Selectivity/Specificity Improvement Strategies in Surface-Enhanced Raman Spectroscopy Analysis
Abstract
:1. Introduction
2. Reaction-SERS Method
2.1. Improving Analyte Affinity with SERS Substrate
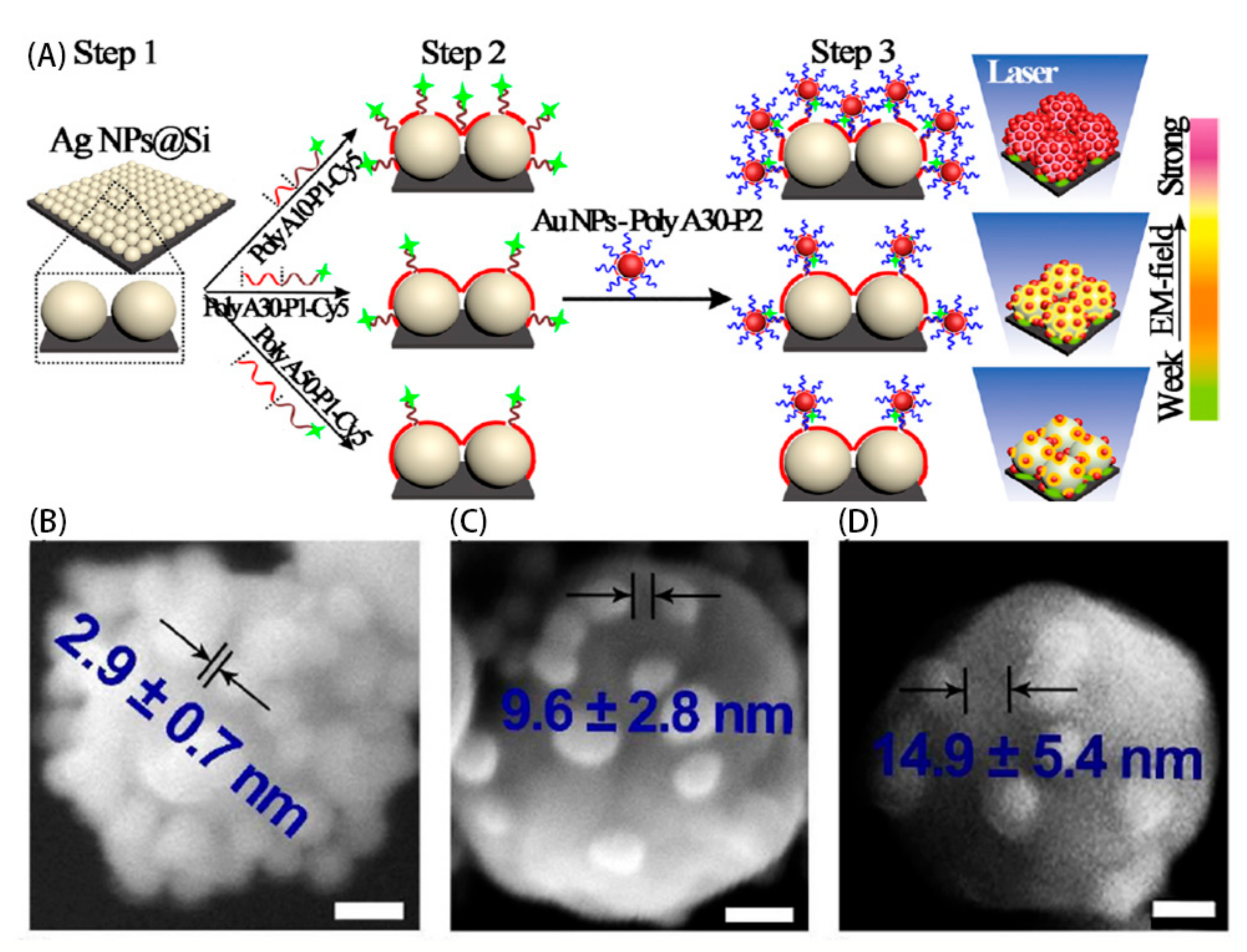
2.2. Increasing the Raman Scattering Cross-Sectional Area
2.3. Reducing the Raman Scattering Cross-Sectional Area
2.4. Application of Reaction-SERS Method
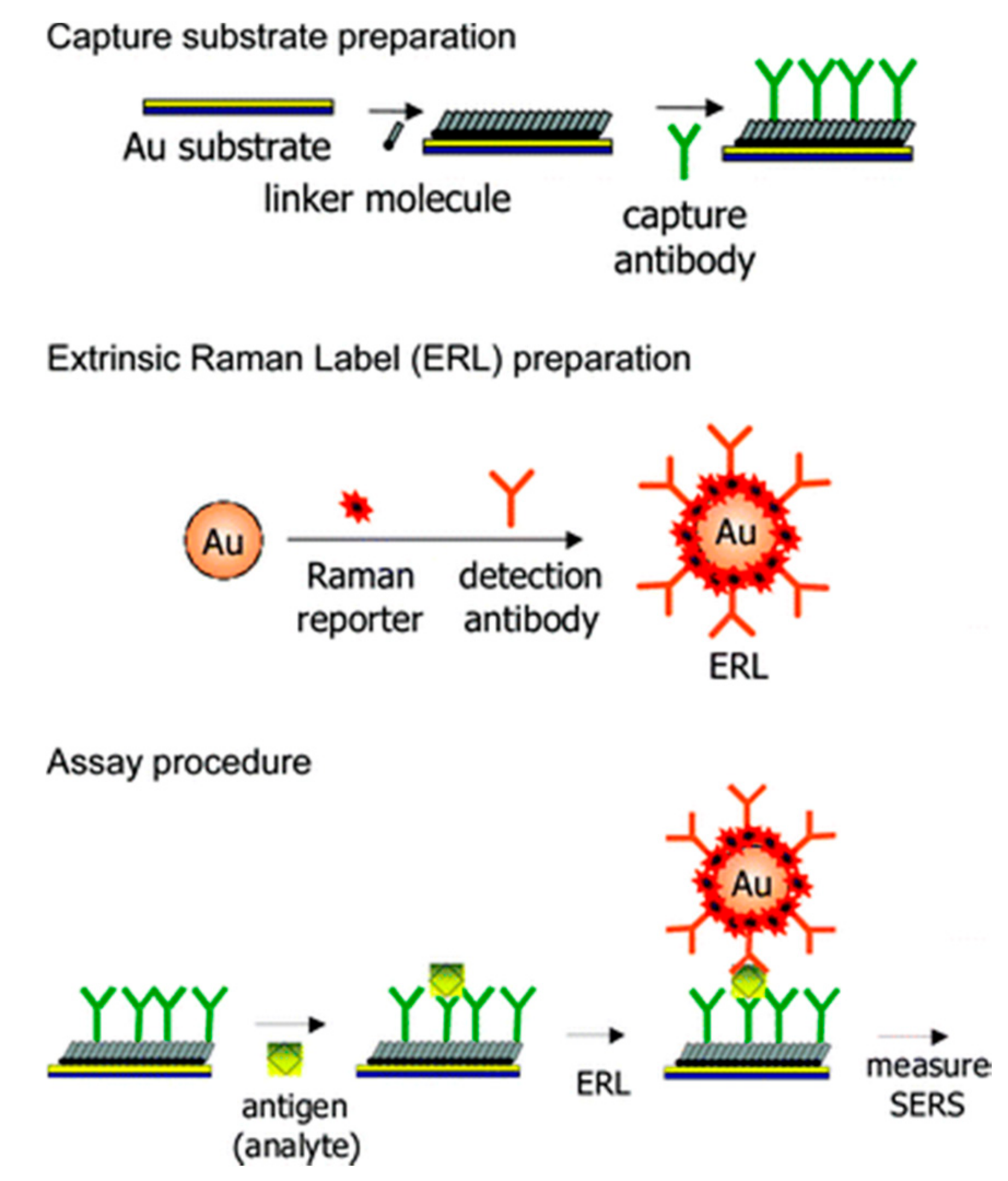
3. Antibody-SERS
4. Aptamer-SERS
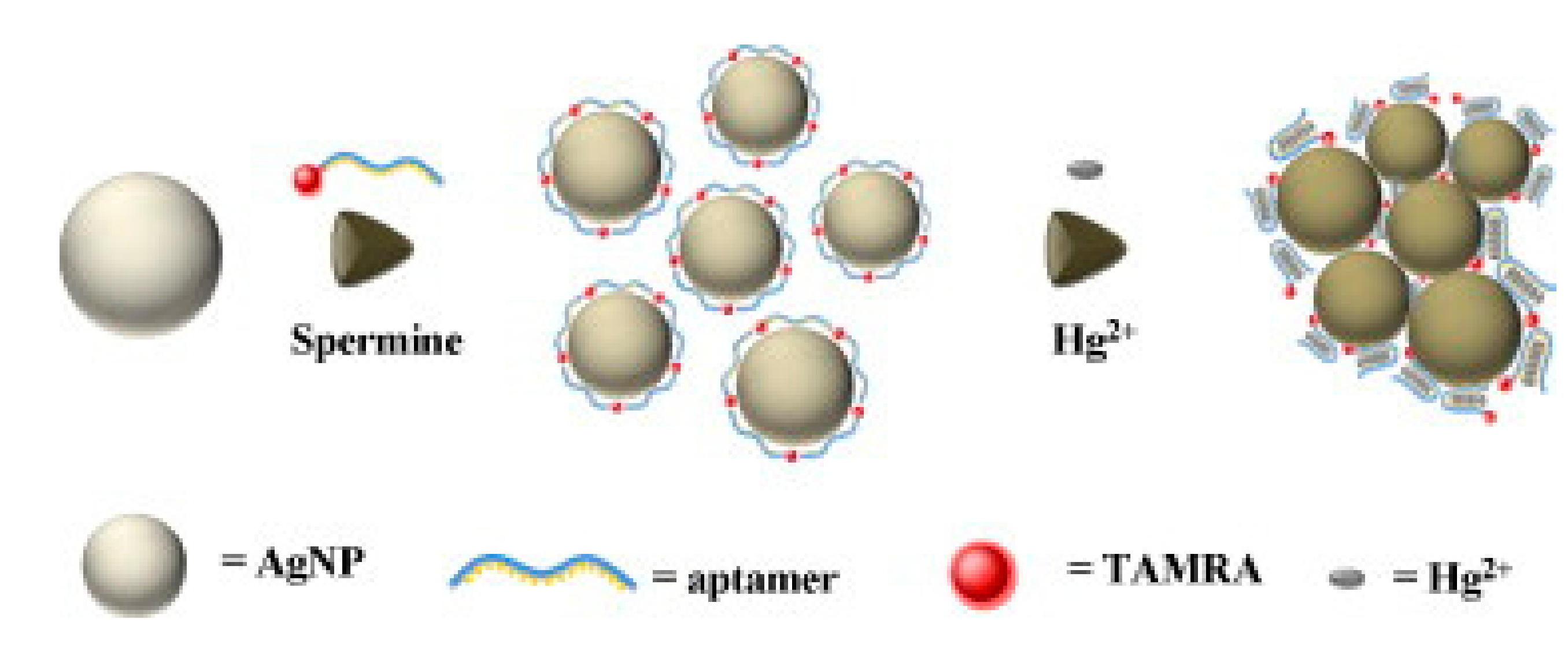
4.1. Metal Ion
4.2. Small Molecule
4.3. Biomacromolecules
4.4. Living Organisms
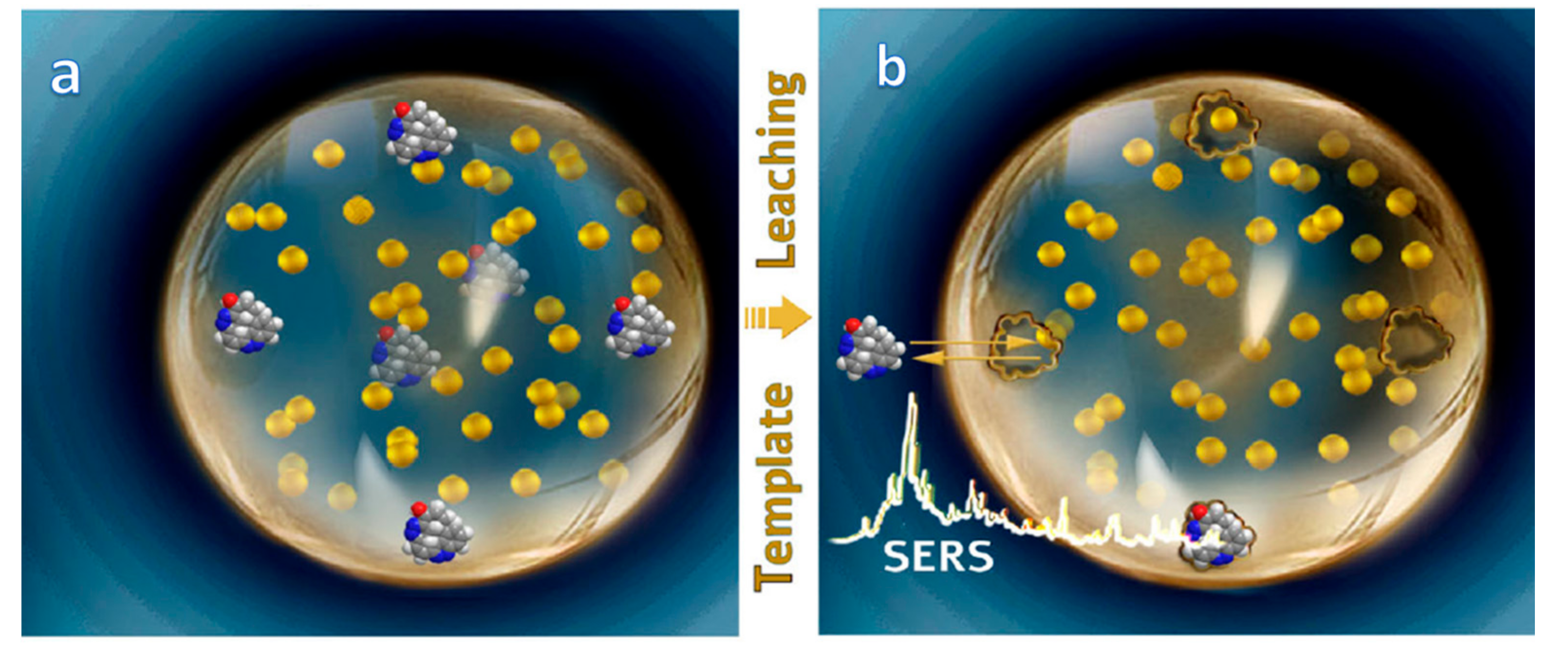
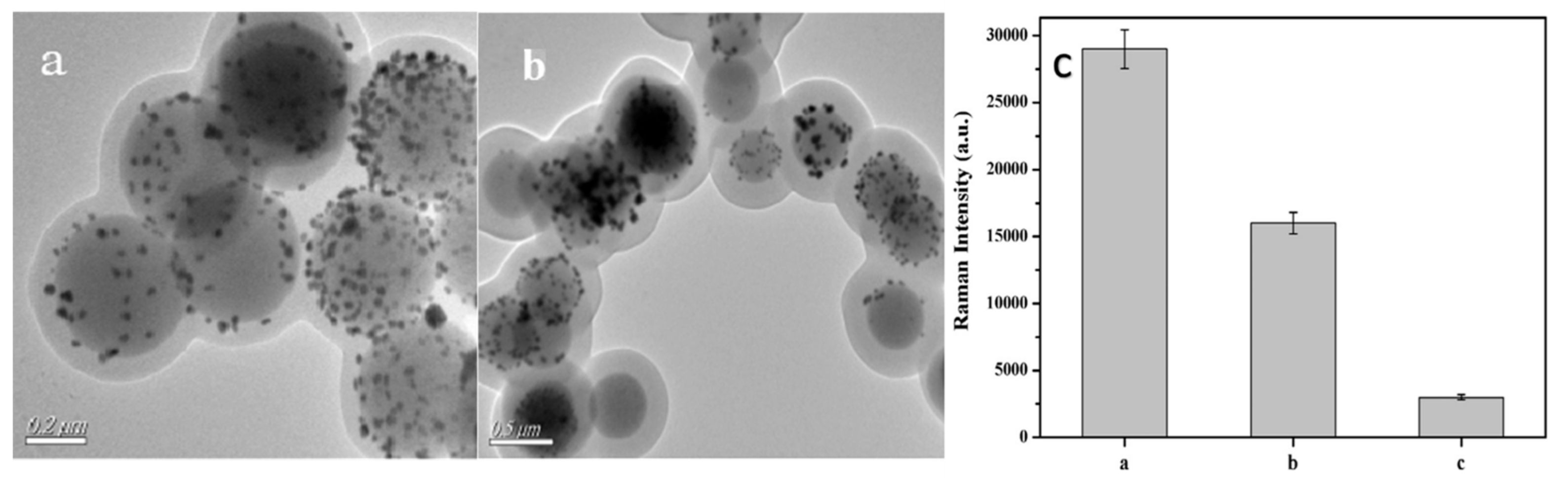
5. MIPs-SERS
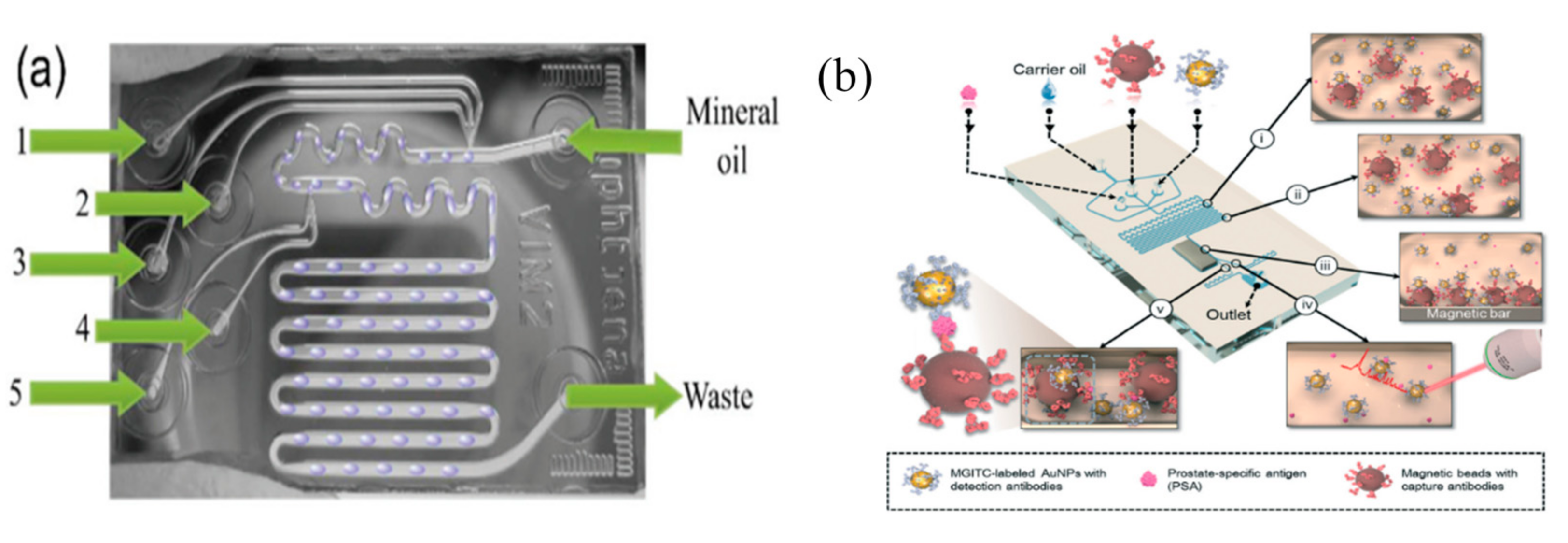
6. Microfluidics-SERS
7. Final Statements
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liao, W.; Lu, X.N. Determination of chemical hazards in foods using surface-enhanced raman spectroscopy coupled with advanced separation techniques. Trends Food Sci. Technol. 2016, 54, 103–113. [Google Scholar] [CrossRef]
- Schlucker, S. Surface-enhanced raman spectroscopy: Concepts and chemical applications. Angew. Chem. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface raman spectroelectrochemistry. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Nie, S. Probing single molecules and single nanoparticles by surface-enhanced raman scattering. Science 1997, 275, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Camden, J.P. Surface-enhanced raman spectroscopy-based approach for ultrasensitive and selective detection of hydrazine. Anal. Chem. 2015, 87, 6460–6464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhan, Y.; Huang, Y.; Li, G. Large-volume constant-concentration sampling technique coupling with surface-enhanced raman spectroscopy for rapid on-site gas analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 183, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Z.; Wang, X.; Jiang, J.; Xu, Y.; Liu, X.; Yan, Y.; Li, C. Preparation of a self-cleanable molecularly imprinted sensor based on surface-enhanced raman spectroscopy for selective detection of r6g. Anal. Bioanal. Chem. 2017, 409, 4627–4635. [Google Scholar] [CrossRef] [PubMed]
- El-Maghrabey, M.; Kishikawa, N.; Kuroda, N. 9,10-phenanthrenequinone as a mass-tagging reagent for ultra-sensitive liquid chromatography-tandem mass spectrometry assay of aliphatic aldehydes in human serum. J. Chromatogr. A 2016, 1462, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Jiang, K.; Liu, D.Q.; Facchine, K.L. Simultaneous quantitation of trace level hydrazine and acetohydrazide in pharmaceuticals by benzaldehyde derivatization with sample ‘matrix matching’ followed by liquid chromatography-mass spectrometry. J. Chromatogr. A 2016, 1462, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Vanhees, I.; Van den Bergh, V.; Schildermans, R.; De Boer, R.; Compernolle, F.; Vinckier, C. Determination of the oxidation products of the reaction between α-pinene and hydroxyl radicals by high-performance liquid chromatography. J. Chromatogr. A 2001, 915, 75–83. [Google Scholar] [CrossRef]
