Parallel Detection of Refractive Index Changes in a Porous Silicon Microarray Based on Digital Images
Abstract
:1. Introduction
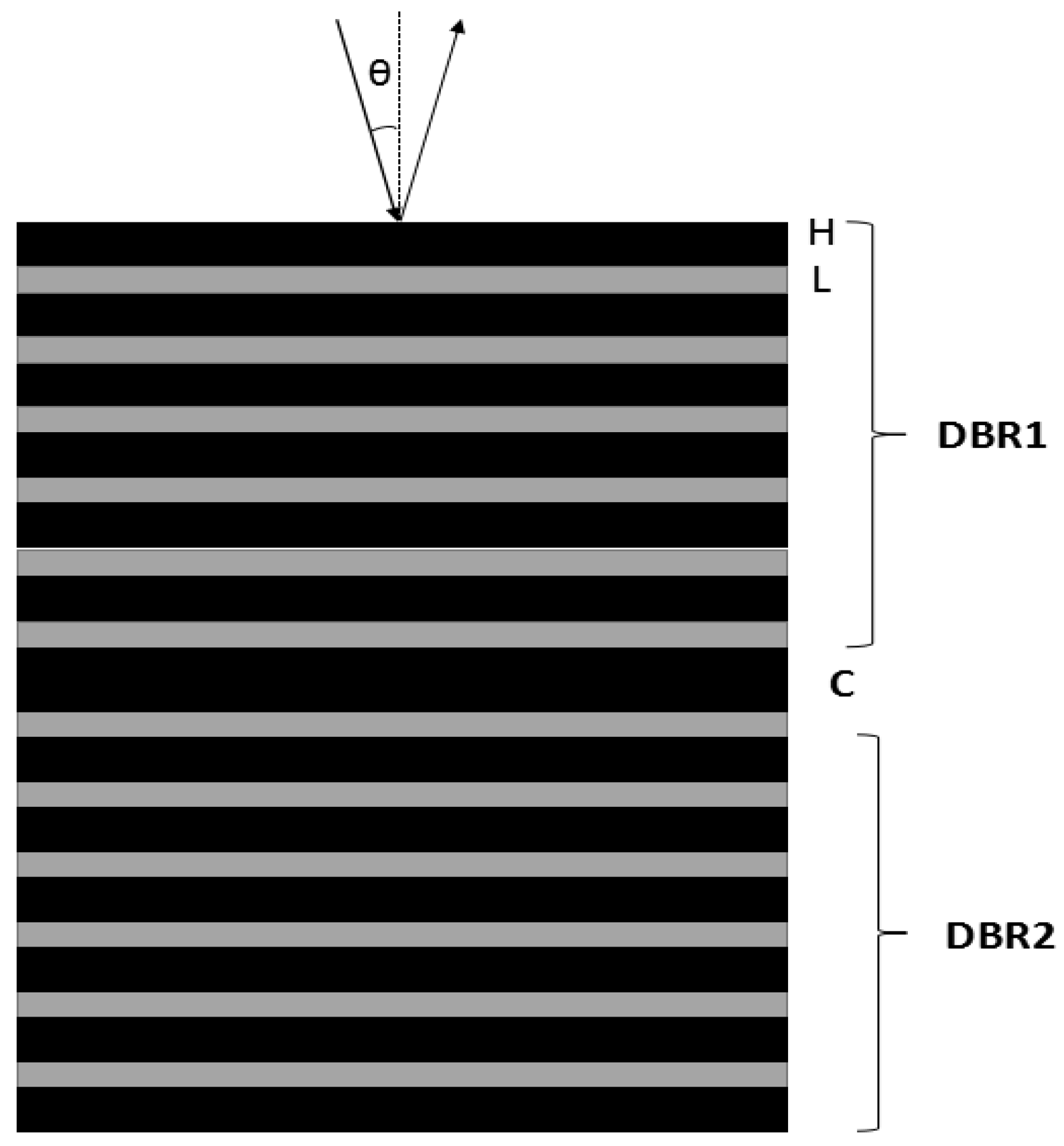
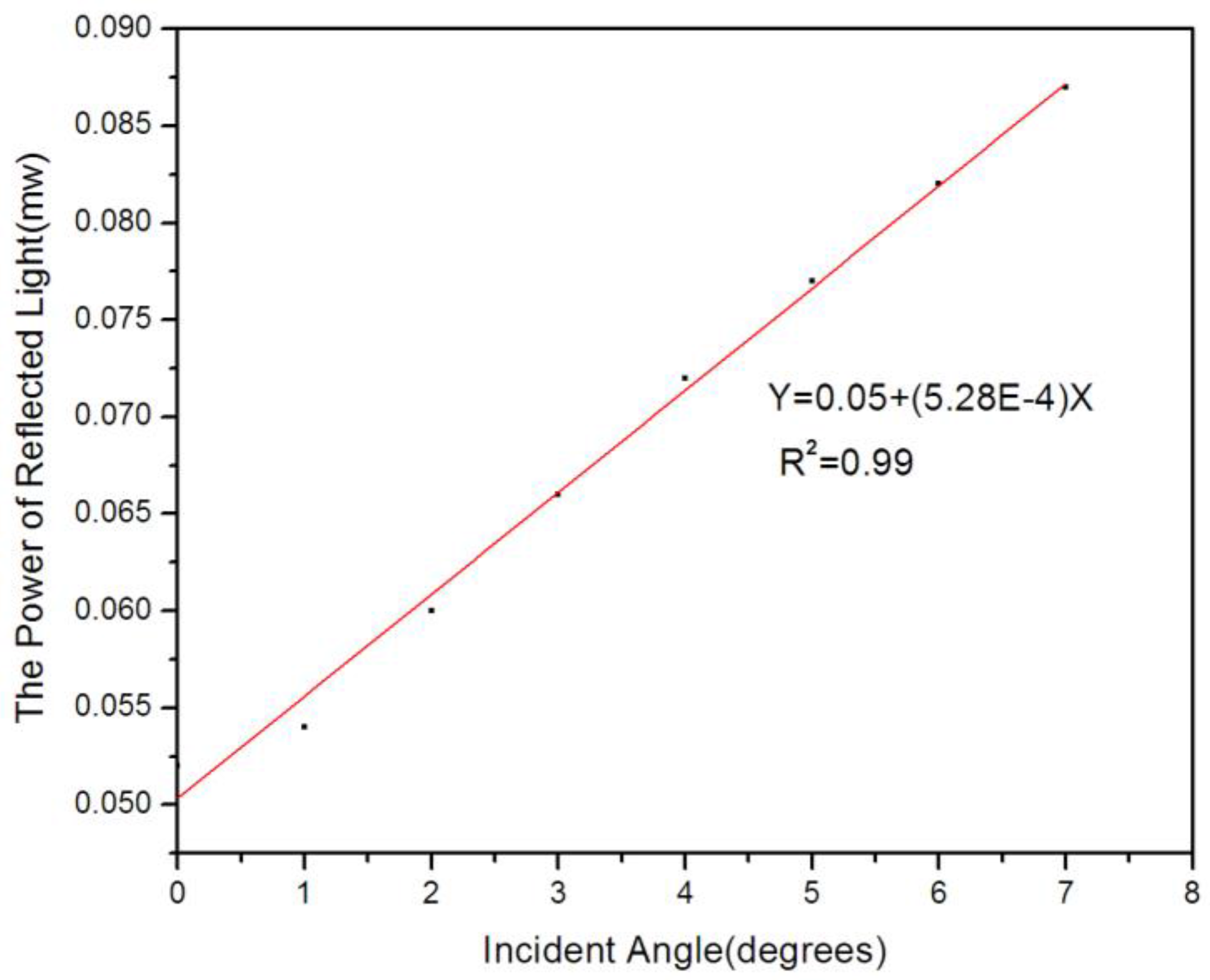
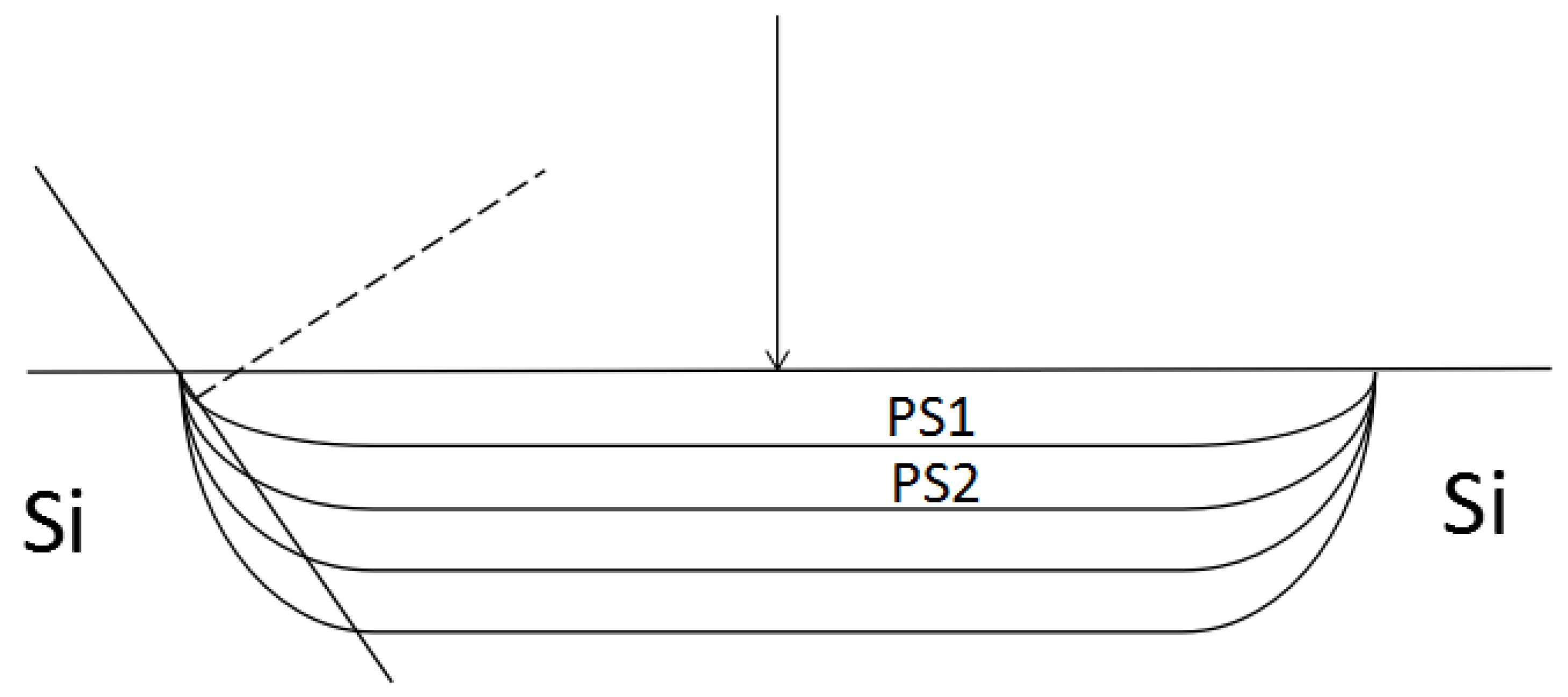
2. Measuring Principle
3. Experiment
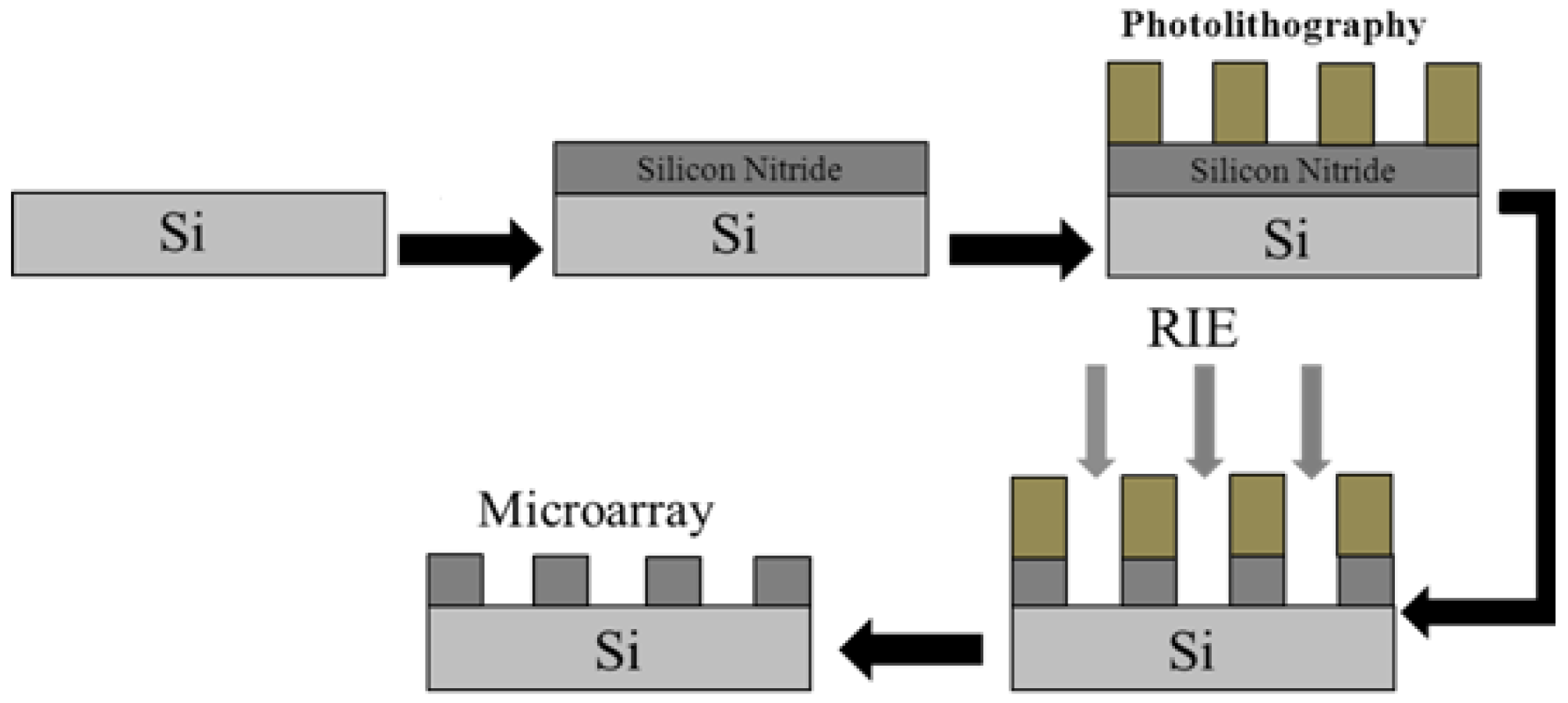
3.1. Fabrication of PSM
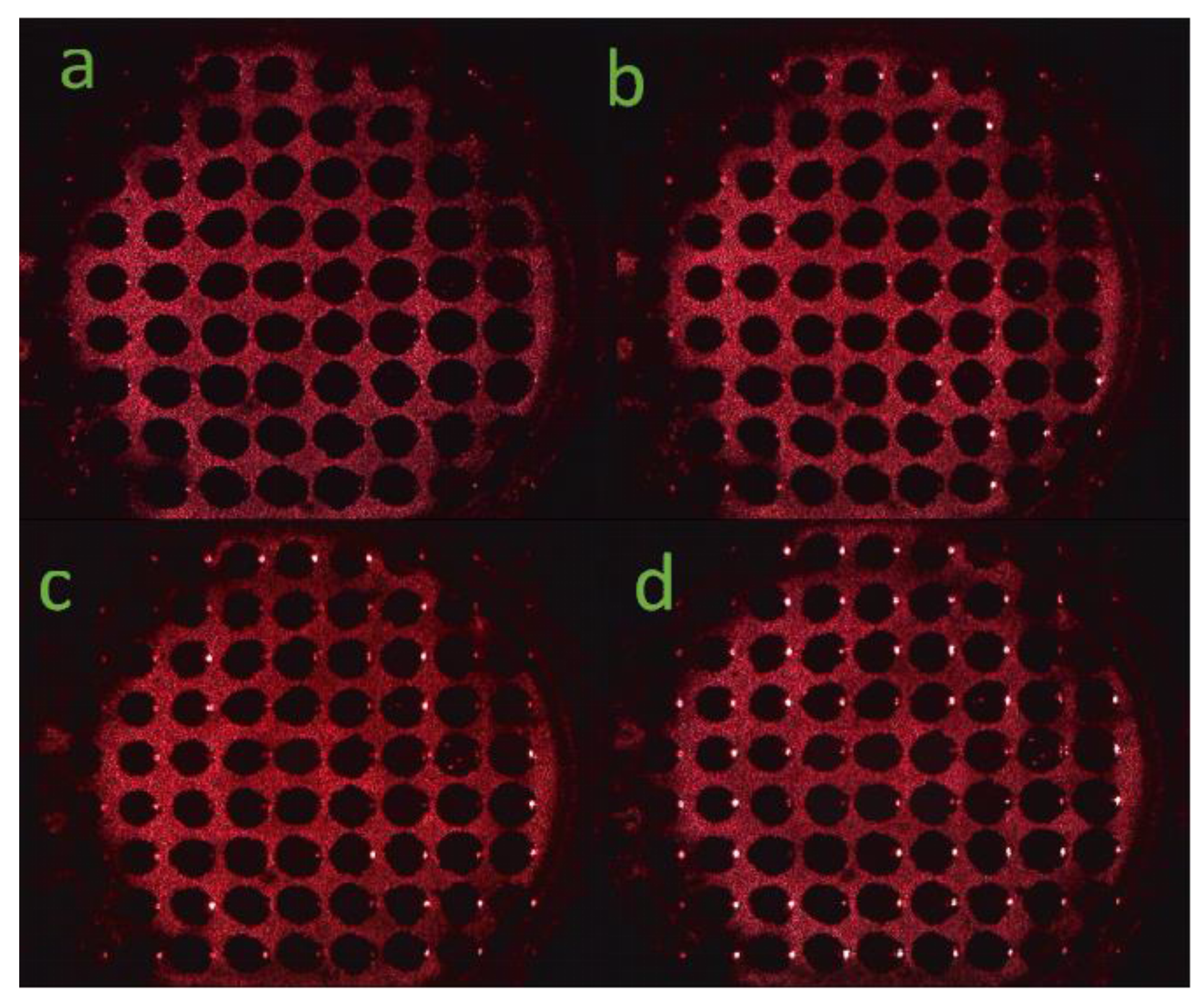
3.2. Detection of the PSM Microarray
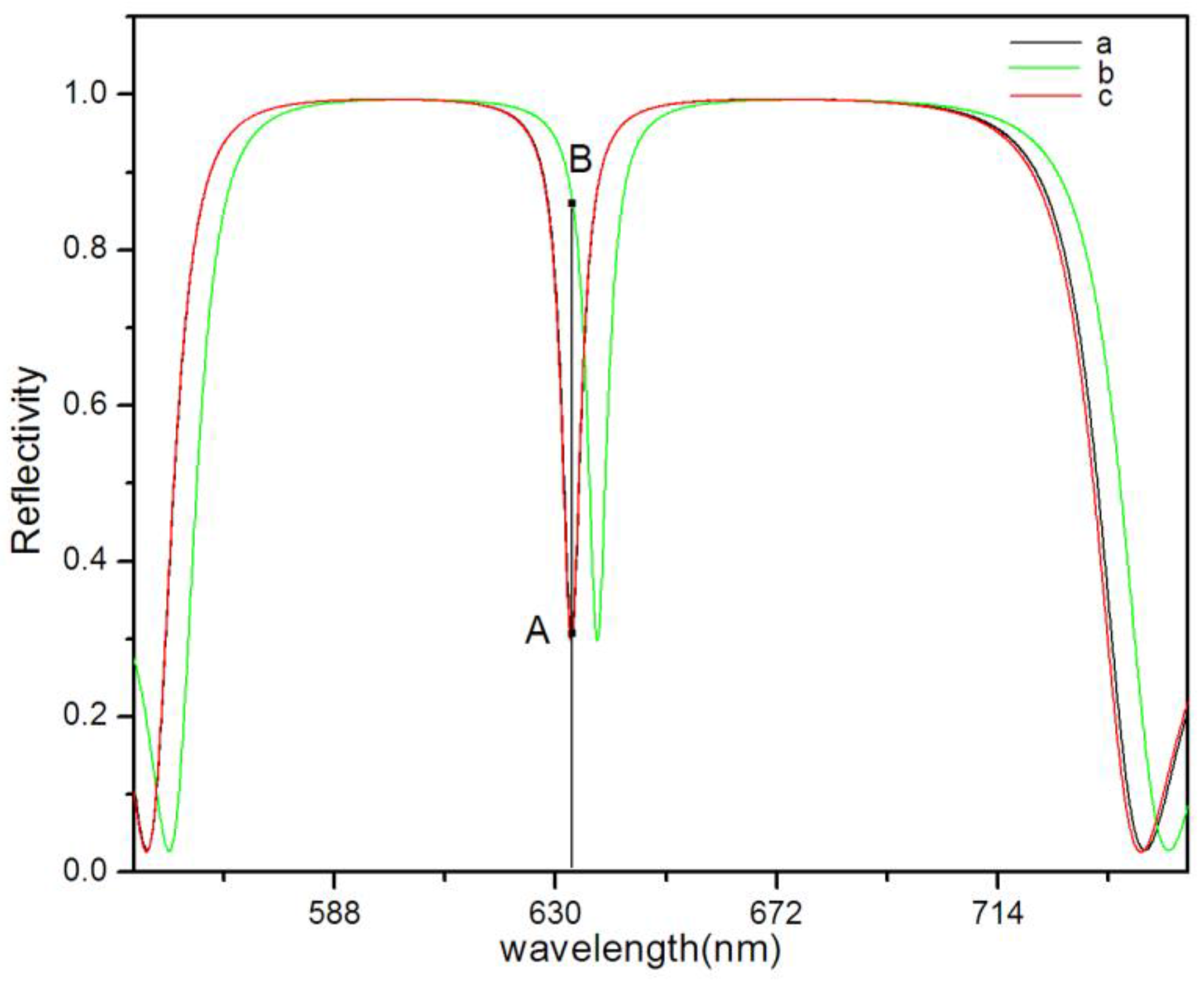
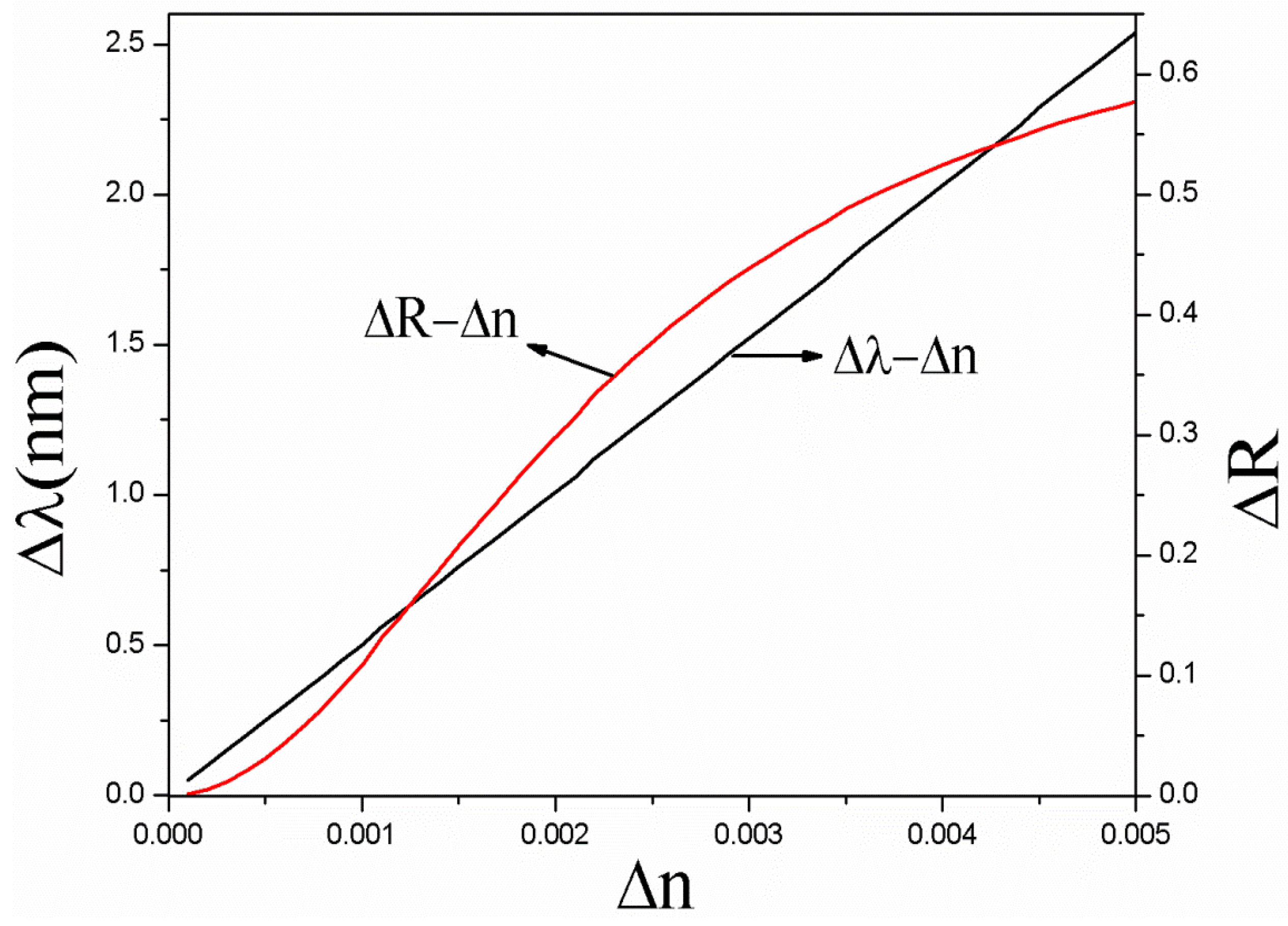
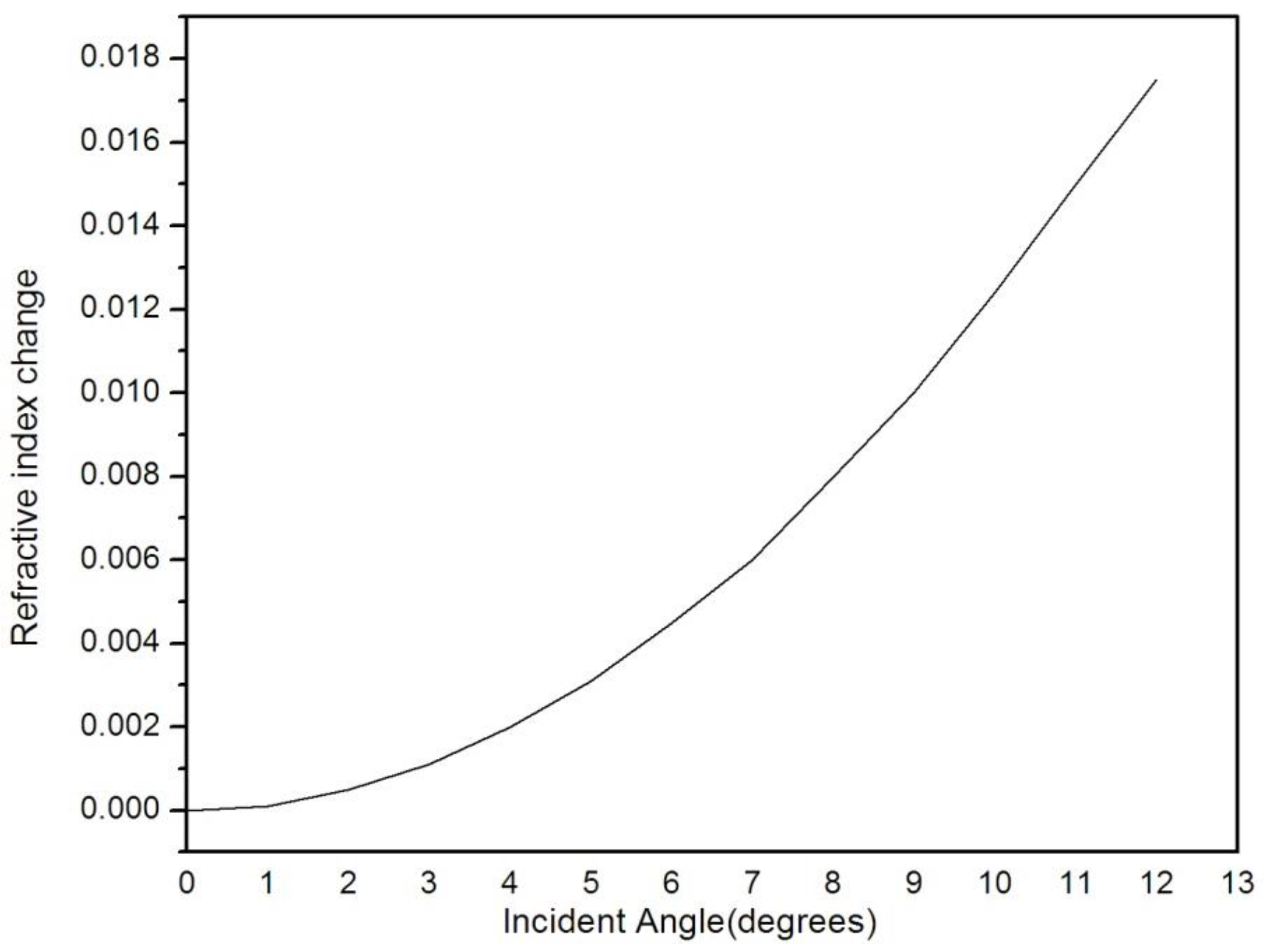
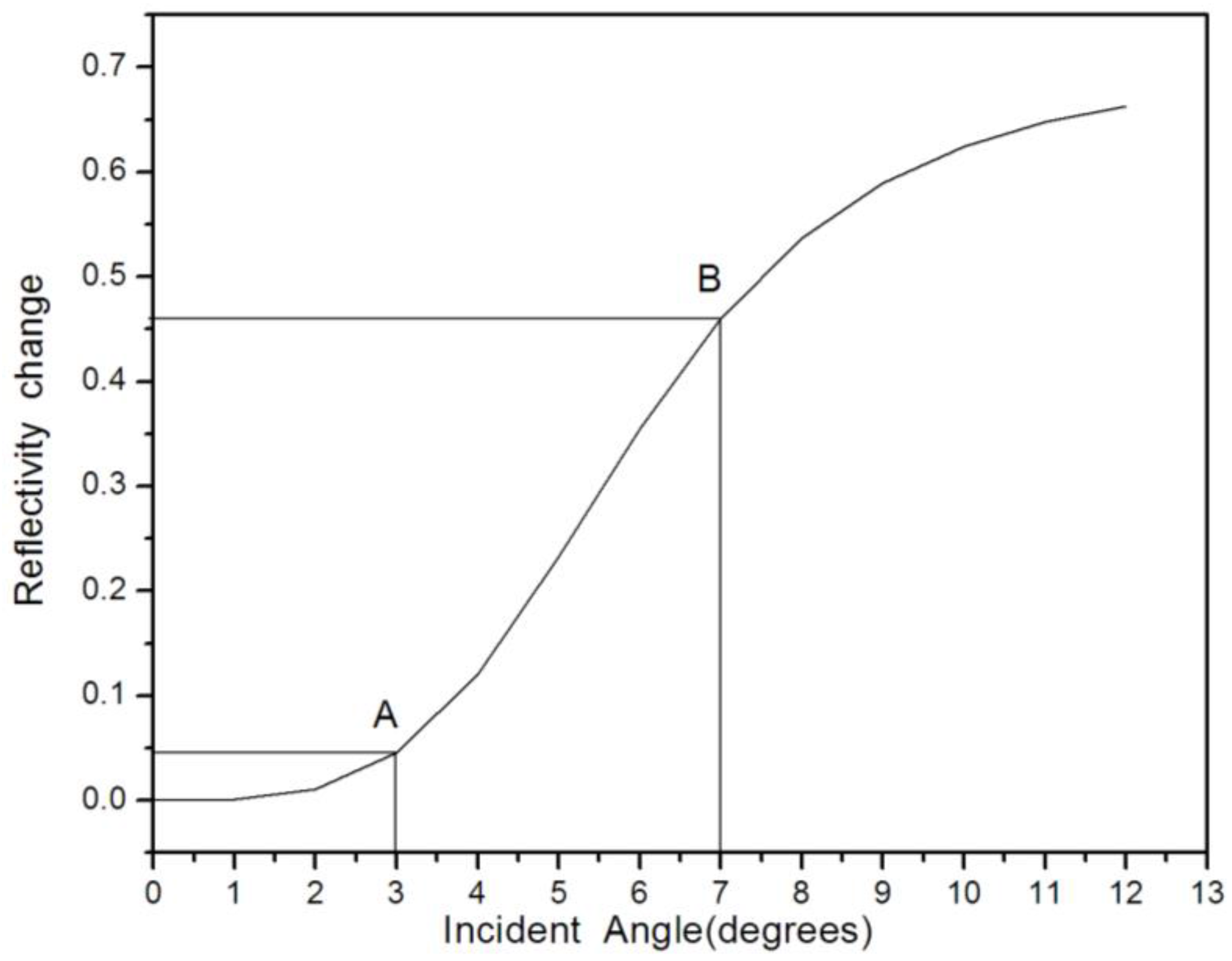
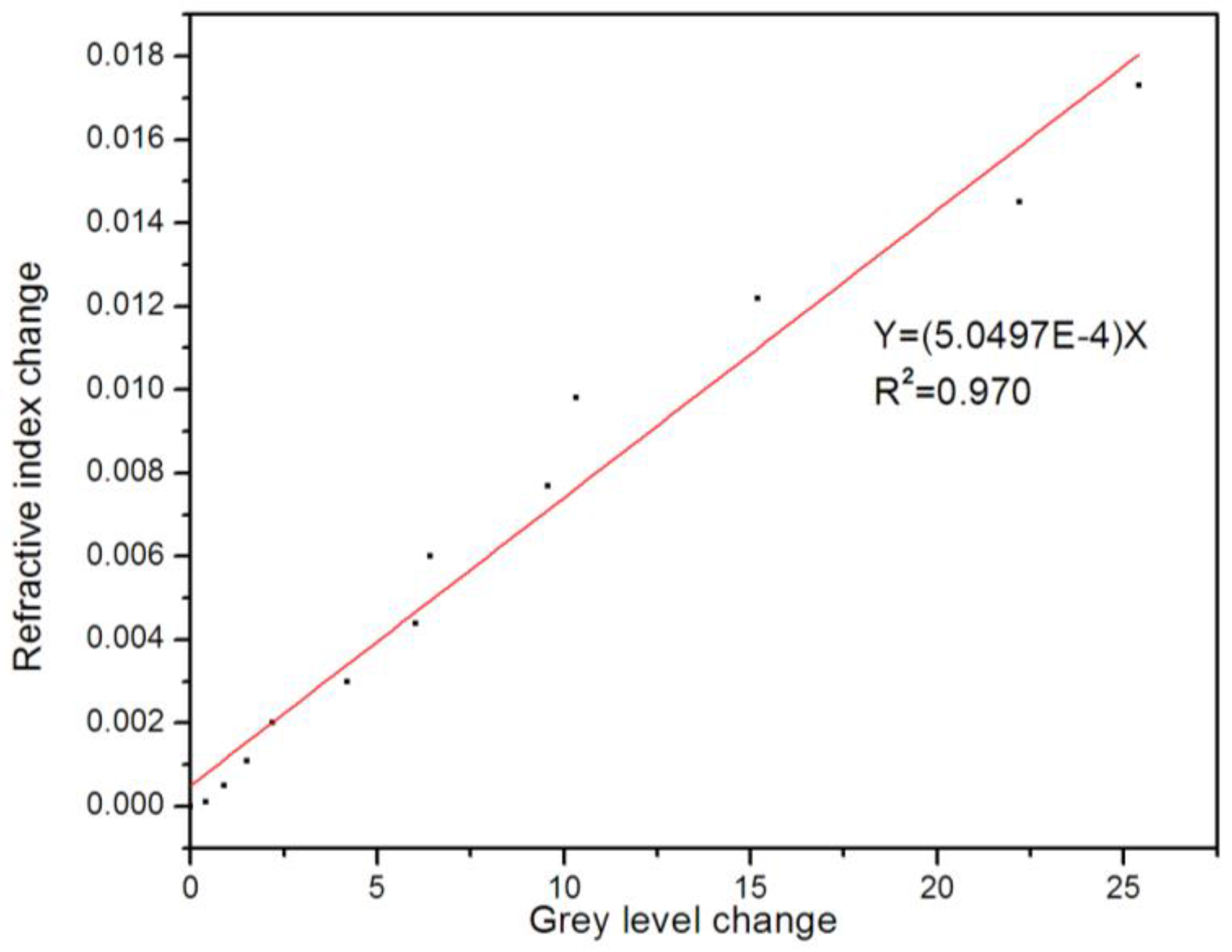
4. Results and Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, C.; Jia, X.-M.; Jiang, C.; Zhuang, G.-N.; Yan, Q.; Xiao, S.