An Exonuclease I-Based Quencher-Free Fluorescent Method Using DNA Hairpin Probes for Rapid Detection of MicroRNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Fluorescence Measurement
2.3. Detection of miRNA
2.4. Optimization of Analysis Conditions
2.5. Quantitative Analysis of miRNA
2.6. Application of the Method in Biological Assays
3. Results and Discussion
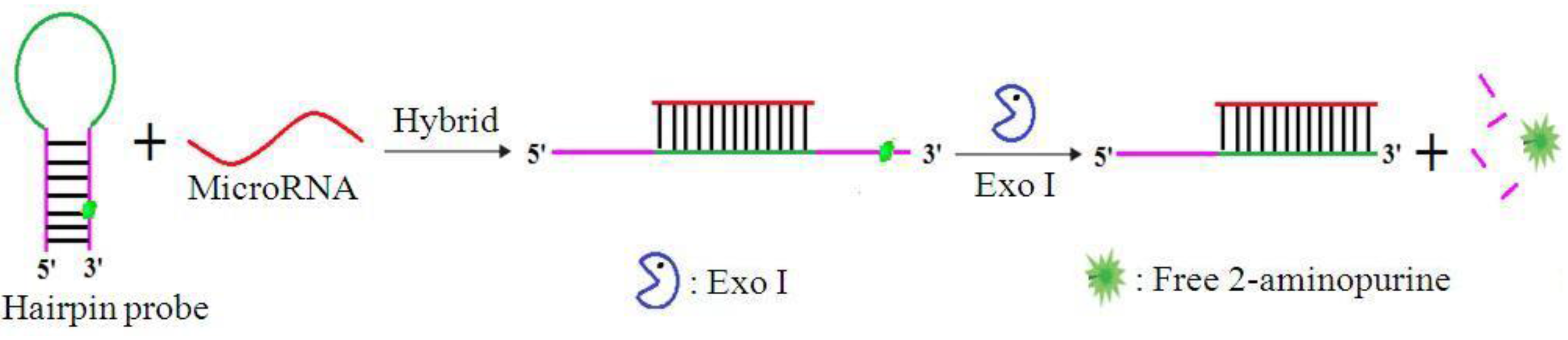
3.1. Designing the Strategy
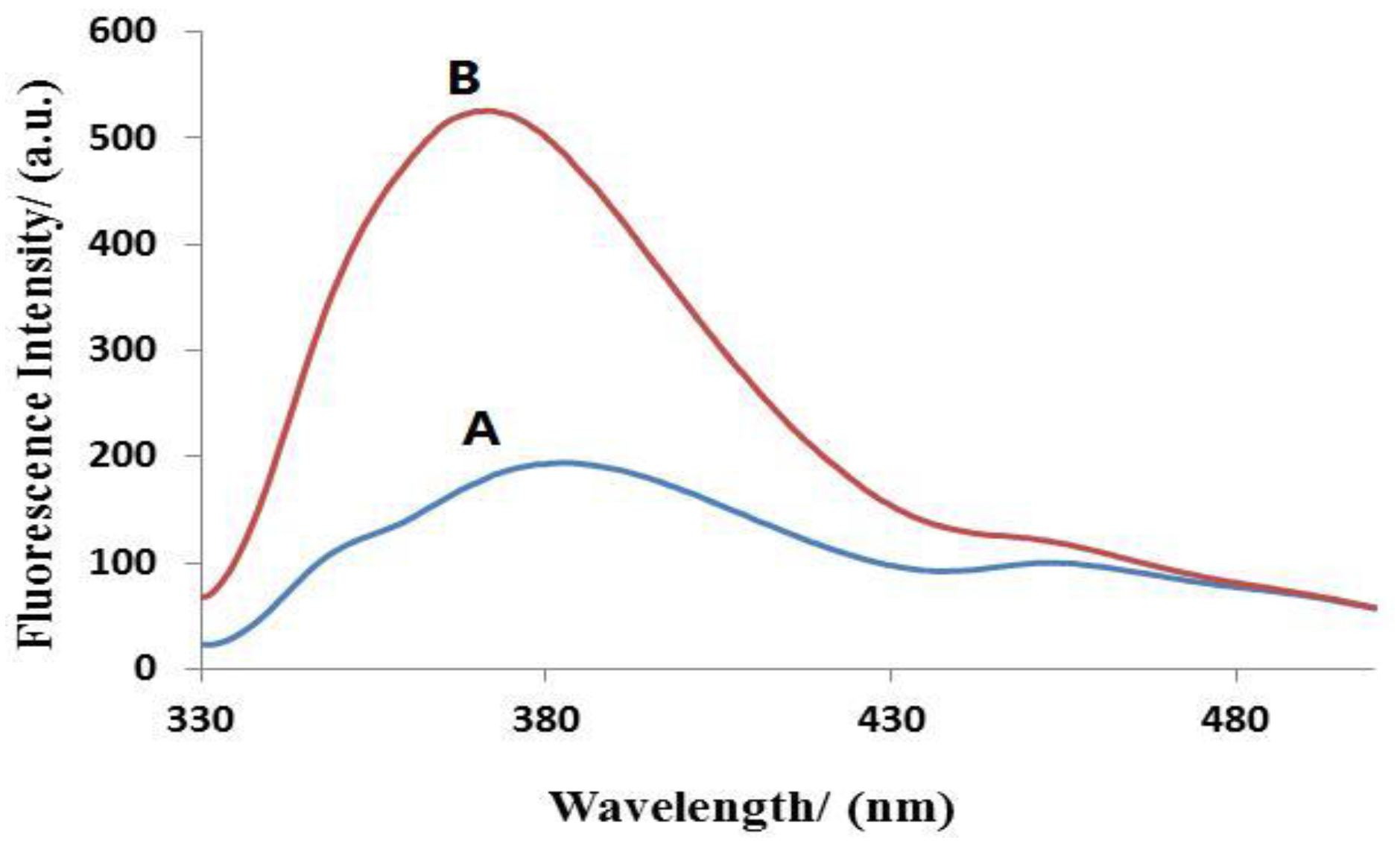
3.2. Validation of miRNA Detection with the Method
3.3. Optimization of Experimental Conditions
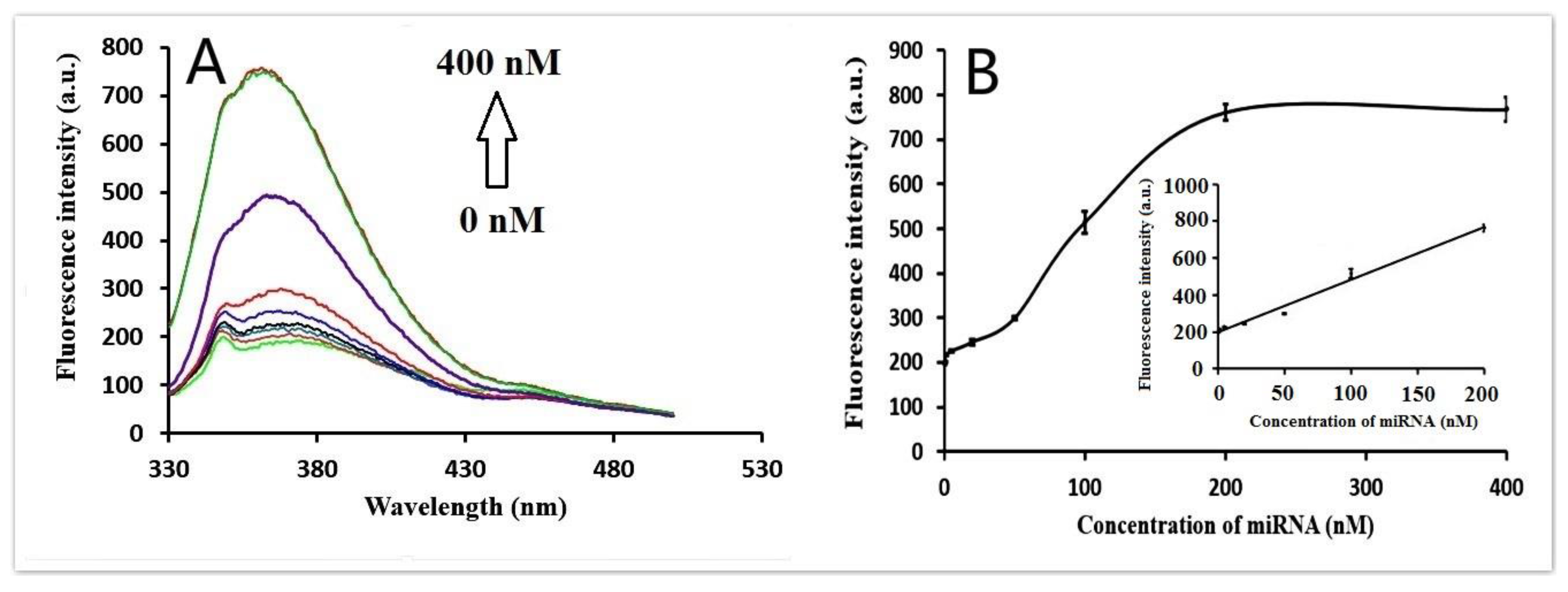
3.4. Quantitative Analysis of miRNA
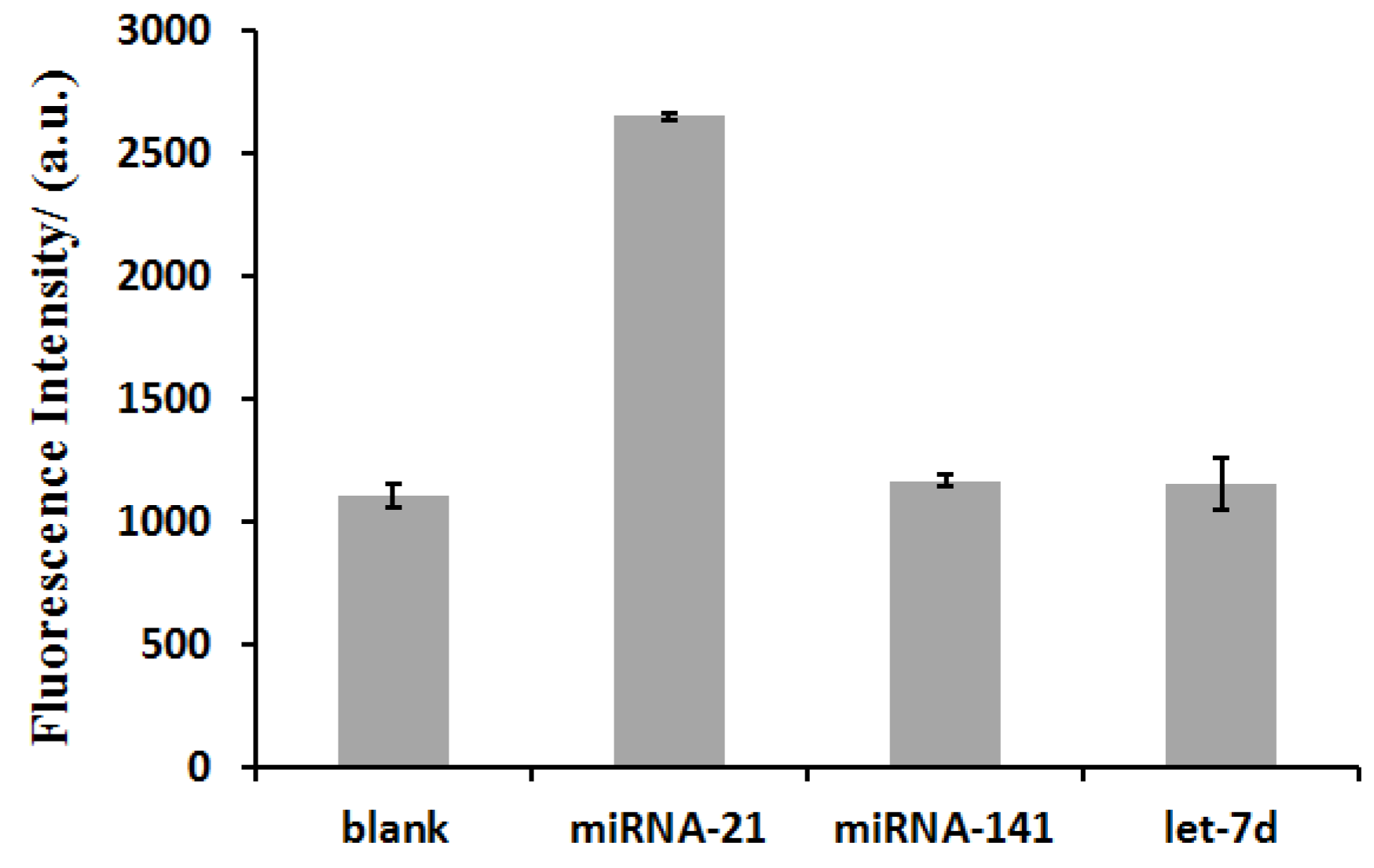
3.5. Selectivity of the miRNA Detection Assay
3.6. Application of the Method in Biological Assays
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, J.; Tan, S.; Kooger, R.; Zhang, C.; Zhang, Y. MicroRNAs as novel biological targets for detection and regulation. Chem. Soc. Rev. 2014, 43, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, J.Q.; Zhou, X. A review: MicroRNA detection methods. Org. Biomol. Chem. 2015, 13, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Viral and cellular messenger RNA targets of viral microRNAs. Nature 2009, 457, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Aviñó, A.; Huertas, C.S.; Lechuga, L.M.; Eritja, R. Sensitive and label-free detection of miRNA-145 by triplex formation. Anal. Bioanal. Chem. 2016, 408, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.B.; Yeung, E.S.; Qi, S.D.; Han, R. Highly sensitive detection of microRNA by chemiluminescence based on enzymatic polymerization. Anal. Bioanal. Chem. 2012, 402, 2217–2220. [Google Scholar] [CrossRef] [PubMed]
- Serpico, D.; Molino, L.; Cosimo, S.D. MicroRNAs in breast cancer development and treatment. Cancer Treat. Rev. 2014, 40, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Srivastava, A.K.; Suman, S.; Kumar, V.; Shukla, Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett. 2015, 369, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Saavedra, E.A.; Lamb, J.; Peck, D.; Cordero, A.S.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Soh, J.H.; Ying, J.Y. A microarray platform for detecting disease-specific circulating miRNA in human serum. Biosens. Bioelectron. 2016, 75, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Válóczi, A.; Hornyik, C.; Varga, N.; Burgyán, J.; Kauppinen1, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.; Liu, S.; Yu, J.H.; Wang, H.Z.; Guo, Y.N.; Huang, J. Ultrasensitive and rapid detection of miRNA with three-way junction structure-based trigger-assisted exponential enzymatic amplification. Biosens. Bioelectron. 2016, 81, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Tang, H.; Wu, D.; Xia, Y.; Wu, M.; Yi, X.; Li, H.; Wang, J. Amplified voltammetric detection of miRNA from serum samples of glioma patients via combination of conducting magnetic microbeads and ferrocene-capped gold nanoparticle/streptavidin conjugates. Biosens. Bioelectron. 2016, 86, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, Z.; Xu, Z.; Yin, H.; Ai, S. MicroRNA-21 detection based on molecular switching by amperometry. New J. Chem. 2012, 36, 1985–1991. [Google Scholar]
- Johnson, B.; Mutharasan, R. Sample preparation-free, real-time detection of microRNA in human serum using piezoelectric cantilever biosensors at attomole level. Anal. Chem. 2012, 84, 10426–10436. [Google Scholar] [CrossRef] [PubMed]
