In Vitro Studies on a Microfluidic Sensor with Embedded Obstacles Using New Antibacterial Synthetic Compounds (1-TDPPO) Mixed Prop-2-en-1-one with Difluoro Phenyl
Abstract
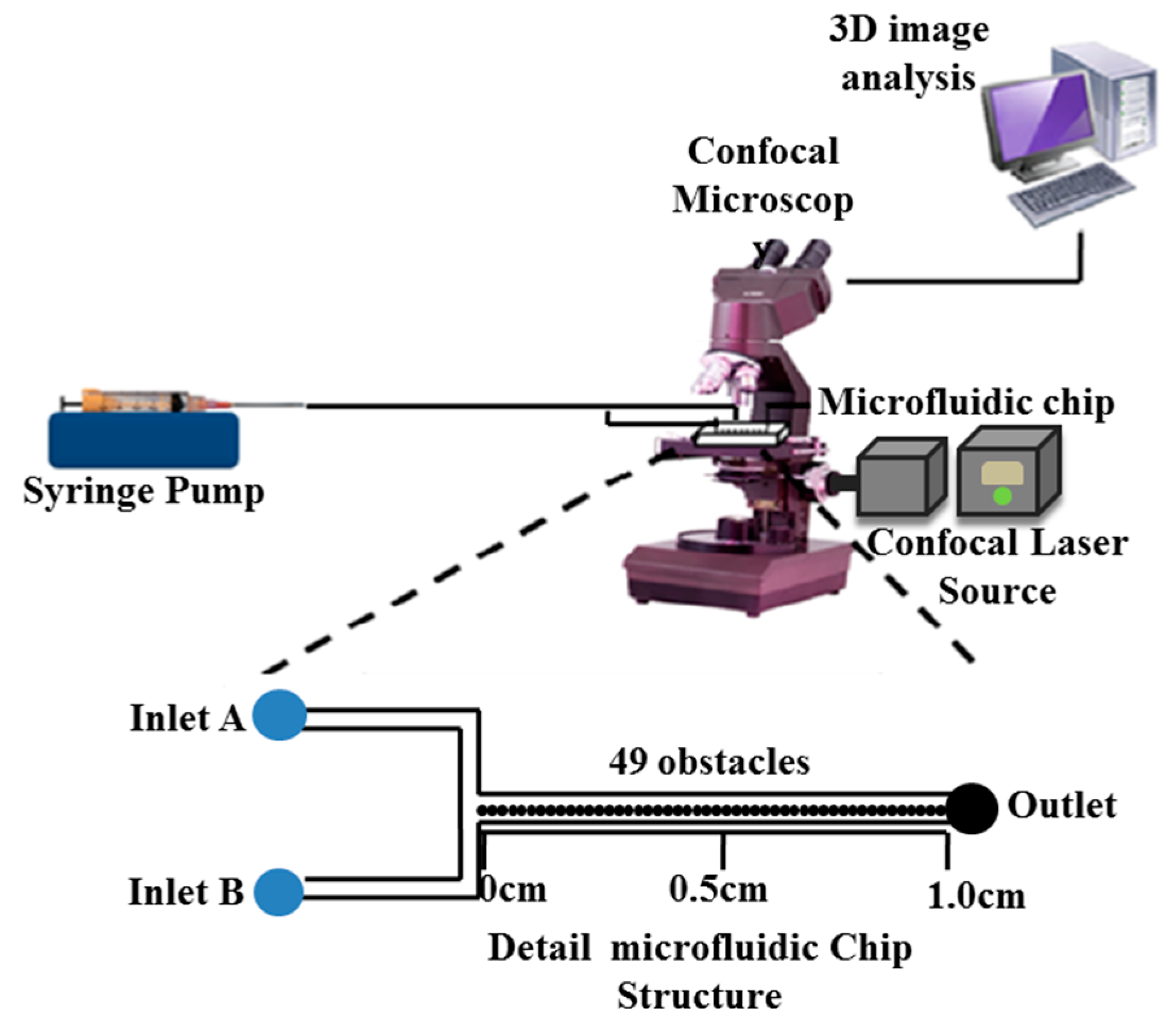
:1. Introduction
2. Results and Discussion
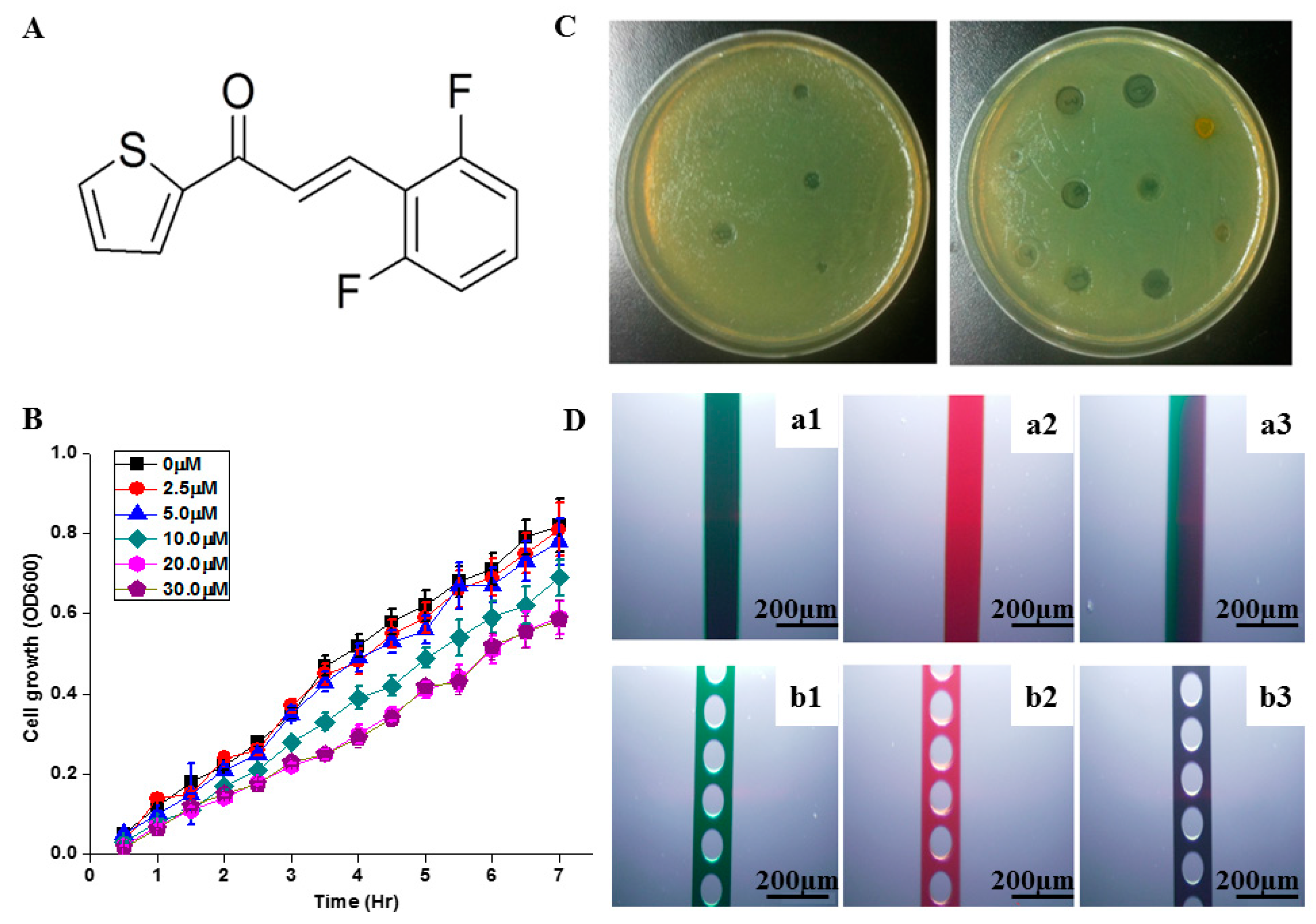
2.1. Selection of Antimicrobial Concentration
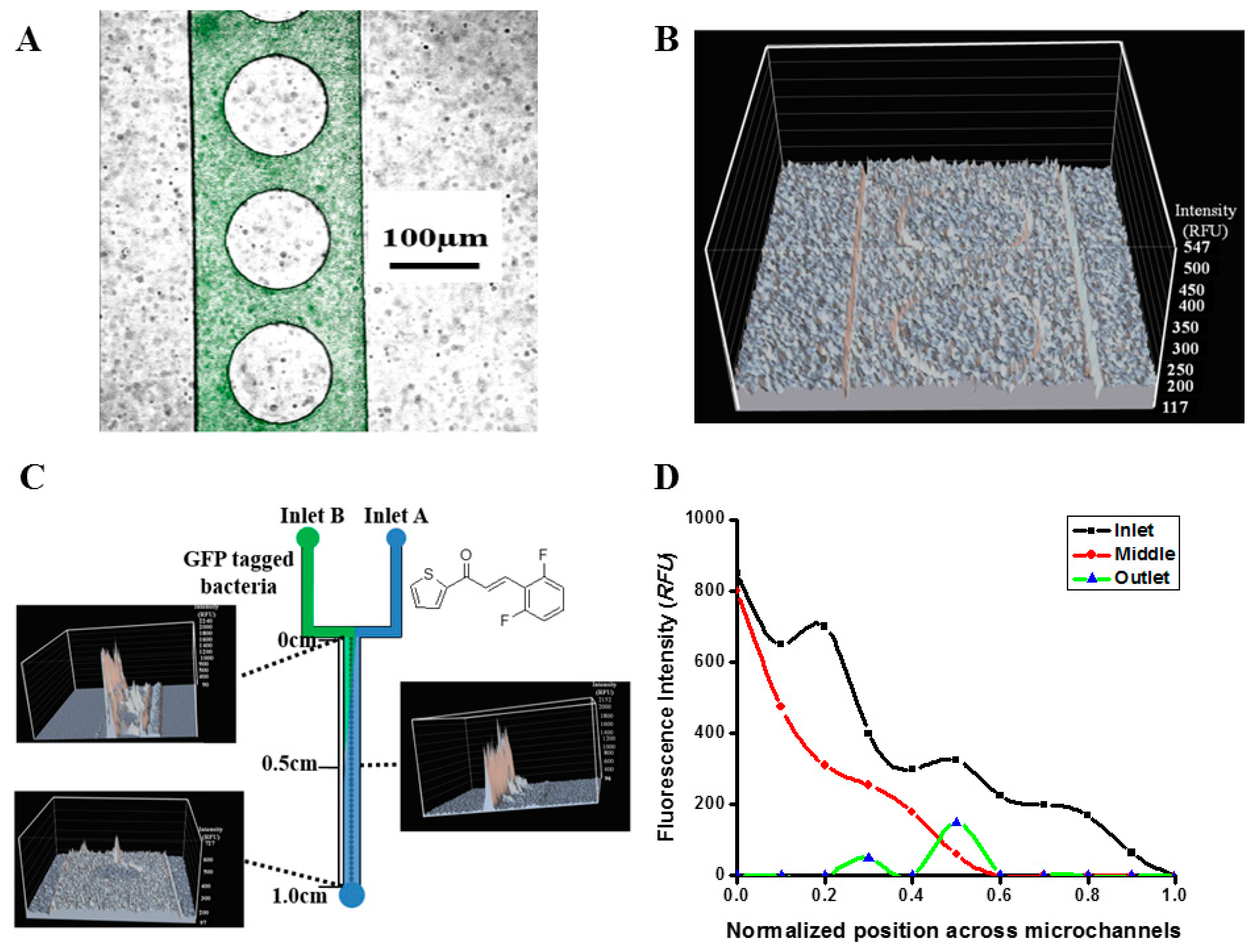
2.2. Real-Time Monitoring of Cell Dynamics
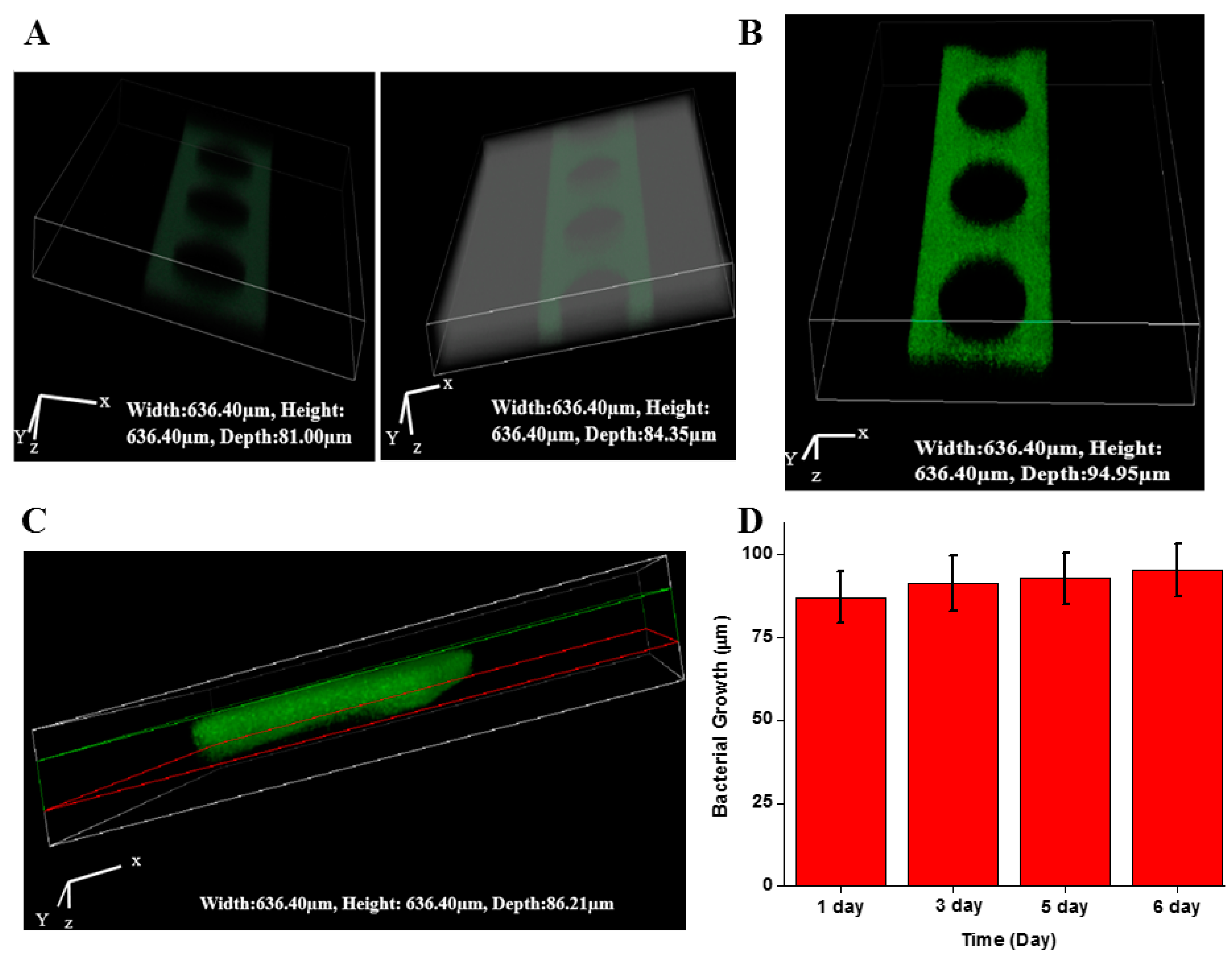
2.3. Biofilm Formation in Microfluidic Sensor
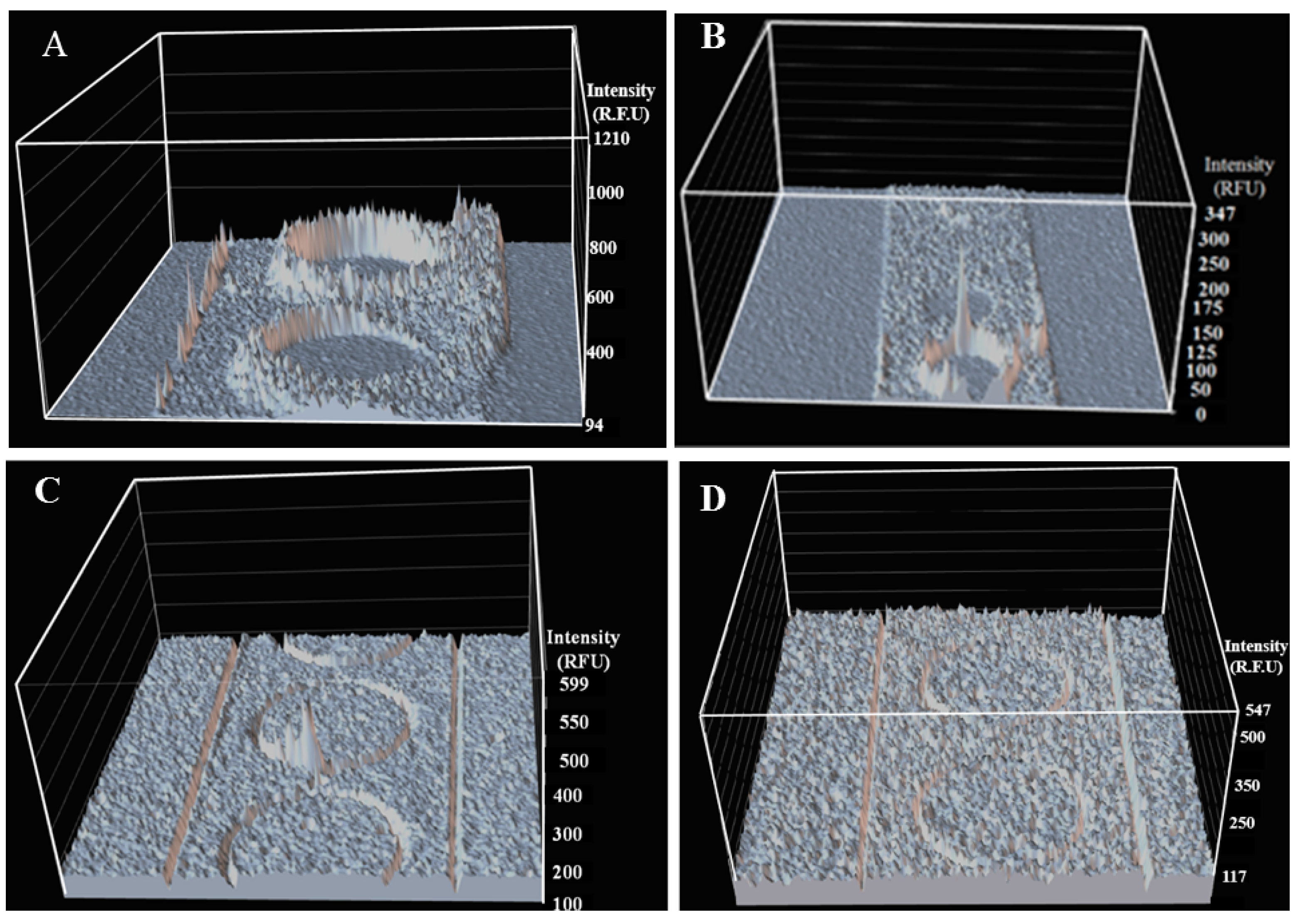
2.4. Biofilm Eradication in Microfluidic Sensor
3. Materials and Methods
3.1. Synthesis of Antimicrobial Agent (1-TDPPO)
3.2. Antimicrobial Activity Using Petri Dish System
3.3. Fabrication of Microfluidic Reactor
3.4. Fluorescence Bio-Imaging
3.5. Experimental Procedures
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lanning, L.M.; Ford, R.M.; Long, T. Bacterial chemotaxis transverse to axial flow in a microfluidic channel. Biotechnol. Bioeng. 2008, 100, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Kato, J.; Kim, H.E.; Takiguchi, N.; Kuroda, A.; Ohtake, H. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J. Biosci. Bioeng. 2008, 106, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Burrows, L.L.; Deber, C.M. Helix induction in antimicrobial peptides by alginate in biofilms. J. Biol. Chem. 2004, 279, 38749–38754. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganism. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.; Brady, K.M.; Greenberg, E.P.; Qi, F.; Yarbrough, D.K.; He, J.; McHardy, I.; Anderson, M.H.; Shi, W. Enhancement of antimicrobial activity against Pseudomonas aeruginosa by coadminstration of G10KHc and Tobramycin. Antimicrob. Agents Chemother. 2006, 50, 3833–3838. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Lieleq, O.; Caldara, M.; Baumqartel, R.; Ribbeck, K. Mechanical robustness of Pseudomonas aeruginosa biofilms. Soft Matter 2001, 7, 3307–3314. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010, 59, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Repo, H.; Kostiala, A.A.; Kosunen, T.U. Use of leucocyte migration under agarose to study spontaneous and directed locomotion of leucocytes. Immunology 1978, 35, 539–548. [Google Scholar] [PubMed]
- Haddox, J.L.; Knowles, I.W.; Sommers, C.I.; Pfister, R.R. Characterization of chemical gradients in the collagen gel-visual chemotaxis assay. J. Immunol. Methods 1994, 171, 1–14. [Google Scholar] [CrossRef]
- Ford, R.M.; Philips, B.R.; Quinn, J.A.; Lauffenburger, D.A. Measurement of bacterial random motility and chemotaxis coefficients: I. Stopped-flow diffusion chamber assay. Biotechnol. Bioeng. 1991, 37, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Law, A.M.J.; Aitken, M.D. Continuous-flow capillary assay for measuring bacterial chemotaxis. Appl. Environ. Microbiol. 2005, 71, 3137–3143. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lee, S.; Kim, J.; Kim, H.; Kim, Y.; Yoo, J.; Chang, W.; Lee, C. Microfluidic monitoring of Pseudomonas aeruginosa chemotaxis under the continuous chemical gradient. Biosens. Bioelectron. 