Rapid Waterborne Pathogen Detection with Mobile Electronics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunoagglutination Assays of Bacterial Detection
2.2. Membrane-Based Filtration for Pathogen Enrichment
2.3. Fabrication and Assembly of Microlens-Embedded Microfluidic Devices
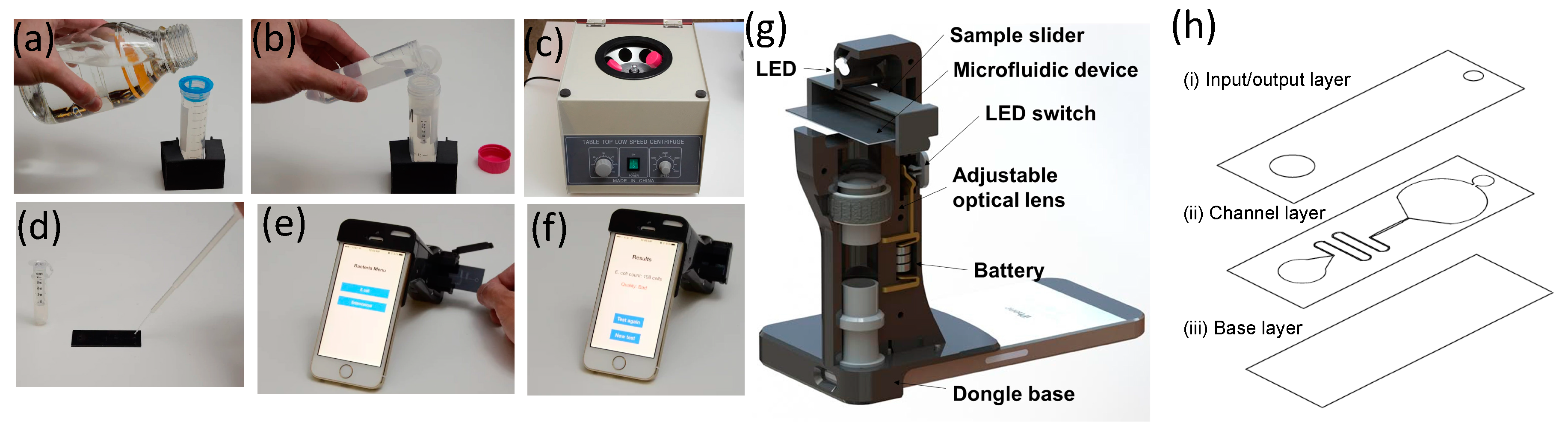
2.4. A Smartphone Dongle for Field Test Imaging
2.5. Protocol Operation
2.6. iOS Application
- (a)
- Turn on the application by clicking the icon;
- (b)
- Choose E. coli on the window menu and power up an LED and CMOS camera of the smartphone;
- (c)
- Insert the sample slider (where the microfluidic device is placed) into the smartphone dongle;
- (d)
- Start the image-acquiring process by clicking the Process button on the screen. Both dark-field scattering images and bright-field transmissive images are captured on the same CMOS camera;
- (e)
- Captured images are analyzed to determine the scattering intensity, which is used for the quantification of bacterium levels.