- Xi, Q.; Zhou, D.; Kan, Y.; Ge, J.; Wu, Z.; Yu, R.; Jiang, J. Highly sensitive and selective strategy for microRNA detection based on WS2 nanosheet mediated fluorescence quenching and duplex-specific nuclease signal amplification. Anal. Chem. 2014, 86, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lin, X.; Lin, N.; Song, Y.; Zhu, Z.; Chen, Z.; Yang, C. Graphene oxide-protected DNA probes for multiplex microRNA analysis in complex biological samples based on a cyclic enzymatic amplification method. Chem. Commun. 2012, 48, 194–196. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhang, C.; Huang, Y.; Zhu, Z.; Chen, X.; Yang, C. Backbone-modified molecular beacons for highly sensitive and selective detection of microRNAs based on duplex specific nuclease signal amplification. Chem. Commun. 2013, 49, 7243–7245. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Zhao, J.; Zhao, L.; Cheng, Y.; Li, Z. A simple molecular beacon with duplex-specific nuclease amplification for detection of microRNA. Analyst 2016, 14, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Lin, X.; Huang, Y.; Zhi, R.P.Z.; Zhou, L.; Yang, C. Highly sensitive and selective detection of miRNA: DNase I-assisted target recycling using DNA probes protected by polydopamine nanospheres. Chem. Commun. 2015, 51, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Li, W.; Liu, X.; Li, F. Label-Free and Enzyme-Free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: A facile, sensitive, and highly specific microRNA assay. Anal. Chem. 2015, 87, 11368–11374. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Shi, H.; He, X.; Wang, K.; He, D.; Guo, Q.; Qing, Z.; Yan, L.; Ye, X.; Li, D.; et al. Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection. Anal. Chem. 2014, 86, 6976–6982. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Cheng, W.; Yan, Y.R.; Zhang, Y.; Yin, Y.B.; Ju, H.X.; Ding, S.J. A colorimetric biosensor for detection of attomolar microRNA with a functional nucleic acid-based amplification machine. Talanta 2016, 146, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, Y.L.; Wang, H.Y.; Wu, J.; Zhu, F.F.; Zou, P. Label-free and enzyme-free colorimetric detection of microRNA by catalyzed hairpin assembly coupled with hybridization chain reaction. Biosens. Bioelectron. 2016, 81, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Y.; Chai, Y.Q.; Yuan, R.; Su, H.L.; Han, J. A novel label-free electrochemical microRNA biosensor using Pd nanoparticles as enhancer and linker. Analyst 2013, 138, 1060–1066. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Electrochemical genosensors for the detection of cancer-related miRNAs. Anal. Bioanal. Chem. 2014, 406, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiang, Y.; Yuan, R.; Chai, Y.Q. Intercalation of quantum dots as the new signal acquisition and amplification platform for sensitive electrochemiluminescent detection of microRNA. Anal. Chim. Acta 2015, 891, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Deng, H.; Shen, W.; Ren, Y. A label-free biosensor for electrochemical detection of femtomolar miRNAs. Anal. Chem. 2013, 85, 1624–1630. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, P.J.; Nix, B.; Tsourkas, A.; Bao, G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004, 32, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.R.; Lee, J.; Yeo, J.; Na, H.K.; Kim, Y.K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N.; et al. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 2013, 7, 5882–5891. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.C.; Liu, Y.Q.; Ye, B.C. One-Step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J. Am. Chem. Soc. 2012, 134, 5064–5067. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, K.; Zhu, X.; Xie, M.H. A one-pot strategy for the sensitive detection of miRNA by catalyst–oligomer-mediated enzymatic amplification-based fluorescence biosensor. Sens. Actuators B Chem. 2016, 223, 586–590. [Google Scholar] [CrossRef]
- Liu, H.S.; Ma, C.B.; Zhou, M.J.; Chen, H.C.; He, H.L.; Wang, K.M. Quencher-free fluorescence strategy for detection of DNA methyltransferase activity based on exonuclease III-assisted signal amplification. Anal. Bioanal. Chem. 2016, 408, 8111–8116. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.C.; Neely, R.K. 2-Aminopurine as a fluorescent probe of DNA conformation and the DNA-enzyme interface. Rev. Biophys. 2015, 48, 244–279. [Google Scholar] [CrossRef] [PubMed]
- Holz, B.; Klimasauskas, S.; Serva1, S.; Weinhold, E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998, 26, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Somsen, J.G.; Hoek, V.A.; Amerongen, V.H. Fluorescence quenching of 2-aminopurine in dinucleotides. Chem. Phys. Lett. 2005, 402, 61–65. [Google Scholar] [CrossRef]
- Zhu, G.; Liang, L.; Zhang, C. Quencher-free fluorescent method for homogeneously sensitive detection of microRNAs in human lung tissues. Anal. Chem. 2014, 86, 11410–11416. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Mutharasan, R. Biosensor-based microRNA detection: Techniques, design, performance, and challenges. Analyst 2014, 139, 1576–1588. [Google Scholar] [CrossRef] [PubMed]
- Hunt, E.; Broyles, D.; Head, T.; Deo, S. MicroRNA Detection: Current Technology and Research Strategies. Annu. Rev. Anal. Chem. 2015, 8, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Liao, R.; He, K.; Chen, C.Y.; Chen, X.M.; Cai, C.Q. Double-strand displacement biosensor and quencher-free fluorescence strategy for rapid detection of microRNA. Anal. Chem. 2016, 88, 4254–4258. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.L.; Huang, K.J.; Chen, Y.X.; Fang, L.X.; Jia, M.P. Au nanoparticles/hollow molybdenum disulfide microcubes based biosensor for microRNA-21 detection coupled with duplex-specific nuclease and enzyme signal amplification. Biosens. Bioelectron. 2017, 89, 989–997. [Google Scholar] [CrossRef] [PubMed]

| Sample | Initial Concentration of miRNA-21 (nM) | Concentration of Recovered miRNA-21 (nM) | Recovery (%) |
|---|---|---|---|
| 1 | 50 | 44.5 | 89 |
| 2 | 100 | 104.4 | 104.4 |
| 3 | 200 | 198.8 | 99.4 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Liu, H.; Wu, K.; Chen, M.; Zheng, L.; Wang, J. An Exonuclease I-Based Quencher-Free Fluorescent Method Using DNA Hairpin Probes for Rapid Detection of MicroRNA. Sensors 2017, 17, 760. https://doi.org/10.3390/s17040760
Ma C, Liu H, Wu K, Chen M, Zheng L, Wang J. An Exonuclease I-Based Quencher-Free Fluorescent Method Using DNA Hairpin Probes for Rapid Detection of MicroRNA. Sensors. 2017; 17(4):760. https://doi.org/10.3390/s17040760
Chicago/Turabian StyleMa, Changbei, Haisheng Liu, Kefeng Wu, Mingjian Chen, Liyang Zheng, and Jun Wang. 2017. "An Exonuclease I-Based Quencher-Free Fluorescent Method Using DNA Hairpin Probes for Rapid Detection of MicroRNA" Sensors 17, no. 4: 760. https://doi.org/10.3390/s17040760