2010, 26, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Shimizu, T.S.; Stocker, R. Microfluidics for bacterial chemotaxis. Integr. Biol. 2010, 2, 604–629. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Mukherjee, A.; Sevgen, S.E.; Sanpitakseree, C.; Lee, J.; Schroeder, C.M.; Kenis, P.J.A. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens. Bioelectron. 2013, 49, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.B.; Cremer, P.S.; Mason, M.D. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 2003, 9, 5449–5454. [Google Scholar] [CrossRef] [PubMed]
- Long, T.; Ford, R. Enhanced transverse migration of bacteria by chemotaxis in a porous T-sensor. Environ. Sci. Technol. 2009, 43, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Englert, D.L.; Manson, M.D.; Jayaraman, A. Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl. Environ. Microbiol. 2009, 75, 4557–4564. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Lee, Y.; Jin, S.; Koo, S.; Lee, C.S.; Yoo, J.Y. Quantitative analysis of single bacterial chemotaxis using a linear concentration gradient microchannel. Biomed. Microdevices 2009, 11, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Papautsky, I.; Gale, B.K.; Mohanty, S.; Ameel, T.A.; Frazier, B.A. Effects of rectangular microchannel aspect ratio on laminar friction constant. Proc. SPIE 1999, 3877, 147–158. [Google Scholar]
- Renfer, A.; Tiwari, M.K.; Tiwari, R.; Alfieri, F.; Brunschwiler, T.; Michel, B.; Poulikakos, D. Microvortex-enhanced heat transfer in 3D-integrated liquid cooling of electronic chip stacks. Int. J. Heat Mass Transfer 2013, 65, 33–43. [Google Scholar] [CrossRef]