3. Results
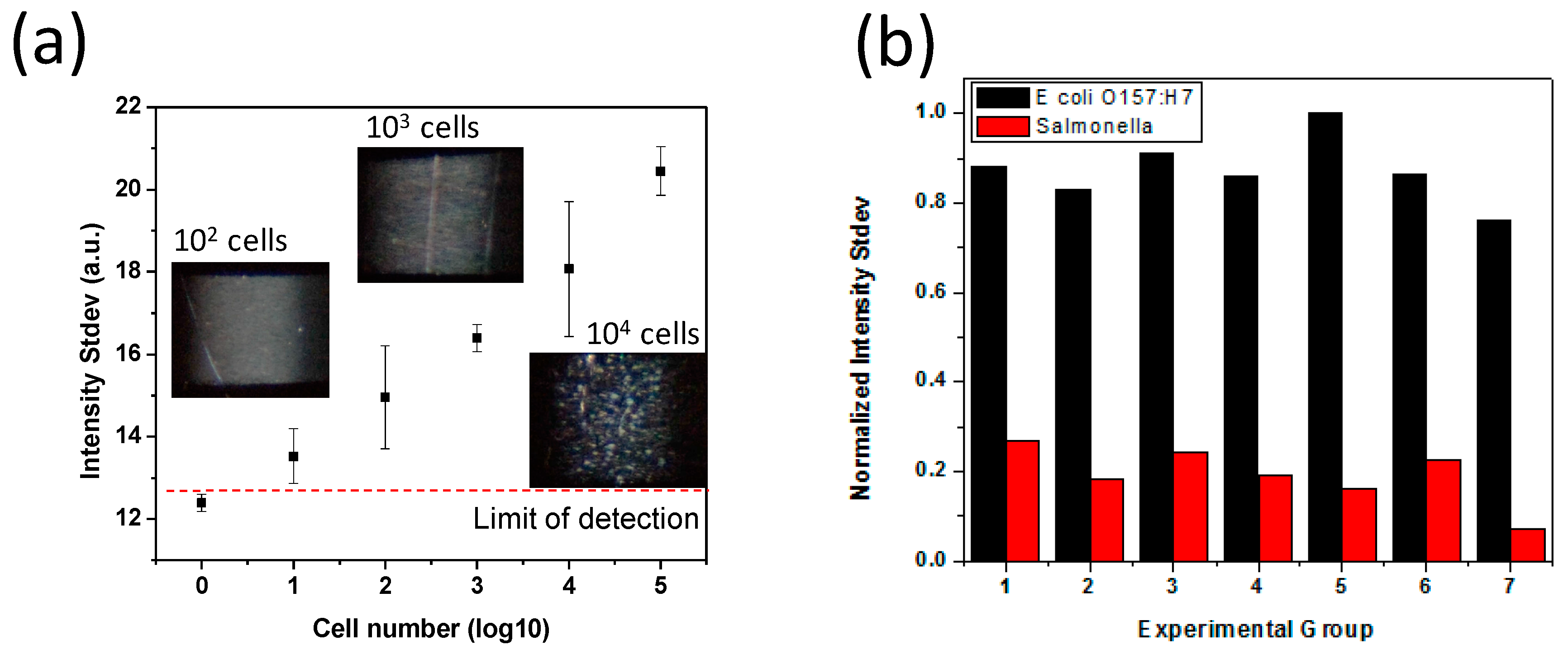
3.1. Scattered Light Detection by Narrow Beam Scanning Technique
3.2. Agglutination Reaction Optimization
3.3. Capillary Driven Microfluidic Device Fabrication
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Global Water, Sanitation and Hygiene. Available online: http://www.cdc.gov/healthywater/global/programs/ (accessed on 20 February 2017).
- Scallan, E.; Griffin, P.M.; Angulo, F.J.; Tauxe, R.V.; Hoekstra, R.M. Foodborne illness acquired in the United States—Unspecified agents. Emerg. Infect. Dis. 2011, 17, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Dusetty, P.; Velazquez, F.R.; Gutierrez-Escolano, A.L.; Ludert, J.E. Evaluation of the second generation of a commercial latex agglutination test for the detection of rotavirus antigens in fecal samples. J. Clin. Virol. 2013, 57, 88–90. [Google Scholar] [CrossRef] [PubMed]
- EPA. 2012 Recreational Water Quality Criteria Documents; EPA: Washington, DC, USA, 2012. [Google Scholar]
- Juhna, T.; Birzniece, D.; Larsson, S.; Zulenkovs, D.; Sharipo, A.; Azevedo, N.F.; Menard-Szczebara, F.; Castagnet, S.; Feliers, C.; Keevil, C.W. Detection of Escherichia coli in biofilms from pipe samples and coupons in drinking water distribution networks. Appl. Environ. Microbiol. 2007, 73, 7456–7464. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, G.; Brassard, J.; Edge, T.A.; Gannon, V.; Gottschall, N.; Jokinen, C.C.; Jones, T.H.; Khan, I.U.; Marti, R.; Sunohara, M.D.; et al. Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired agricultural watersheds under controlled and conventional tile drainage management. Appl. Environ. Microbiol. 2014, 80, 3708–3720. [Google Scholar] [CrossRef] [PubMed]
- Edge, T.A.; El-Shaarawi, A.; Gannon, V.; Jokinen, C.; Kent, R.; Khan, I.U.; Koning, W.; Lapen, D.; Miller, J.; Neumann, N.; et al. Investigation of an Escherichia coli environmental benchmark for waterborne pathogens in agricultural watersheds in Canada. J. Environ. Qual. 2012, 41, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.H.; Croft, R.; Atwill, E.R.; Wade, S.; Stehman, S. Waterborne pathogens in agricultural watersheds. Tech. Note 2000, 2, 1–64. [Google Scholar]
- Kim, K.; Myung, H. Sensor node for remote monitoring of waterborne disease-causing bacteria. Sensors 2015, 15, 10569–10579. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Kim, B. Lab-on-a-chip pathogen sensors for food safety. Sensors 2012, 12, 10713–10741. [Google Scholar] [CrossRef] [PubMed]
- Fratamico, P.M. Comparison of culture, polymerase chain reaction (PCR), TaqMan Salmonella, and Transia Card Salmonella assays for detection of Salmonella spp. in naturally-contaminated ground chicken, ground turkey, and ground beef. Mol. Cell. Probes 2003, 17, 215–221. [Google Scholar] [CrossRef]
- Melendez, J.H.; Frankel, Y.M.; An, A.T.; Williams, L.; Price, L.B.; Wang, N.Y.; Lazarus, G.S.; Zenilman, J.M. Real-time PCR assays compared to culture-based approaches for identification of aerobic bacteria in chronic wounds. Clin. Microbiol. Infect. 2010, 16, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.W.; Devenish, J.; Lutze-Wallace, C.L.; Milnes, D.; Robertson, R.H.; Berlie-Surujballi, G. Evaluation of a monoclonal antibody-based enzyme-linked immunosorbent assay for detection of Campylobacter fetus in bovine preputial washing and vaginal mucus samples. Vet. Microbiol. 2004, 103, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fu, Y.; Fang, W.; Li, Y. Electrochemical Impedance Immunosensor Based on Self-Assembled Monolayers for Rapid Detection of Escherichia coli O157:H7 with Signal Amplification Using Lectin. Sensors 2015, 15, 19212–19224. [Google Scholar] [CrossRef] [PubMed]
- Uddin, S.M.; Ibrahim, F.; Sayad, A.A.; Thiha, A.; Pei, K.X.; Mohktar, M.S.; Hashim, U.; Cho, J.; Thong, K.L. A portable automatic endpoint detection system for amplicons of loop mediated isothermal amplification on microfluidic compact disk platform. Sensors 2015, 15, 5376–5389. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sikora, U.; Ozcan, A. Quantum dot enabled detection of Escherichia coli using a cell-phone. Analyst 2012, 137, 2541–2544. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-Y.; Park, T.S. Smartphone Detection of Escherichia coli from Field Water Samples on Paper Microfluidics. IEEE Sens. J. 2015, 15, 1902–1907. [Google Scholar]