- Bunkoed, O.; Thavarungkul, P.; Thammakhet, C.; Kanatharana, P. A simple and high collection efficiency sampling method for monitoring of carbonyl compounds in a workplace environment. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2012, 47, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Brandao, P.F.; Ramos, R.M.; Almeida, P.J.; Rodrigues, J.A. Determination of carbonyl compounds in cork agglomerates by gdme-hplc-uv: Identification of the extracted compounds by hplc-ms/ms. J. Agric. Food Chem. 2017, 65, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, H.; Luo, J.; Li, L.; Zhao, R.; Zhang, R.; Liu, G. New method for hplc separation and fluorescence detection of malonaldehyde in normal human plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2006, 832, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Giera, M.; Kloos, D.P.; Raaphorst, A.; Mayboroda, O.A.; Deelder, A.M.; Lingeman, H.; Niessen, W.M. Mild and selective labeling of malondialdehyde with 2-aminoacridone: Assessment of urinary malondialdehyde levels. Analyst 2011, 136, 2763–2769. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-M.; Huang, Y.-Q.; Liu, Z.; Lin, C.-Q.; Ma, X.; Liu, J.-M. Design of an ultra-sensitive gold nanorod colorimetric sensor and its application based on formaldehyde reducing Ag+. RSC Adv. 2015, 5, 99944–99950. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, Z.; Zheng, C.; Lee, Y.I.; Hou, X.; Wu, L.; Tian, Y. Derivatization reaction-based surface-enhanced raman scattering (SERS) for detection of trace acetone. Talanta 2016, 155, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Liang, F.; Wang, D.; Yang, Q.; Ding, Y.; Yu, Y.; Gao, D.; Song, D.; Wang, X. Ultrasensitive determination of formaldehyde in environmental waters and food samples after derivatization and using silver nanoparticle assisted SERS. Microchim. Acta 2014, 182, 863–869. [Google Scholar] [CrossRef]
- Oklejas, V.; Uibel, R.H.; Horton, R.; Harris, J.M. Electric-field control of the tautomerization and metal ion binding reactivity of 8-hydroxyquinoline immobilized to an electrode surface. Anal. Chem. 2008, 80, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhao, C.; Ma, Y.; Li, G. Rapid analysis of trace volatile formaldehyde in aquatic products by derivatization reaction-based surface enhanced raman spectroscopy. Analyst 2014, 139, 3614–3621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Haputhanthri, R.; Ansar, S.M.; Vangala, K.; De Silva, H.I.; Sygula, A.; Saebo, S.; Pittman, C.U., Jr. Ultrasensitive detection of malondialdehyde with surface-enhanced raman spectroscopy. Anal. Bioanal. Chem. 2010, 398, 3193–3201. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Promthaveepong, K.; Li, N. Chemical sensing on a single SERS particle. ACS Sens. 2017, 2, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, W.; Wang, J.; Luo, D.; Qiao, X.; Qin, X.; Wang, T. Ultrasensitive surface-enhanced raman scattering sensor of gaseous aldehydes as biomarkers of lung cancer on dendritic ag nanocrystals. Anal. Chem. 2017, 89, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Weatherston, J.D.; Worstell, N.C.; Wu, H.J. Quantitative surface-enhanced raman spectroscopy for kinetic analysis of aldol condensation using ag-au core-shell nanocubes. Analyst 2016, 141, 6051–6060. [Google Scholar] [CrossRef] [PubMed]