- Yeom, J.; Agonafer, D.D.; Han, J.H.; Shannon, M.A. Low Reynolds numbers flow across an array of cylindrical microposts in a microchannel and figure-of-merit analysis of micropost- filled microreactors. J. Micromech. Microeng. 2009, 19, 065025. [Google Scholar] [CrossRef]
- Mitsos, A.; Barton, P.I. Microfabricated Power Generation Devices: Design and Technology; Wiley-VCH: Weinheim, Germany, 2009; pp. 183–184. [Google Scholar]
- Roth, R.; Lenk, G.; Cobry, K.; Woias, P. Heat transfer in freestanding microchannels within-line and staggered pin fin structures with clearance. Int. J. Heat Mass Transfer 2013, 67, 1–15. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Peles, Y. Flow boiling heat transfer on micro pin fins entrenched in a microchannel. J. Heat Transfer 2010, 132, 041007. [Google Scholar] [CrossRef]
- Bhagat, A.A.S.; Peterson, E.T.K.; Papautsky, I. A passive planar micromixer with obstructions for mixing at low Reynolds numbers. J. Micromech. Microeng. 2007, 17, 1017–1024. [Google Scholar] [CrossRef]
- Bessoth, F.G.; deMello, A.J.; Manz, A. Microstructure for efficient continuous mixing. Anal. Commun. 1999, 36, 213–215. [Google Scholar] [CrossRef]
- Wu, J.; Wu, X.; Lin, F. Recent developments in microfluidics-based chemotaxis studies. Lab Chip 2013, 13, 2484–2499. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.A.; Costa, L.; Terekhov, A.; Jowhar, D.; Hofmeister, W.; Janetopoulos, C. On-chip open microfluidic devices for chemotaxis studies. Microsc. Microanal. 2012, 18, 816–828. [Google Scholar] [CrossRef] [PubMed]