- Park, T.S.; Li, W.; McCracken, K.E.; Yoon, J.Y. Smartphone quantifies Salmonella from paper microfluidics. Lab Chip 2013, 13, 4832–4840. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Tang, T.H.; Chen, Y.; Citartan, M.; Lakshmipriya, T. Bacterial detection: From microscope to smartphone. Biosens. Bioelectron. 2014, 60, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, H.; Asghar, W.; Inci, F.; Yuksekkaya, M.; Jahangir, M.; Zhang, M.H.; Durmus, N.G.; Gurkan, U.A.; Kuritzkes, D.R.; Demirci, U. Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Sci. Rep. 2015, 5, 8719. [Google Scholar] [CrossRef] [PubMed]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.E.; Daugherity, E.K.; Altier, C.; Maurer, K.J. Efficacy and limitations of an ATP-based monitoring system. J. Am. Assoc. Lab Anim. Sci. 2010, 49, 190–195. [Google Scholar] [PubMed]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Bashir, R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol. Adv. 2008, 26, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Rehman, A.; Sims, M.; Zeng, X. Antimicrobial Susceptibility Assays Based on the Quantification of Bacterial Lipopolysaccharides via a Label Free Lectin Biosensor. Anal. Chem. 2015, 87, 4385–4393. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Rehman, A.; Liu, H.; Zhang, J.; Zhu, S.; Zeng, X. Glycosylation of Quinone-Fused Polythiophene for Reagentless and Label-Free Detection of E. coli. Anal. Chem. 2015, 87, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.F.; Yen, T.M.; Han, Y.; Chiu, Y.J.; Lin, E.Y.; Lo, Y.H. A light-sheet microscope compatible with mobile devices for label-free intracellular imaging and biosensing. Lab Chip 2014, 14, 3341–3348. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J. Genetic-Determinants Coding for Fimbriae and Adhesins of Extraintestinal Escherichia-coli. Curr. Top. Microbiol. Immunol. 1990, 151, 1–27. [Google Scholar] [PubMed]
- Normark, S.; Lark, D.; Hull, R.; Norgren, M.; Baga, M.; Ohanley, P.; Schoolnik, G.; Falkow, S. Genetics of Digalactoside-Binding Adhesin from a Uropathogenic Escherichia-Coli Strain. Infect. Immun. 1983, 41, 942–949. [Google Scholar] [PubMed]
- Sack, R.B. Human Diarrheal Disease Caused by Enterotoxigenic Escherichia-Coli. Annu. Rev. Microbiol. 1975, 29, 333–353. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Abe, K. Topological structural analysis of digitized binary images by border following. Computer Vision, Graphics, and Image Processing. Comput. Vis. Graph. Image Process. 1985, 30, 32–46. [Google Scholar] [CrossRef]
- Wang, H.; Yuan, X.; Zhang, L. Latex agglutination: Diagnose the early cryptococcus neoformans test of capsular polysaccharide antigen. Pak. J. Pharm. Sci. 2015, 28 (Suppl. 1), 307–311. [Google Scholar] [PubMed]
- Trinh, D.Q.; Ogawa, H.; Bui, V.N.; Nguyen, T.T.; Gronsang, D.; Baatartsogt, T.; Kizito, M.K.; AboElkhair, M.; Yamaguchi, S.; Nguyen, V.K.; et al. Development of a blocking latex agglutination test for the detection of antibodies to chicken anemia virus. J. Virol. Methods 2015, 221, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Rocha, L.B.; Santos, A.R.; Munhoz, D.D.; Cardoso, L.T.; Luz, D.E.; Andrade, F.B.; Horton, D.S.; Elias, W.P.; Piazza, R.M. Development of a rapid agglutination latex test for diagnosis of enteropathogenic and enterohemorrhagic Escherichia coli infection in developing world: Defining the biomarker, antibody and method. PLoS Negl. Trop. Dis. 2014, 8, e3150. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Walther, T.; Molinaro, S.; Li, X.; Xia, G.; Wieser, A.; Peters, G.; Peschel, A.; Becker, K. Bacteriophage-based latex agglutination test for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 3394–3398. [Google Scholar] [CrossRef] [PubMed]
