Discrimination of Transgenic Maize Kernel Using NIR Hyperspectral Imaging and Multivariate Data Analysis
Abstract
:1. Introduction
2. Materials and Methods
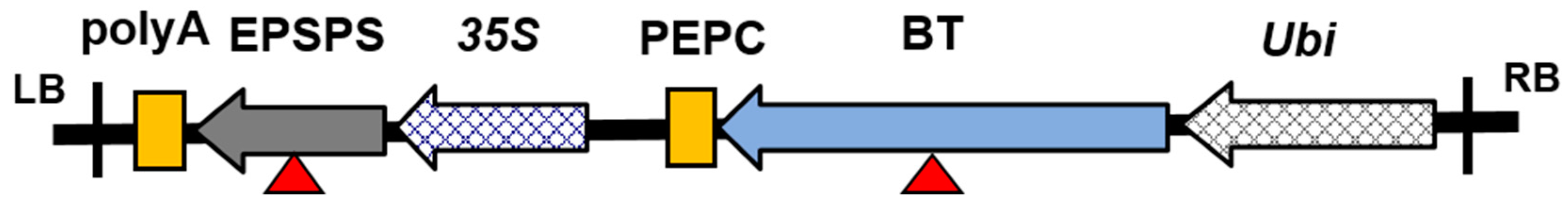
2.1. Maize Samples
2.2. Near-Infrared Hyperspectral Imaging
2.3. Spectral Collection and Pretreatment
2.4. Multivariate Chemometrics Analysis
2.5. Software Tools
3. Results and Discussion
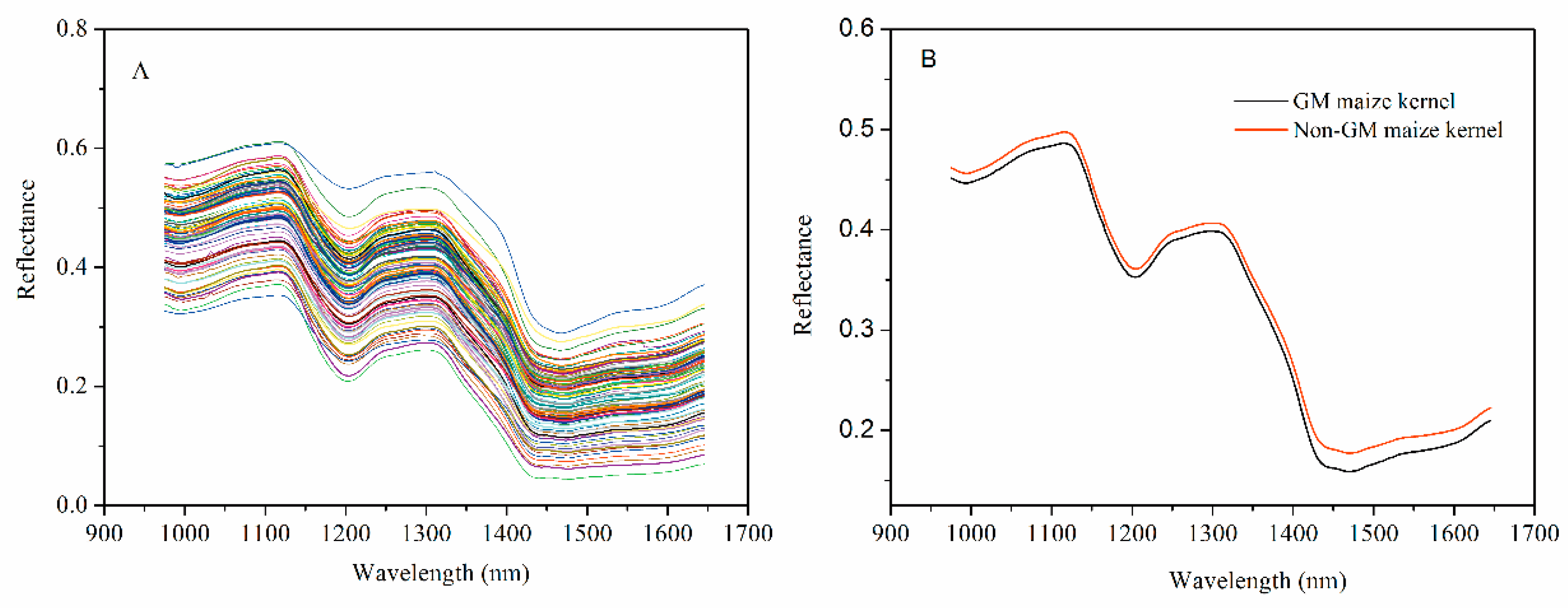
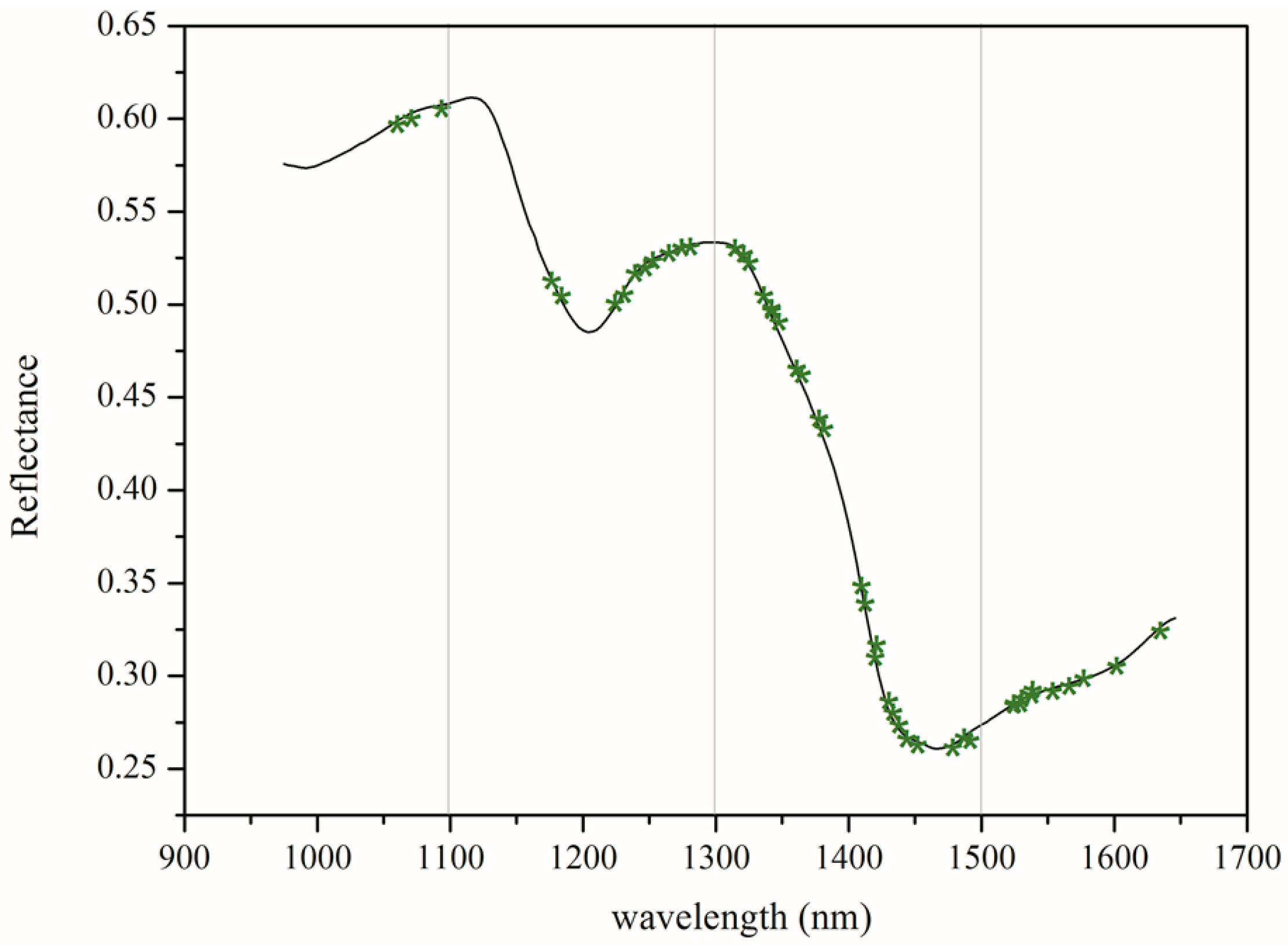
3.1. Spectroscopic Analysis
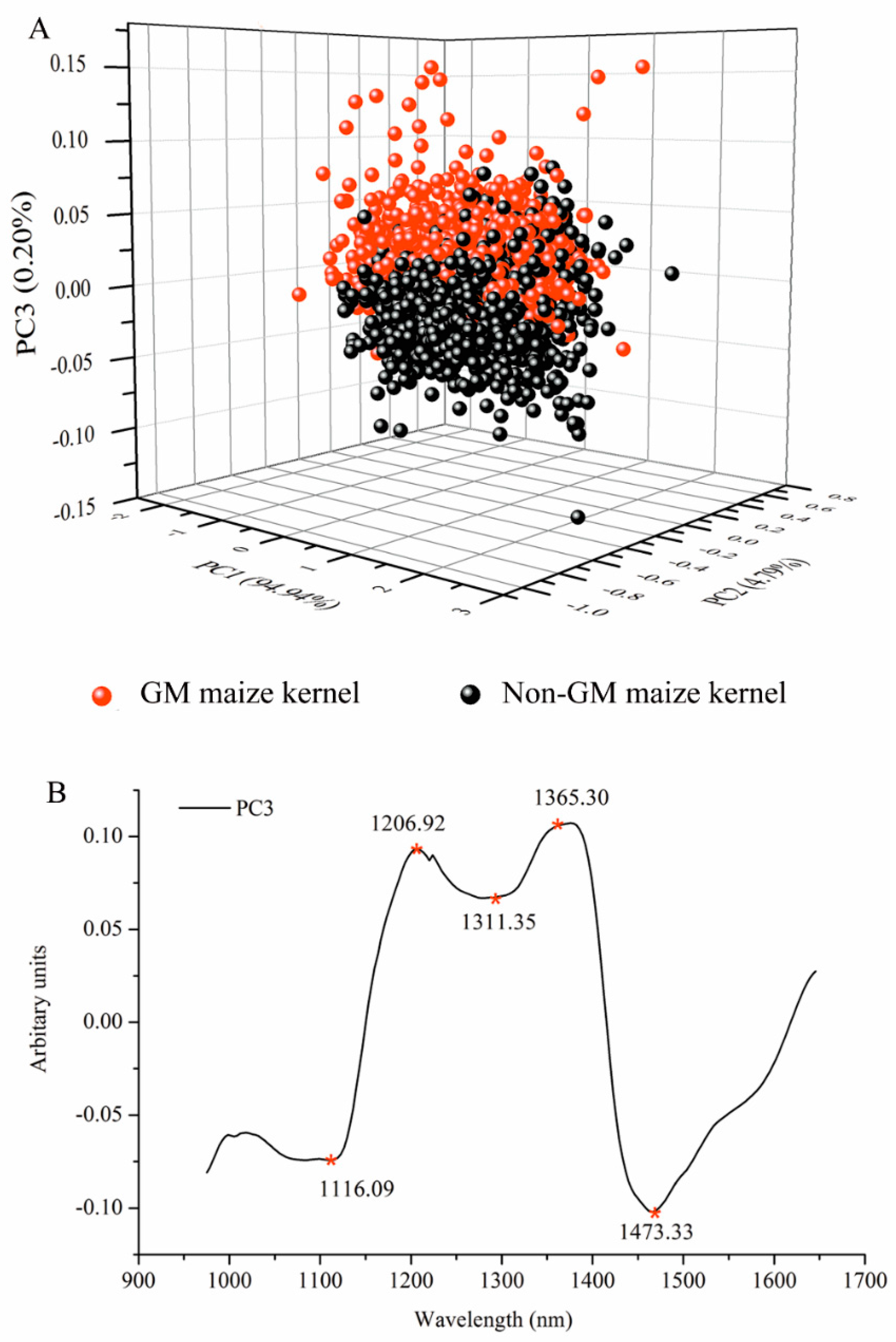
3.2. Spectral Analysis by Principal Component Analysis
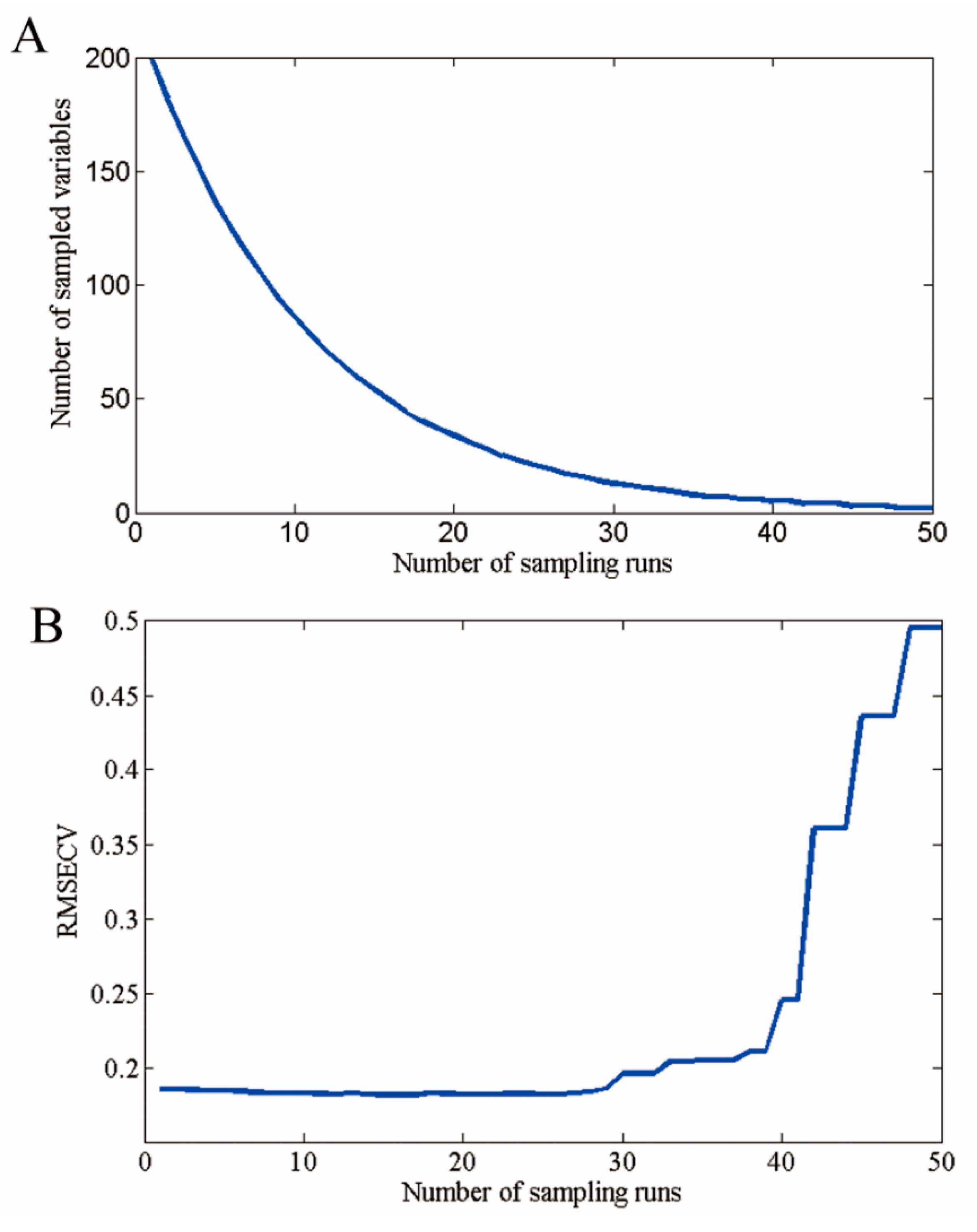
3.3. Selection of Optimal Wavelengths
3.4. Classification Analysis by the Discrimination Model
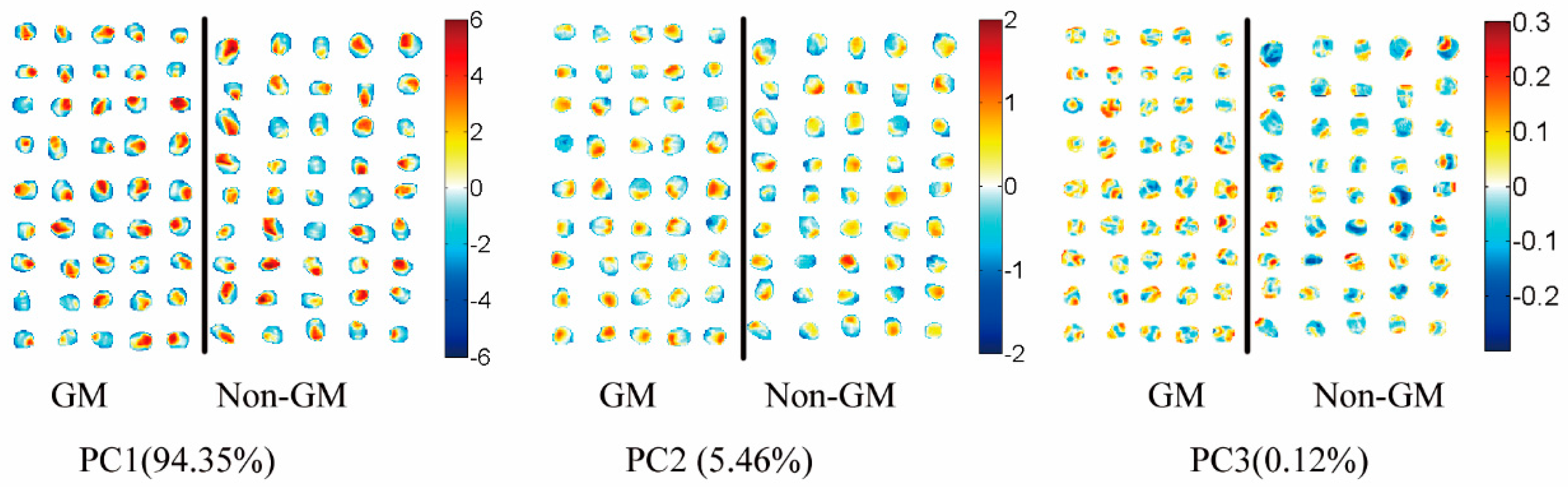
3.5. Transgenic Maize Kernel Visualization
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef]
- Rensburg, J.B.J.V. First report of field resistance by the stem borer, Busseola fusca (Fuller) to Bt-transgenic maize. S. Afr. J. Plant Soil 2007, 24, 147–151. [Google Scholar] [CrossRef]
- Vermerris, W.; Saballos, A.; Ejeta, G.; Mosier, N.S.; Ladisch, M.R.; Carpita, N.C.; Albrecht, B.; Bernardo, R.; Godshalk, E.B. Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci. 2007, 47 (Suppl. 3), S142. [Google Scholar] [CrossRef]
- Aluru, M. Generation of transgenic maize with enhanced provitamin A content. J. Exp. Bot. 2015, 23, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Agapito-Tenfen, S.Z.; Vilperte, V.; Benevenuto, R.F.; Rover, C.M.; Traavik, T.I.; Nodari, R.O. Effect of stacking insecticidal cry and herbicide tolerance epsps transgenes on transgenic maize proteome. BMC Plant Biol. 2014, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Nap, J.P.; Metz, P.L.J.; Escaler, M.; Conner, A.J. The release of genetically modified crops into the environment. Plant J. Cell Mol. Biol. 2003, 33, 19–46. [Google Scholar] [CrossRef]
- Taverniers, I.; Van, B.E.; De, L.M. Cloned plasmid DNA fragments as calibrators for controlling GMOs: Different real-time duplex quantitative PCR methods. Anal. Bioanal. Chem. 2004, 378, 1198–1207. [Google Scholar] [CrossRef] [PubMed]
- Brunnert, H.J.; Spener, F.; Börchers, T. PCR-ELISA for the CaMV-35S promoter as a screening method for genetically modified Roundup Ready soybeans. Eur. Food Res. Technol. 2001, 213, 366–371. [Google Scholar] [CrossRef]
- Huang, X.; Zhai, C.; You, Q.; Chen, H. Potential of cross-priming amplification and DNA-based lateral-flow strip biosensor for rapid on-site GMO screening. Anal. Bioanal. Chem. 2014, 406, 4243–4249. [Google Scholar] [CrossRef] [PubMed]
- Tengs, T.; Kristoffersen, A.B.; Berdal, K.G.; Thorstensen, T.; Butenko, M.A.; Nesvold, H.; Holstjensen, A. Microarray-based method for detection of unknown genetic modifications. BMC Biotechnol. 2007, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alishahi, A.; Farahmand, H.; Prieto, N.; Cozzolino, D. Identification of transgenic foods using NIR spectroscopy: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 75, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Ying, Y.; Ying, T.; Yu, H.; Fu, X. Discrimination of transgenic tomatoes based on visible/near-infrared spectra. Anal. Chim. Acta 2007, 584, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Chen, J.; Pan, T.; Wang, J.; Cao, G. Vis-NIR wavelength selection for non-destructive discriminant analysis of breed screening of transgenic sugarcane. Anal. Methods 2014, 6, 8810–8816. [Google Scholar] [CrossRef]
- Dolores, G.M.M.; Juan, G.O.; Francisco, B. Effective Identification of Low-Gliadin Wheat Lines by Near Infrared Spectroscopy (NIRS): Implications for the Development and Analysis of Foodstuffs Suitable for Celiac Patients. PLoS ONE 2016, 11, e0152292. [Google Scholar]
- Liu, C.; Wei, L.; Lu, X.; Wei, C.; Yang, J.; Lei, Z. Nondestructive determination of transgenic Bacillus thuringiensis rice seeds (Oryza sativa L.) using multispectral imaging and chemometric methods. Food Chem. 2014, 153, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Geladi, P.; Fox, G.; Manley, M. Maize kernel hardness classification by near infrared (NIR) hyperspectral imaging and multivariate data analysis. Anal. Chim. Acta 2009, 653, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.J.; Kucheryavskiy, S. Classification of maize kernels using NIR hyperspectral imaging. Food Chem. 2016, 209, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Investigation of fungal development in maize kernels using NIR hyperspectral imaging and multivariate data analysis. J. Cereal Sci. 2012, 55, 272–278. [Google Scholar] [CrossRef]
- Burger, J.; Geladi, P. Hyperspectral NIR imaging for calibration and prediction: A comparison between image and spectrometer data for studying organic and biological samples. Analyst 2006, 131, 1152–1160. [Google Scholar] [CrossRef] [PubMed]
- Grusche, S. Basic slit spectroscope reveals three-dimensional scenes through diagonal slices of hyperspectral cubes. Appl. Opt. 2014, 53, 4594–4603. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, D.; Pu, H.; Sun, D.W.; Gao, W.; Xiong, Z. Use of Hyperspectral Imaging to Discriminate the Variety and Quality of Rice. Food Anal. Methods 2015, 8, 515–523. [Google Scholar] [CrossRef]
- Min, H.; Wan, X.; Min, Z.; Zhu, Q. Detection of insect-damaged vegetable soybeans using hyperspectral transmittance image. J. Food Eng. 2013, 116, 45–49. [Google Scholar]
- Sun, J.; Lu, X.; Mao, H.; Wu, X.; Gao, H. Quantitative Determination of Rice Moisture Based on Hyperspectral Imaging Technology and BCC-LS-SVR Algorithm. J. Food Process Eng. 2016, 40, e12446. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Liu, F.; Nie, P.; He, Y. Rice seed cultivar identification using near-infrared hyperspectral imaging and multivariate data analysis. Sensors 2013, 13, 8916–8927. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Zhuang, Y. Determination of chlorogenic acid in plant samples by using near-infrared spectrum with wavelet transform preprocessing. Anal. Sci. 2004, 20, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Rinnan, Å.; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Herrman, T.J.; Lingenfelser, J.; Jackson, D.S. Classification and prediction of maize hardness-associated properties using multivariate statistical analyses. J. Cereal Sci. 2005, 41, 85–93. [Google Scholar] [CrossRef]
- Manley, M.; Mcgoverin, C.M.; Engelbrecht, P.; Geladi, P. Influence of grain topography on near infrared hyperspectral images. Talanta 2012, 89, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Wold, J.P.; Jakobsen, T.; Krane, L. Atlantic Salmon Average Fat Content Estimated by Near-Infrared Transmittance Spectroscopy. J. Food Sci. 1996, 61, 74–77. [Google Scholar] [CrossRef]
- Rodríguez-Pulido, F.J.; Barbin, D.F.; Sun, D.W.; Gordillo, B.; González-Miret, M.L.; Heredia, F.J. Grape seed characterization by NIR hyperspectral imaging. Postharvest Biol. Technol. 2013, 76, 74–82. [Google Scholar] [CrossRef]
- Almeida, M.R.; Fidelis, C.H.; Barata, L.E.; Poppi, R.J. Classification of Amazonian rosewood essential oil by Raman spectroscopy and PLS-DA with reliability estimation. Talanta 2013, 117, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Chapelle, O.; Haffner, P.; Vapnik, V.N. Support vector machines for histogram-based image classification. IEEE Trans. Neural Netw. 1999, 10, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Burges, C.J.C. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Gwirtz, J.A.; Garcia-Casal, M.N. Processing maize flour and corn meal food products. Ann. N. Y. Acad. Sci. 2014, 1312, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Osborne, B.G.; Fearn, T.; Hindle, P.H. Practical Nir Spectroscopy with Applications in Food & Beverage Analysis; Longman Scientific & Technical: Harlow, UK, 1993. [Google Scholar]
- Workman, J.; Weyer, L. Practical Guide to Interpretive Near-Infrared Spectroscopy; CRC Press, Inc.: Boca Raton, FL, USA, 2007. [Google Scholar]
- Ishikawa, D.; Murayama, K.; Awa, K.; Genkawa, T.; Komiyama, M.; Kazarian, S.G.; Ozaki, Y. Application of a newly developed portable NIR imaging device to monitor the dissolution process of tablets. Anal. Bioanal. Chem. 2013, 405, 9401–9409. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, T.; Downey, G.; Kelly, J.D.; O’Donnell, C. Geographical classification of honey samples by near-infrared spectroscopy: A feasibility study. J. Agric. Food Chem. 2007, 55, 9128–9134. [Google Scholar] [CrossRef] [PubMed]
- Daszykowski, M.; Wrobel, M.S.; Czarnik-Matusewicz, H.; Walczak, B. Near-infrared reflectance spectroscopy and multivariate calibration techniques applied to modelling the crude protein, fibre and fat content in rapeseed meal. Analyst 2008, 133, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Cséfalvayová, L.; Pelikan, M.; Kralj, C.I.; Kolar, J.; Strlic, M. Use of genetic algorithms with multivariate regression for determination of gelatine in historic papers based on FT-IR and NIR spectral data. Talanta 2010, 82, 1784–1790. [Google Scholar] [CrossRef] [PubMed]



| Methods | PLS-DA | ||
|---|---|---|---|
| Parameter 1 | Calibration Set | Prediction Set | |
| Raw | 9 | 98.50% | 97.86% |
| WT | 9 | 99.43% | 98.71% |
| SNV | 9 | 99.50% | 95.00% |
| MSC | 9 | 99.36% | 94.57% |
| Methods | PLS-DA | SVM | ||||
|---|---|---|---|---|---|---|
| Parameter 1 | Calibration Set | Prediction Set | Parameter 1 | Calibration Set | Prediction Set | |
| Full spectra | 9 | 99.43% | 98.71% | (256, 0.0625) | 98.5% | 97.00% |
| Optimal wavelengths | 8 | 99.35% | 99.00% | (256, 16) | 99.14% | 98.29% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Zhao, Y.; Zhang, C.; Cheng, P.; He, Y. Discrimination of Transgenic Maize Kernel Using NIR Hyperspectral Imaging and Multivariate Data Analysis. Sensors 2017, 17, 1894. https://doi.org/10.3390/s17081894
Feng X, Zhao Y, Zhang C, Cheng P, He Y. Discrimination of Transgenic Maize Kernel Using NIR Hyperspectral Imaging and Multivariate Data Analysis. Sensors. 2017; 17(8):1894. https://doi.org/10.3390/s17081894
Chicago/Turabian StyleFeng, Xuping, Yiying Zhao, Chu Zhang, Peng Cheng, and Yong He. 2017. "Discrimination of Transgenic Maize Kernel Using NIR Hyperspectral Imaging and Multivariate Data Analysis" Sensors 17, no. 8: 1894. https://doi.org/10.3390/s17081894