- Yawata, Y.; Toda, K.; Setoyama, E.; Fukuda, J.; Suzuki, H.; Uchiyama, H.; Nomura, N. Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. J. Biosci. Bioeng. 2010, 110, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Takagi, R.; Fukuda, J.; Nagata, K.; Yawata, Y.; Nomura, N.; Suzuki, H. A microfluidicmicrobial culture device for rapid determination of the minimum inhibitory concentration of antibiotics. Analyst 2013, 138, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Park, S.J.; Ko, S.Y.; Park, J.O.; Park, S. Development of bacteria-based microrobot using biocompatible poly(ethylene glycol). Biomed. Microdevices 2012, 14, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Z.Z.; Li, Q.L. Microfluidic techniques for dynamic single-cell analysis. Microchim. Acta 2010, 168, 177–195. [Google Scholar] [CrossRef]
- Polzer, H.; Haasters, F.; Prall, W.C.; Saller, M.M.; Volkmer, E.; Drosse, I.; Mutschler, W.; Schieker, M. Quantification of fluorescence intensity of labeled human mesenchymal stem cells and cell counting of unlabeled cells in phase-contrast imaging: An open-source-based algorithm. Tissue Eng. Part C Methods 2010, 16, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, S.; Hassannia, B.; Delrue, I.; Goossens, V.; Wiernicki, B.; Dondelinger, Y.; Bertrnad, M.J.M.; Krysko, D.V.; Vuylsteke, M.; Vandenabeele, P.; et al. A real-time fluorometric method for the simultaneous detection of cell death type and rate. Nat. Protocols 2016, 11, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- Krishnanjaneyulu, I.S.; Saravanan, G.; Vamsi, J.; Supriya, P.; Bhavana, J.U.; Kumar, M.V.S. Synthesis, characterization