- Hardy, M.; Doherty, M.D.; Krstev, I.; Maier, K.; Moller, T.; Muller, G.; Dawson, P. Detection of low-concentration contaminants in solution by exploiting chemical derivatization in surface-enhanced raman spectroscopy. Anal. Chem. 2014, 86, 9006–9012. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.J.; Sun, J.J.; Song, H.; Tian, J.Z.; Li, D.W.; Long, Y.T. Real-time sensing of o-phenylenediamine oxidation on gold nanoparticles. Sensors 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Li, D.-W.; Cui, J.; Cao, Y.; Long, Y.-T. In situ monitoring of palladacycle-mediated carbonylation by surface-enhanced raman spectroscopy. RSC Adv. 2015, 5, 97734–97737. [Google Scholar] [CrossRef]
- Huang, Y.F.; Wu, D.Y.; Zhu, H.P.; Zhao, L.B.; Liu, G.K.; Ren, B.; Tian, Z.Q. Surface-enhanced raman spectroscopic study of p-aminothiophenol. Phys. Chem. Chem. Phys. PCCP 2012, 14, 8485–8497. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Guo, X.; Wu, Y.; Wang, H.; Fu, S.; Wen, Y.; Yang, H. Facile and label-free detection of lung cancer biomarker in urine by magnetically assisted surface-enhanced raman scattering. ACS Appl. Mater. Interfaces 2014, 6, 20985–20993. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, L.; Li, L.; Tian, Y. A single nanoprobe for ratiometric imaging and biosensing of hypochlorite and glutathione in live cells using surface-enhanced raman scattering. Anal. Chem. 2016, 88, 9518–9523. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Hu, K.; Sun, J.J.; Qu, L.L.; Li, D.W. Sers nanoprobes for the monitoring of endogenous nitric oxide in living cells. Biosens. Bioelectron. 2016, 85, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Hoebe, R.A.; Van Oven, C.H.; Gadella, T.W., Jr.; Dhonukshe, P.B.; Van Noorden, C.J.; Manders, E.M. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat. Biotechnol. 2007, 25, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Herrman, T.J.; Bisrat, Y.; Murray, S.C. Feasibility of surface-enhanced raman spectroscopy for rapid detection of aflatoxins in maize. J. Agric. Food Chem. 2014, 62, 4466–4474. [Google Scholar] [CrossRef] [PubMed]
- Bontempi, N.; Biavardi, E.; Bordiga, D.; Candiani, G.; Alessandri, I.; Bergese, P.; Dalcanale, E. Probing lysine mono-methylation in histone h3 tail peptides with an abiotic receptor coupled to a non-plasmonic resonator. Nanoscale 2017, 9, 8639–8646. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.D.; Lipert, R.J.; Siperko, L.M.; Wang, G.; Narayanan, R. Sers as a bioassay platform: Fundamentals, design, and applications. Chem. Soc. Rev. 2008, 37, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Rohr, T.E.; Cotton, T.; Fan, N.; Tarcha, P.J. Immunoassay employing surface-enhanced raman spectroscopy. Anal. Biochem. 1989, 182, 388–398. [Google Scholar] [CrossRef]
- Grubisha, D.S.; Lipert, R.J.; Park, H.Y.; Driskell, J.; Porter, M.D. Femtomolar detection of prostate-specific antigen: An immunoassay based on surface-enhanced raman scattering and immunogold labels. Anal. Chem. 2003, 75, 5936–5943. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.C.; Jin, R.; Nam, J.M.; Thaxton, C.S.; Mirkin, C.A. Raman dye-labeled nanoparticle probes for proteins. J. Am. Chem. Soc. 2003, 125, 14676–14677. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ji, X.; Xu, W.; Li, X.; Wang, L.; Bai, Y.; Zhao, B.; Ozaki, Y. Immunoassay using probe-labelling immunogold nanoparticles with silver staining enhancement via surface-enhanced raman scattering. Analyst 2004, 129, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cushing, S.K.; Zhang, J.; Lankford, J.; Aguilar, Z.P.; Ma, D.; Wu, N. Shape-dependent surface-enhanced raman scattering in gold-raman probe-silica sandwiched nanoparticles for biocompatible applications. Nanotechnology 2012, 23, 115501. [Google Scholar] [CrossRef] [PubMed]
- Driskell, J.D.; Kwarta, K.M.; Lipert, R.J.; Porter, M.D.; Neill, J.D.; Ridpath, J.F. Low-level detection of viral pathogens by a surface-enhanced raman scattering based immunoassay. Anal. Chem. 2005, 77, 6147–6154. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Xu, M.M.; Fang, C.W.; Jin, Q.; Yuan, Y.X.; Yao, J.L. Improving the sensitivity of immunoassay based on MBA-embedded Au@SiO2 nanoparticles and surface enhanced raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 175, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, P.; Song, I.; Cha, J.M.; Lee, S.H.; Kim, B.; Suh, K.Y. Guided three-dimensional growth of functional cardiomyocytes on polyethylene glycol nanostructures. Langmuir ACS J. Surfaces Colloids 2006, 22, 5419–5426. [Google Scholar] [CrossRef] [PubMed]
- Chuong, T.T.; Pallaoro, A.; Chaves, C.A.; Li, Z.; Lee, J.; Eisenstein, M.; Stucky, G.D.; Moskovits, M.; Soh, H.T. Dual-reporter SERS-based biomolecular assay with reduced false-positive signals. Pro. Natl. Acad. Sci. USA 2017, 114, 9056–9061. [Google Scholar] [CrossRef] [PubMed]
- Chon, H.; Lee, S.; Son, S.W.; Oh, C.H.; Choo, J. Highly sensitive immunoassay of lung cancer marker carcinoembryonic antigen using surface-enhanced raman scattering of hollow gold nanospheres. Anal. Chem. 2009, 81, 3029–3034. [Google Scholar] [CrossRef] [PubMed]
- Chon, H.; Lim, C.; Ha, S.M.; Ahn, Y.; Lee, E.K.; Chang, S.I.; Seong, G.H.; Choo, J. On-chip immunoassay using surface-enhanced raman scattering of hollow gold nanospheres. Anal. Chem. 2010, 82, 5290–5295. [Google Scholar] [CrossRef] [PubMed]
- Guven, B.; Basaran-Akgul, N.; Temur, E.; Tamer, U.; Boyaci, I.H. Sers-based sandwich immunoassay using antibody coated magnetic nanoparticles for escherichia coli enumeration. Analyst 2011, 136, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xu, L.; Zhang, F.; Song, Z.; Hu, Y.; Ji, Y.; Shen, J.; Li, B.; Lu, H.; Yang, H. A promising magnetic SERS immunosensor for sensitive detection of avian influenza virus. Biosens. Bioelectron. 2017, 89, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Phillips, J.A.; Xiong, X.; Meng, L.; Van Simaeys, D.; Chen, H.; Martin, J.; Tan, W. Nucleic acid aptamers for biosensors and bio-analytical applications. Analyst 2009, 134, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Brody, E.N.; Willis, M.C.; Smith, J.D.; Jayasena, S.; Zichi, D.; Gold, L. The use of aptamers in large arrays for molecular diagnostics. Mol. Diagn. J. Devoted Underst. Hum. Dis. Clin. Appl. Mol. Biol. 1999, 4, 381–388. [Google Scholar] [CrossRef]