-J. DNA microarray fabricated on poly(acrylic acid) brushes-coated porous silicon by in situ rolling circle amplification. Analyst 2012, 137, 4539–4545. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, P.C.H. Microfluidic DNA microarray analysis: A review. Anal. Chim. Acta 2011, 687, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Karakach, T.K.; Flight, R.M.; Douglas, S.E.; Wentzell, P.D. An introduction to DNA microarrays for gene expression analysis. Chemom. Intell. Lab. Syst. 2010, 104, 28–52. [Google Scholar] [CrossRef]
- Manzano, M.; Cecchini, F.; Fontanot, M.; Iacumin, L.; Comi, G.; Melpignano, P. OLED-based DNA biochip for Campylobacter spp. detection in poultry meat samples. Biosens. Bioelectron. 2015, 66, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.Z.; Zhao, C.; Melbourne, W. Biochip immunofluorescence microscopy as a new diagnostic tool for autoimmune blistering skin diseases. Br. J. Dermatol. 2015, 173, 7–8. [Google Scholar]
- Zhou, Z.; Xu, L.; Wu, S.; Su, B. A novel biosensor array with a wheel-like pattern for glucose, lactate and choline based on electrochemiluminescence imaging. Analyst 2014, 139, 4934–4939. [Google Scholar] [CrossRef] [PubMed]
- Tavares, A.J.; Noor, O.; Krull, U.J. Label-free detection of nucleic acid hybridization in an electrokinetically controlled biochip using quantum dots as donors in fluorescence resonance energy transfer. In Proceedings of the 248th National Meeting of the American-Chemical-Society, San Francisco, CA, USA, 10–14 August 2014. [Google Scholar]
- Yeh, C.-H.; Chang, Y.-H.; Lin, H.-P. A newly developed optical biochip for bacteria detection hybridization. Sens. Actuators B Chem. 2012, 161, 1168–1175. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, L.; Wang, X.; Shi, H.; Chong, X.; Ma, S.; Ji, Y.; Guo, J.; Ma, H.; He, Y. Polarization Interferometry Based Wavelength-Interrogation Surface Plasmon Resonance Imager for Analysis of Microarrays. J. Biomed. Opt. 2012, 17, 036002. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, X.; Zhao, M.; Qi, P.; Zhong, J. Quick and Label-Free Detection for Coumaphos by Using Surface Plasmon Resonance Biochip. PLoS ONE 2014, 9, e104689. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, S.; Dai, J.; Lü, H.; Jin, K.; Yang, G. Label-free high-throughput and real-time detections of protein interactions by oblique-incidence reflectivity difference method. Sci. China Phys. Mech. Astron. 2014, 57, 615–618. [Google Scholar] [CrossRef]
- Sharma, P.; Sharan, P. An Analysis and Design of Photonic Crystal-Based Biochip for Detection of Glycosuria. IEEE Sens. J. 2015, 15, 5569–5575. [Google Scholar] [CrossRef]
- Zhao, Z.; Hui, M.; Liu, M.; Dong, L.; Liu, X.; Zhao, Y. Centroid shift analysis of microlens array detector in interference imaging system. Opt. Commun. 2015, 354, 132–139. [Google Scholar] [CrossRef]
- Shang, Y.; Zhao, W.; Xu, E.; Tong, C.; Wu, J. FTRIFS biosensor based on double layer porous silicon as a LC detector for target molecule screening from complex samples. Biosens. Bioelectron. 2010, 25, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.; Lamberti, A.; Rendina, I.; Coppola, G.; Gioffrè, M.; Iodice, M.; Casalino, M.; De Tommasi, E.; De Stefano, L. Fabrication and characterization of a porous silicon based microarray for label-free optical monitoring of biomolecular interactions. J. Appl. Phys. 2010, 107, 014513. [Google Scholar] [CrossRef]
- Syshchyk, O.; Skryshevsky, V.A.; Soldatkin, O.O.; Soldatkin, A.P. Enzyme biosensor systems based on porous silicon photoluminescence for detection of glucose, urea and heavy metals. Biosens. Bioelectron. 2015, 66, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Han, H.; Xiao, S. High hydrosilylation efficiency of porous silicon SiHx species produced by Pt-assisted chemical etching for biochip fabrication. Sci. China Chem. 2013, 56, 1152–1163. [Google Scholar] [CrossRef]
- Pei, J.; Tang, Y.; Xu, N.; Lu, W.; Xiao, S.; Liu, J. Covalently derivatized NTA microarrays on porous silicon for multi-mode detection of His-tagged proteins. Sci. China Chem. 2011, 54, 526–535. [Google Scholar] [CrossRef]
- Chen, W.; Jia, Z.; Li, P.; Lv, G.; Lv, X. Refractive index change detection based on porous silicon microarray. Appl. Phys. B 2016, 122, 120. [Google Scholar] [CrossRef]
- Pavesi, L. Porous silicon dielectric multilayers and microcavities. La Rivista del Nuovo Cimento 1997, 20, 1–76. [Google Scholar] [CrossRef]
- Stefano, L.D.; Moretti, L.; Rendina, I.; Rossi, A.M. Porous silicon microcavities for optical hydrocarbons detection. Sens. Actuators A Phys. 2003, 104, 179–182. [Google Scholar] [CrossRef]
- Stefano, L.D.; Rendina, I.; Moretti, L.; Rossi, A.M. Optical sensing of flammable substances using porous silicon microcavities. Mater. Sci. Eng. B 2003, 100, 271–274. [Google Scholar] [CrossRef]
- Li, H.F.; Han, H.M.; Wu, Y.G.; Xiao, S.J. Biological functionalization and patterning of porous silicon prepared by Pt-assisted chemical etching. Appl. Surf. Sci. 2010, 256, 4048–4051. [Google Scholar] [CrossRef]






© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Jia, Z.; Li, P.; Wen, H.; Lv, G.; Huang, X. Parallel Detection of Refractive Index Changes in a Porous Silicon Microarray Based on Digital Images. Sensors 2017, 17, 750. https://doi.org/10.3390/s17040750
Li C, Jia Z, Li P, Wen H, Lv G, Huang X. Parallel Detection of Refractive Index Changes in a Porous Silicon Microarray Based on Digital Images. Sensors. 2017; 17(4):750. https://doi.org/10.3390/s17040750
Chicago/Turabian StyleLi, Chuanxi, Zhenhong Jia, Peng Li, Hao Wen, Guodong Lv, and Xiaohui Huang. 2017. "Parallel Detection of Refractive Index Changes in a Porous Silicon Microarray Based on Digital Images" Sensors 17, no. 4: 750. https://doi.org/10.3390/s17040750