and antimicrobial activity of some novel benzimidazole derivaties. J. Adv. Pharm. Technol. Res. 2014, 5, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Konduru, N.K.; Owais, M. Design, synthesis and antimicrobial activities of novel ferrocenyl and organic chalcone based sulfones and bis-sulfones. Arabian J. Chem. 2015. [Google Scholar] [CrossRef]
- Vankadari, S.R.; Mandala, D.; Pochampalli, J.; Tigulla, P.; Valery, A.; Thampu, R. Synthesis, evaluation of antimicrobial activity, and molecular modeling of novel 2-((4-(2H-benxo[d] [1,2,3] triazol-2-yl)piperidin-1-yl)methyl)-5-substituted phenyl-1,3,4-oxadiazoles. Med. Chem. Res. 2013, 22, 5912–5919. [Google Scholar] [CrossRef]
- Qu, Y.; Istivan, T.S.; Daley, A.J.; Rouch, D.A.; Deighton, M.A. Comparison of various antimicrobial agents as catheter lock solutions: Preference for ethanol in eradication of coagulase-negative staphylococcal biofilms. J. Med. Microbiol. 2009, 58, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, S.A.; Levy, F.; Walker, E.S. Hand sanitizer alert. Emerg. Infect. Dis. 2006, 12, 527–529. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, G.; Russell, A.D. Antiseptics and Disinfectants: Activity, Action, and Resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [PubMed]
- Kim, J.; Park, H.; Chung, S. Microfluidic approaches to bacterial biofilm formation. Molecules 2012, 17, 9818–9834. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Cho, I.H.; Jeong, B.C.; Lee, S.H. Strategies to minimize antibiotic resistance. Int. J. Environ. Res. Public Health 2013, 10, 4274–4305. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.; Wang, X.; Curtiss, R.; Castillo-Chavez, C. Evaluating the efficacy of antimicrobial cycling programmes and patient isolation on dual resistance in hospitals. J. Biol. Dyn. 2011, 5, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Roh, C.; Lee, J.; Kang, C. The deformation of Polydimethylsiloxane (PDMS) microfluidic channels filled with embedded circular obstacles under certain circumstances. Molecules 2016, 21, 798. [Google Scholar] [CrossRef] [PubMed]
- Orland, I.J.; Burstyn, Y.; Bar-Matthews, M.; Kozdon, R.; Ayalon, A.; Matthews, A.; Valley, J.W. Seasonal climate signals (1990–2008) in a modern Soreq Cave stalagmite as revealed by high-resolution geochemical analysis. Chem. Geol. 2014, 363, 322–333. [Google Scholar] [CrossRef]
- Xie, L.; Altindal, T.; Chattopadhyay, S.; Wu, X. Bacterial flagellum as a propeller and as a rudder for efficient chemitaxis. Proc. Natl. Acad. Sci. USA 2011, 108, 2246–2251. [Google Scholar] [CrossRef] [PubMed]
- Samadi, A.; Zhang, C.; Chen, J.; Reihani, S.N.S.; Chen, Z. Evaluating the toxic effect of an antimicrobial agent on single bacterial cells with optical tweezers. Biomed. Opt. Express 2015, 6, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Rutala, W.A.; Weber, D.J. Guideline for Disinfection and Sterilization in Healthcare Facilities, CDC 2008. Available online: http://www.cdc.gov/hicpac/pdf/guidelines/Disinfection_Nov_2008.pdf (accessed on 29 December 2009).