- Fang, X.; Tan, W. Aptamers generated from cell-selex for molecular medicine: A chemical biology approach. Acc. Chem. Res. 2010, 43, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, K.; Li, Y.; Ji, J.; Liu, B. High-resolution and universal visualization of latent fingerprints based on aptamer-functionalized core-shell nanoparticles with embedded SERS reporters. ACS Appl. Mater. Interfaces 2016, 8, 14389–14395. [Google Scholar] [CrossRef] [PubMed]
- Willner, I.; Zayats, M. Electronic aptamer-based sensors. Angew. Chem. 2007, 46, 6408–6418. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Rothberg, L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 14036–14039. [Google Scholar] [CrossRef] [PubMed]
- Demers, L.M.; Mirkin, C.A.; Mucic, R.C.; Reynolds, R.A.; Letsinger, R.L.; Elghanian, R.; Viswanadham, G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 2000, 72, 5535–5541. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Li, F.; Wan, Y.; Wei, M.; Liu, H.; Su, Y.; Chen, N.; Huang, Q.; Fan, C. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA-gold nanoparticle nanoconjugates. J. Am. Chem. Soc. 2012, 134, 11876–11879. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, J.; Suri, S.; Sooter, L.J.; Ma, D.; Wu, N. Detection of adenosine triphosphate with an aptamer biosensor based on surface-enhanced raman scattering. Anal. Chem. 2012, 84, 2837–2842. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Sefah, K.; Bamrungsap, S.; Chang, H.T.; Tan, W. Selective photothermal therapy for mixed cancer cells using aptamer-conjugated nanorods. Langmuir ACS J. Surfaces Colloids 2008, 24, 11860–11865. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lee, K.; Irudayaraj, J. Sers aptasensor from nanorod-nanoparticle junction for protein detection. Chem. Commun. 2010, 46, 613–615. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lamont, E.; Veeregowda, B.; Sreevatsan, S.; Haynes, C.L.; Diez-Gonzalez, F.; Labuza, T.P. Aptamer-based surface-enhanced raman scattering detection of ricin in liquid foods. Chem. Sci. 2011, 2, 1579. [Google Scholar] [CrossRef]
- Zheng, J.; Jiao, A.; Yang, R.; Li, H.; Li, J.; Shi, M.; Ma, C.; Jiang, Y.; Deng, L.; Tan, W. Fabricating a reversible and regenerable raman-active substrate with a biomolecule-controlled DNA nanomachine. J. Am. Chem. Soc. 2012, 134, 19957–19960. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Irudayaraj, J. A SERS dnazyme biosensor for lead ion detection. Chem. Commun. 2011, 47, 4394–4396. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Chen, X.; Liu, R.; Liu, B.; Jiang, C. Target induced aggregation of modified Au@Ag nanoparticles for surface enhanced raman scattering and its ultrasensitive detection of arsenic(iii) in aqueous solution. RSC Adv. 2015, 5, 77755–77759. [Google Scholar] [CrossRef]
- Song, L.; Mao, K.; Zhou, X.; Hu, J. A novel biosensor based on Au@Ag core-shell nanoparticles for SERS detection of arsenic(iii). Talanta 2016, 146, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Q.; Chen, L.X. Aptameric SERS sensor for Hg2+ analysis using silver nanoparticles. Chin. Chem. Lett. 2009, 20, 1475–1477. [Google Scholar] [CrossRef]
- Zhu, Y.; Jiang, X.; Wang, H.; Wang, S.; Wang, H.; Sun, B.; Su, Y.; He, Y. A poly adenine-mediated assembly strategy for designing surface-enhanced resonance raman scattering substrates in controllable manners. Anal. Chem. 2015, 87, 6631–6638. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhang, L.; Zhang, H.; Wu, T.; Du, Y. A selective surface-enhanced raman scattering sensor for mercury(ii) based on a porous polymer material and the target-mediated displacement of a t-rich strand. J. Appl. Spectrosc. 2017, 84, 225–230. [Google Scholar] [CrossRef]
- Pang, S.; Yang, T.; He, L. Review of surface enhanced raman spectroscopic (SERS) detection of synthetic chemical pesticides. TrAC Trends Anal. Chem. 2016, 85, 73–82. [Google Scholar] [CrossRef]
- Pang, S.; Labuza, T.P.; He, L. Development of a single aptamer-based surface enhanced raman scattering method for rapid detection of multiple pesticides. Analyst 2014, 139, 1895–1901. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chao, J.; Qu, X.; Zhang, H.; Zhu, D.; Su, S.; Aldalbahi, A.; Wang, L.; Pei, H. Probing cellular molecules with polya-based engineered aptamer nanobeacon. ACS Appl. Mater. Interfaces 2017, 9, 8014–8020. [Google Scholar] [CrossRef] [PubMed]
- Groopman, J.D.; Croy, R.G.; Wogan, G.N. In vitro reactions of aflatoxin b1-adducted DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 5445–5449. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Tang, L.; Song, D.; Song, S.; Ma, W.; Xu, L.; Kuang, H.; Wu, X.; Liu, L.; Chen, X.; et al. A SERS-active sensor based on heterogeneous gold nanostar core-silver nanoparticle satellite assemblies for ultrasensitive detection of aflatoxinb1. Nanoscale 2016, 8, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
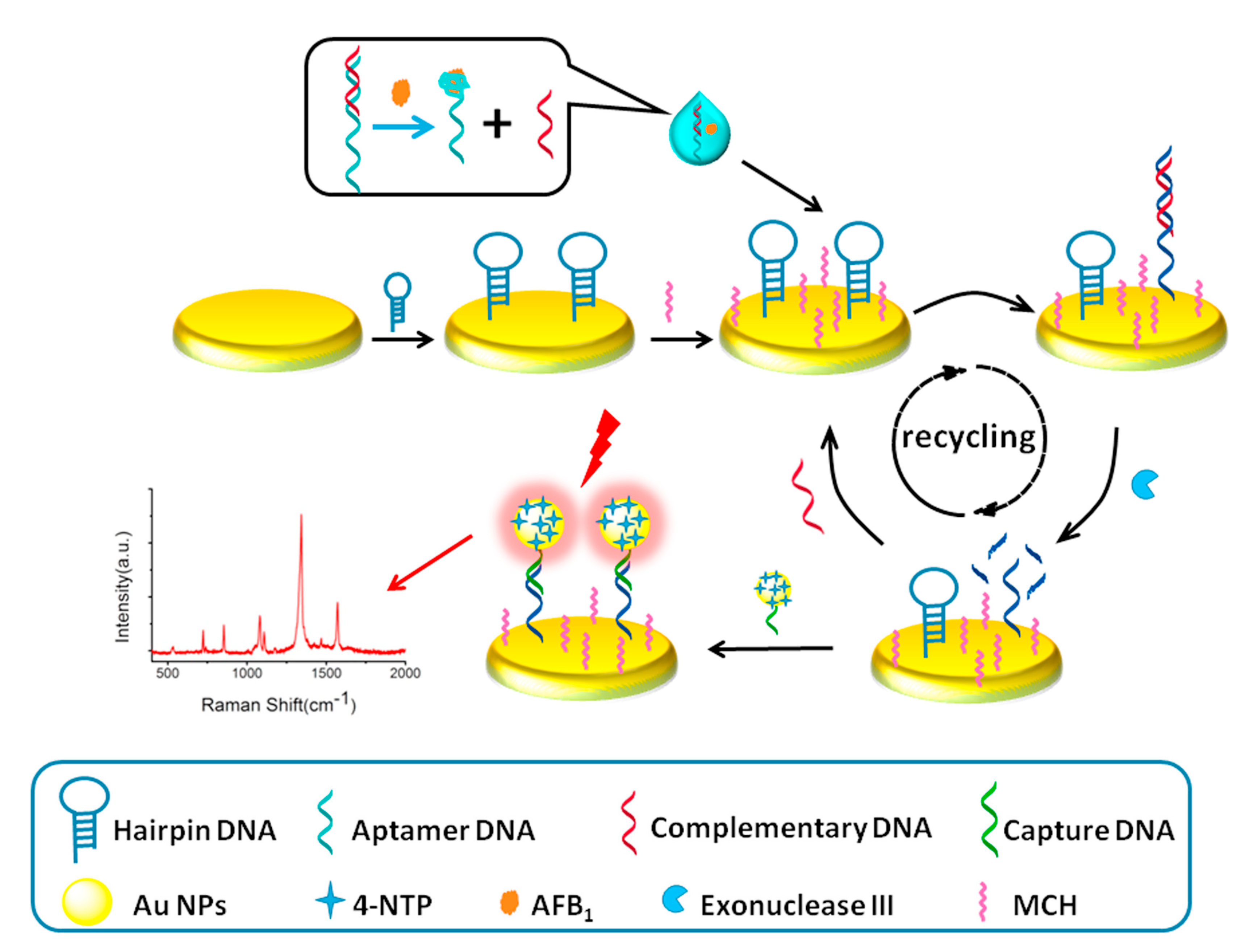
- Li, Q.; Lu, Z.; Tan, X.; Xiao, X.; Wang, P.; Wu, L.; Shao, K.; Yin, W.; Han, H. Ultrasensitive detection of aflatoxin b1 by SERS aptasensor based on exonuclease-assisted recycling amplification. Biosens. Bioelectron. 2017, 97, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Hu, Y.; Dong, N.; Zhu, G.; Zhu, T.; Jiang, N. A novel SERS-based magnetic aptasensor for prostate specific antigen assay with high sensitivity. Biosens. Bioelectron. 2017, 94, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Wu, X.; Xu, L.; Liu, L.; Ma, W.; Kuang, H.; Xu, C. Ultrasensitive detection of prostate-specific antigen and thrombin based on gold-upconversion nanoparticle assembled pyramids. Small 2017, 13, 1603944. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Fu, P.; Ma, W.; Xu, L.; Kuang, H.; Xu, C. Sers-active silver nanoparticle trimers for sub-attomolar detection of alpha fetoprotein. RSC Adv. 2015, 5, 73395–73398. [Google Scholar] [CrossRef]
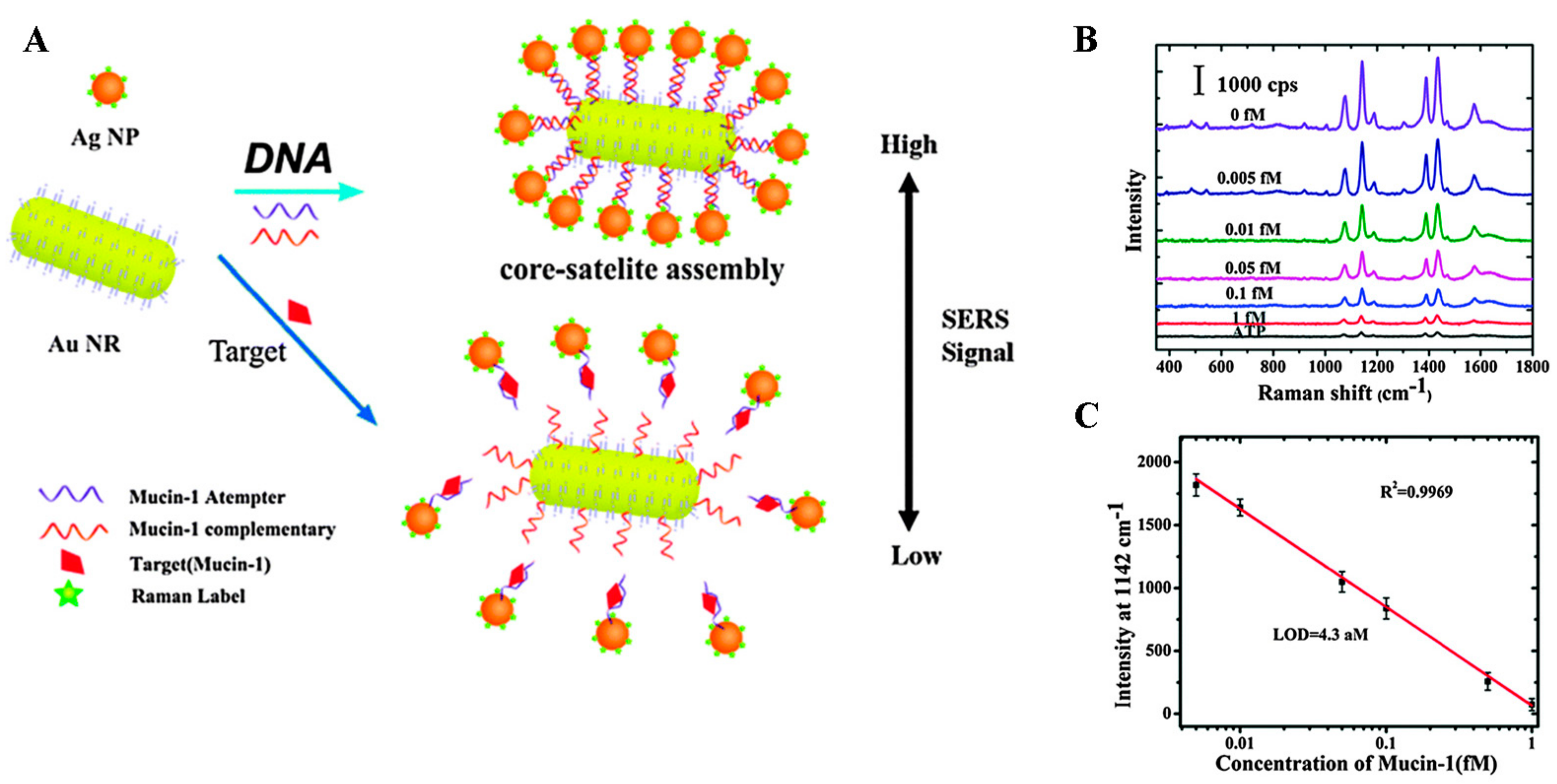
- Feng, J.; Wu, X.; Ma, W.; Kuang, H.; Xu, L.; Xu, C. A SERS active bimetallic core-satellite nanostructure for the ultrasensitive detection of mucin-1. Chem. Commun. 2015, 51, 14761–14763. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, D.; Norbel, L.; Shen, J.; Zhao, Z.; Dou, Y.; Peng, T.; Shi, J.; Mathur, S.; Fan, C.; et al. Multicolor gold-silver nano-mushrooms as ready-to-use SERS probes for ultrasensitive and multiplex DNA/miRNA detection. Anal. Chem. 2017, 89, 2531–2538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, X.; Liu, Y.; Duan, N.; Wu, S.; Wang, Z.; Xu, B. Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of salmonella typhimurium and staphylococcus aureus. Biosens. Bioelectron. 2015, 74, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Zavaleta, C.; de la Zerda, A.; Liu, Z.; Keren, S.; Cheng, Z.; Schipper, M.; Chen, X.; Dai, H.; Gambhir, S.S. Noninvasive raman spectroscopy in living mice for evaluation of tumor targeting with carbon nanotubes. Nano Lett. 2008, 8, 2800–2805. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Qu, H.; Liang, L.; Zhang, J.; Zhang, B.; Huang, D.; Xu, S.; Liang, C.; Xu, W. Tracing the therapeutic process of targeted aptamer/drug conjugate on cancer cells by surface-enhanced raman scattering spectroscopy. Anal. Chem. 2017, 89, 2844–2851. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Hoon, D.; Ambrosi, A.; Nitti, D.; Rossi, C.R. The prognostic value of circulating tumor cells in patients with melanoma: A systematic review and meta-analysis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 4605–4613. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xia, Y.; Huang, Y.; Li, J.; Ruan, H.; Chen, T.; Luo, L.; Shen, Z.; Wu, A. Improved SERS-active nanoparticles with various shapes for ctc detection without enrichment process with supersensitivity and high specificity. ACS Appl. Mater. Interfaces 2016, 8, 19928–19938. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, R.; Gao, M.; Zhang, X. A rapid and simple method for efficient capture and accurate discrimination of circulating tumor cells using aptamer conjugated magnetic beads and surface-enhanced raman scattering imaging. Anal. Bioanal. Chem. 2015, 407, 8883–8892. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Yu, Z.; Yang, T. Surface-enhanced raman spectroscopic analysis of phorate and fenthion pesticide in apple skin using silver nanoparticles. Appl. Spectrosc. 2014, 68, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Wulff, G.; Sarhan, A. Macromolecular colloquium. Angew. Chem. Int. Ed. Engl. 1972, 11, 334–342. [Google Scholar]
- Gao, D.; Zhang, Z.; Wu, M.; Xie, C.; Guan, G.; Wang, D. A surface functional monomer-directing strategy for highly dense imprinting of tnt at surface of silica nanoparticles. J. Am. Chem. Soc. 2007, 129, 7859–7866. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.; Mosbach, K. Molecularly imprinted polymers and their use in biomimetic sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Shahar, T.; Sicron, T.; Mandler, D. Nanosphere molecularly imprinted polymers doped with gold nanoparticles for high selectivity molecular sensors. Nano Res. 2017, 10, 1056–1063. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Hu, Y.; Ma, L.; Lu, X. Development of molecularly imprinted polymers-surface-enhanced raman spectroscopy/colorimetric dual sensor for determination of chlorpyrifos in apple juice. Sens. Actuators B Chem. 2017, 241, 750–757. [Google Scholar] [CrossRef]
- Ji, W.; Zhang, M.; Wang, T.; Wang, X.; Zheng, Z.; Gong, J. Molecularly imprinted solid-phase extraction method based on sh-au modified silica gel for the detection of six sudan dyes in chili powder samples. Talanta 2017, 165, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lu, X. Rapid detection of melamine in tap water and milk using conjugated “one-step” molecularly imprinted polymers-surface enhanced raman spectroscopic sensor. J. Food Sci. 2016, 81, N1272–N1280. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Feng, S.; Chen, Z.; Li-Chan, E.C.; Grant, E.; Lu, X. Detection and quantification of chloramphenicol in milk and honey using molecularly imprinted polymers: Canadian penny-based SERS nano-biosensor. J. Food Sci. 2014, 79, N2542–N2549. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Gao, F.; Chen, Z.; Grant, E.; Kitts, D.D.; Wang, S.; Lu, X. Determination of alpha-tocopherol in vegetable oils using a molecularly imprinted polymers-surface-enhanced raman spectroscopic biosensor. J. Agric. Food Chem. 2013, 61, 10467–10475. [Google Scholar] [CrossRef] [PubMed]
- Ashley, J.; Shahbazi, M.A.; Kant, K.; Chidambara, V.A.; Wolff, A.; Bang, D.D.; Sun, Y. Molecularly imprinted polymers for sample preparation and biosensing in food analysis: Progress and perspectives. Biosens. Bioelectron. 2017, 91, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, R.; Guan, G.; Jiang, C.; Wang, S.; Zhang, Z. Surface-enhanced raman scattering sensor for theophylline determination by molecular imprinting on silver nanoparticles. Analyst 2011, 136, 4152–4158. [Google Scholar] [CrossRef] [PubMed]
- Matsui, J.; Kato, T.; Takeuchi, T.; Suzuki, M.; Yokoyama, K.; Tamiya, E.; Karube, I. Molecular recognition in continuous polymer rods prepared by a molecular imprinting technique. Anal. Chem. 2002, 65, 2223–2224. [Google Scholar] [CrossRef]
- Ou, J.; Hu, L.; Hu, L.; Li, X.; Zou, H. Determination of phenolic compounds in river water with on-line coupling bisphenol a imprinted monolithic precolumn with high performance liquid chromatography. Talanta 2006, 69, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.W.; Qiao, F.X.; Liu, G.Y. Characteristic of theophylline imprinted monolithic column and its application for determination of xanthine derivatives caffeine and theophylline in green tea. J. Chromatogr. A 2006, 1134, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Titirici, M.-M.; Sellergren, B. Thin molecularly imprinted polymer films via reversible addition−fragmentation chain transfer polymerization. Chem. Mater. 2006, 18, 1773–1779. [Google Scholar] [CrossRef]
- Sergeyeva, T.A.; Matuschewski, H.; Piletsky, S.A.; Bendig, J.; Schedler, U.; Ulbricht, M. Molecularly imprinted polymer membranes for substance-selective solid-phase extraction from water by surface photo-grafting polymerization. J. Chromatogr. A 2001, 907, 89–99. [Google Scholar] [CrossRef]
- Sulitzky, C.; Rückert, B.; Hall, A.J.; Lanza, F.; Unger, K.; Sellergren, B. Grafting of molecularly imprinted polymer films on silica supports containing surface-bound free radical initiators. Macromolecules 2002, 35, 79–91. [Google Scholar] [CrossRef]
- Jia, H.; Yin, R.; Zhong, X.; