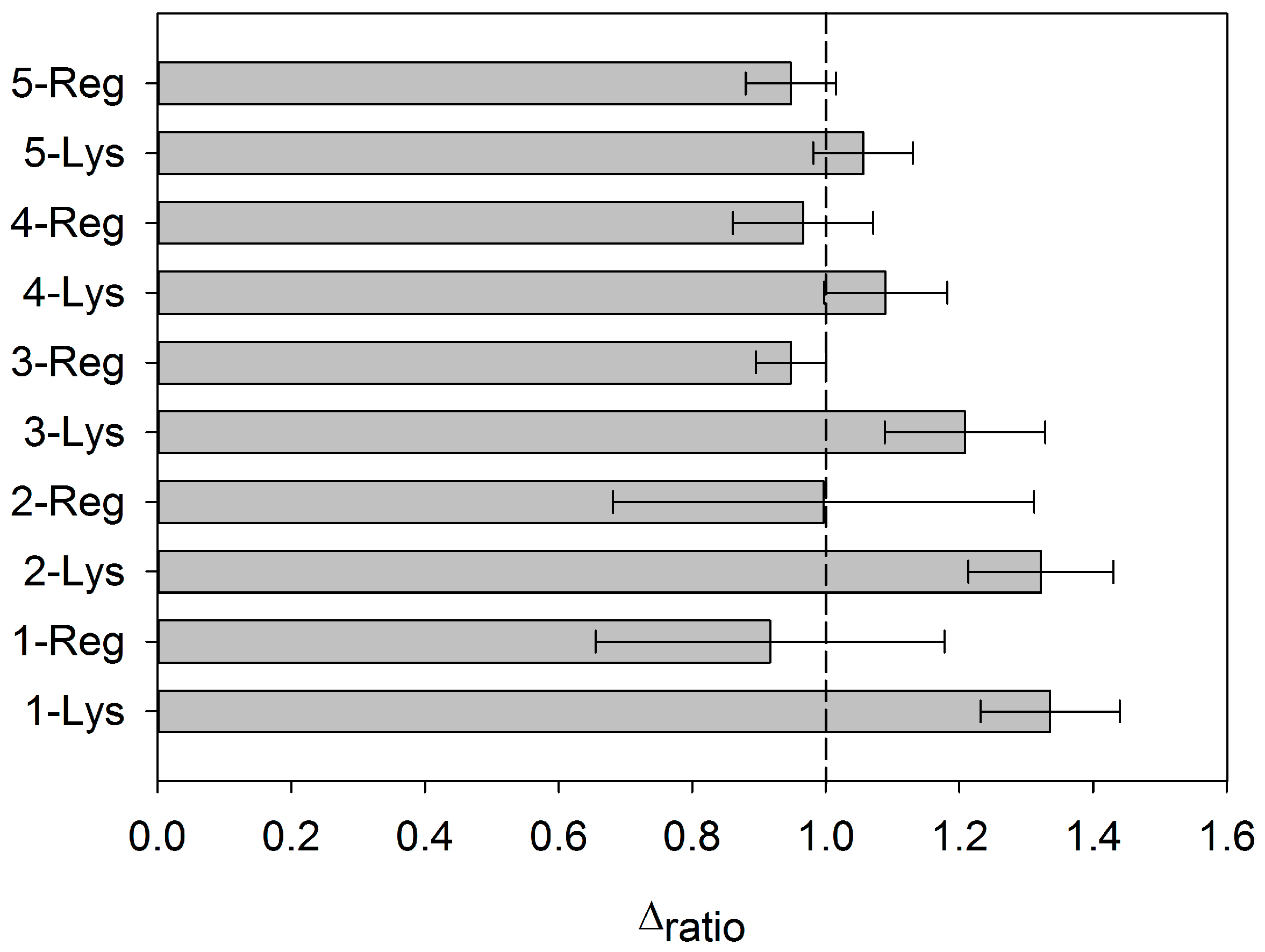
3.4. Regeneration of the Aptasensor
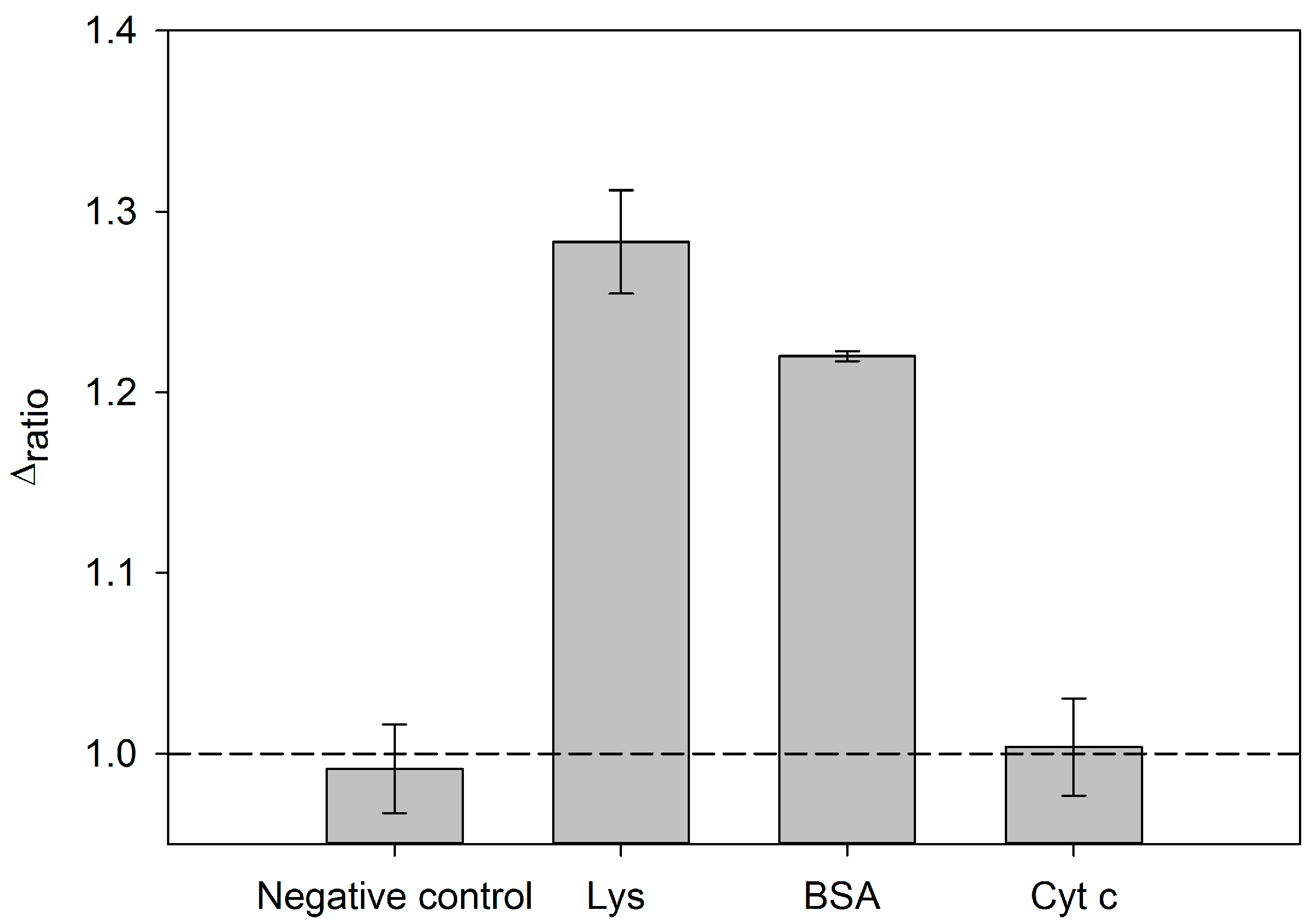
3.5. Specificity of the Aptasensor
3.6. Application of the Aptasensors in Spiked Wine Samples
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical Biosensors: Recommended Definitions and Classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Mascini, M. Aptamers in Bioanalysis; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical Applications of Aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, Antibody scFv, and Antibody Fab’ Fragments: An Overview and Comparison of Three of the Most Versatile Biosensor Biorecognition Elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An Emerging Class of Therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, C.; Pacios, M.; del Valle, M. A Reusable Impedimetric Aptasensor for Detection of Thrombin Employing a Graphite-Epoxy Composite Electrode. Sensors 2012, 12, 3037–3048. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, C.; Arcay, E.; del Valle, M. Label-Free Impedimetric Aptasensor Based on Epoxy-Graphite Electrode for the Recognition of Cytochrome c. Sens. Actuators B Chem. 2014, 191, 860–865. [Google Scholar] [CrossRef]
- Velasco-Garcia, M.N.; Missailidis, S. New Trends in Aptamer-Based Electrochemical Biosensors. Genes Ther. Mol. Biol. 2009, 13, 1–9. [Google Scholar]
- Gu, M.B.; Kim, H.-S. Biosensors Based on Aptamers and Enzymes; Springer: Berlin, Germany, 2014. [Google Scholar]
- Bonanni, A.; del Valle, M. Use of Nanomaterials for Impedimetric DNA Sensors: A Review. Anal. Chim. Acta 2010, 678, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Barsoukov, E.; Macdonald, J.R. Impedance Spectroscopy: Theory, Experiment, and Applications, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Schindler, M.; Assaf, Y.; Sharon, N.; Chipman, D.M. Mechanism of Lysozyme Catalysis: Role of Ground-State Strain in Subsite D in Hen Egg-White and Human Lysozymes. Biochemistry 1977, 16, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, A.; Wang, Q.; Li, M.; Boukherroub, R.; Szunerits, S. Aptamer-Based Electrochemical Sensing of Lysozyme. Chemosensors 2016, 4, 1–20. [Google Scholar] [CrossRef]
- Organisation Internationale de la Vigne et du Vin (OIV). Code International des Pratiques Oenologiques; OIV—Organisation Internationale de la Vigne et du Vin: Paris, France, 2016. [Google Scholar]
- Porstmann, B.; Jung, K.; Schmechta, H.; Evers, U.; Pergande, M.; Porstmann, T.; Kramm, H.J.; Krause, H. Measurement of Lysozyme in Human Body Fluids: Comparison of Various Enzyme Immunoassay Techniques and Their Diagnostic Application. Clin. Biochem. 1989, 22, 349–355. [Google Scholar] [CrossRef]
- Pascual, R.S.; Gee, J.B.L.; Finch, S.C. Usefulness of Serum Lysozyme Measurement in Diagnosis and Evaluation of Sarcoidosis. N. Engl. J. Med. 1973, 289, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, A.; Gaspar, S.; Mihai, I.; Tache, A.; Litescu, S.C. Development of a Label-Free Aptasensor for Monitoring the Self-Association of Lysozyme. Analyst 2013, 138, 3530–3537. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, C.; Hayat, A.; Mishra, R.K.; Vasilescu, A.; del Valle, M.; Marty, J.L. Label Free Aptasensor for Lysozyme Detection: A Comparison of the Analytical Performance of Two Aptamers. Bioelectrochemistry 2015, 105, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Lisdat, F.; Schäfer, D. The Use of Electrochemical Impedance Spectroscopy for Biosensing. Anal. Bioanal. Chem. 2008, 391, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, C.; Lukic, S.; del Valle, M. Aptamer-Antibody Sandwich Assay for Cytochrome c Employing an MWCNT Platform and Electrochemical Impedance. Microchim. Acta 2015, 182, 2045–2053. [Google Scholar] [CrossRef]
- Bonanni, A.; Pividori, M.I.; del Valle, M. Application of the Avidin–biotin Interaction to Immobilize DNA in the Development of Electrochemical Impedance Genosensors. Anal. Bioanal. Chem. 2007, 389, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Bonanni, A.; Esplandiu, M.J.; Pividori, M.I.; Alegret, S.; del Valle, M. Impedimetric Genosensors for the Detection of DNA Hybridization. Anal. Bioanal. Chem. 2006, 385, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Bélanger, D.; Pinson, J. Electrografting: A Powerful Method for Surface Modification. Chem. Soc. Rev. 2011, 40, 3995–4048. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, C.; del Valle, M. A Comparison of Four Protocols for the Immobilization of an Aptamer on Graphite Composite Electrodes. Microchim. Acta 2014, 181, 355–363. [Google Scholar] [CrossRef]
- Yang, X.; Qian, J.; Jiang, L.; Yan, Y.; Wang, K.; Liu, Q.; Wang, K. Ultrasensitive Electrochemical Aptasensor for Ochratoxin A Based on Two-Level Cascaded Signal Amplification Strategy. Bioelectrochemistry 2014, 96, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Willner, I. Probing Biomolecular Interactions at Conductive and Semiconductive Surfaces by Impedance Spectroscopy: Routes to Impedimetric Immunosensors, DNA-Sensors, and Enzyme Biosensors. Electroanalysis 2003, 15, 913–947. [Google Scholar] [CrossRef]
- Radi, A.-E.; Acero Sánchez, J.L.; Baldrich, E.; O’Sullivan, C.K. Reusable Impedimetric Aptasensor. Anal. Chem. 2005, 77, 6320–6323. [Google Scholar] [CrossRef] [PubMed]
| Aptasensor | Spike Lys (µM) | Δratio | Found Lys (µM) | Recovery (%) |
|---|---|---|---|---|
| Apt Lys | 0 | 1.19 | 0 | - |
| 1 | 1.25 | 0.77 | 77 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz-Aguayo, D.; Del Valle, M. Label-Free Aptasensor for Lysozyme Detection Using Electrochemical Impedance Spectroscopy. Sensors 2018, 18, 354. https://doi.org/10.3390/s18020354
Ortiz-Aguayo D, Del Valle M. Label-Free Aptasensor for Lysozyme Detection Using Electrochemical Impedance Spectroscopy. Sensors. 2018; 18(2):354. https://doi.org/10.3390/s18020354
Chicago/Turabian StyleOrtiz-Aguayo, Dionisia, and Manel Del Valle. 2018. "Label-Free Aptasensor for Lysozyme Detection Using Electrochemical Impedance Spectroscopy" Sensors 18, no. 2: 354. https://doi.org/10.3390/s18020354