- Wendakoon, C.; Calderon, P.; Gagnon, D. Evaluation of selected medicinal plants extracted in different ethanol concentrations for antibacterial activity against human pathogens. J. Med. Active Plants 2012, 1, 60–68. [Google Scholar]
| 0 min | 5 min | 7 min | 10 min | 30 min | 40 min | |
|---|---|---|---|---|---|---|
| 1-TDPPO (20 µM) with microstrctures | 771 ± 44 | 196 ± 16 | 0 | 0 | 0 | 0 |
| 1-TDPPO (20 µM) without microstrctures | 731 ± 39 | 512 ± 33 | 357 ± 28 | 258 ± 20 | 0 | 0 |
| Ethanol (20%) with microstrctures | 753 ± 48 | 531 ± 26 | 433 ± 25 | 366 ± 23 | 211 ± 12 | 0 |
| Ethanol (20%) without microstrctures | 767 ± 57 | 681 ± 48 | 598 ± 34 | 511 ± 43 | 458 ± 37 | 0 |
| Ethanol (90%) with microstrctures | 758 ± 36 | 302 ± 17 | 178 ± 13 | 0 | 0 | 0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roh, C.; Lee, J.; Kinger, M.; Kang, C. In Vitro Studies on a Microfluidic Sensor with Embedded Obstacles Using New Antibacterial Synthetic Compounds (1-TDPPO) Mixed Prop-2-en-1-one with Difluoro Phenyl. Sensors 2017, 17, 803. https://doi.org/10.3390/s17040803
Roh C, Lee J, Kinger M, Kang C. In Vitro Studies on a Microfluidic Sensor with Embedded Obstacles Using New Antibacterial Synthetic Compounds (1-TDPPO) Mixed Prop-2-en-1-one with Difluoro Phenyl. Sensors. 2017; 17(4):803. https://doi.org/10.3390/s17040803
Chicago/Turabian StyleRoh, Changhyun, Jaewoong Lee, Mayank Kinger, and Chankyu Kang. 2017. "In Vitro Studies on a Microfluidic Sensor with Embedded Obstacles Using New Antibacterial Synthetic Compounds (1-TDPPO) Mixed Prop-2-en-1-one with Difluoro Phenyl" Sensors 17, no. 4: 803. https://doi.org/10.3390/s17040803