- Deneke, Y.; Sabarinath, T.; Gogia, N.; Lalsiamthara, J.; Viswas, K.N.; Chaudhuri, P. Evaluation of recombinant LigB antigen-based indirect ELISA and latex agglutination test for the serodiagnosis of bovine leptospirosis in India. Mol. Cell. Probes 2014, 28, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.G.; Bi, X.L.; Wu, J.H.; Xu, H.; Liao, W.Q. Disseminated cryptococcal lymphadenitis with negative latex agglutination test. Chin. Med. J. 2012, 125, 2393–2396. [Google Scholar] [PubMed]
- Silveira-Gomes, F.; Marques-da-Silva, S.H. Effects of pretreating serum samples on the performance of a latex agglutination test for serodiagnosis of paracoccidioidomycosis. Clin. Vaccine Immunol. 2012, 19, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Yao, L.; Ding, M.; Wang, D.J.; Chen, H.C.; Liu, Z.F. Development of latex agglutination test for rapid detection of antibodies against Bovine herpesvirus 1 and Bovine herpesvirus 5 in cattle. J. Vet. Diagn. Investig. 2012, 24, 1162–1165. [Google Scholar] [CrossRef] [PubMed]
- Silveira-Gomes, F.; Sarmento, D.N.; Pinto, T.M.; Pimentel, R.F.; Nepomuceno, L.B.; Espirito Santo, E.P.; Mesquita-da-Costa, M.; Camargo, Z.P.; Marques-da-Silva, S.H. Development and evaluation of a latex agglutination test for the serodiagnosis of paracoccidioidomycosis. Clin. Vaccine Immunol. 2011, 18, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Andriesse, G.I.; Elberts, S.; Vrolijk, A.; Verhulst, C.; Kluytmans, J.A. Evaluation of a fourth-generation latex agglutination test for the identification of Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hu, S.; Yu, X. Development of a latex agglutination test for detecting antibodies against avian influenza virus based on matrix 1 protein expressed in vitro. Avian Dis. 2010, 54, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Sanz, J.C.; Culebras, E.; Rios, E.; Rodriguez-Avial, I.; Wilhelmi, I.; Ramos, B.; Ordobas, M.; Picazo, J.J. Direct serogrouping of Streptococcus pneumoniae strains in clinical samples by use of a latex agglutination test. J. Clin. Microbiol. 2010, 48, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, Y.; Kim, M.; Jee, Y.; Cheon, D.S.; Jeong, H.S.; Ko, G. Development of a latex agglutination test for norovirus detection. J. Microbiol. 2010, 48, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Narang, N.; Fratamico, P.M.; Tillman, G.; Pupedis, K.; Cray, W.C., Jr. Performance comparison of a fliC(h7) real-time PCR assay with an H7 latex agglutination test for confirmation of the H type of Escherichia coli O157:H7. J. Food Prot. 2009, 72, 2195–2197. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.-F.; Chen, Y.-C.; Wang, W.-C.; Kucknoor, A.S.; Lin, C.-J.; Lo, Y.-H.; Yao, C.-W.; Lian, I. Rapid Waterborne Pathogen Detection with Mobile Electronics. Sensors 2017, 17, 1348. https://doi.org/10.3390/s17061348
Wu T-F, Chen Y-C, Wang W-C, Kucknoor AS, Lin C-J, Lo Y-H, Yao C-W, Lian I. Rapid Waterborne Pathogen Detection with Mobile Electronics. Sensors. 2017; 17(6):1348. https://doi.org/10.3390/s17061348
Chicago/Turabian StyleWu, Tsung-Feng, Yu-Chen Chen, Wei-Chung Wang, Ashwini S. Kucknoor, Che-Jen Lin, Yu-Hwa Lo, Chun-Wei Yao, and Ian Lian. 2017. "Rapid Waterborne Pathogen Detection with Mobile Electronics" Sensors 17, no. 6: 1348. https://doi.org/10.3390/s17061348





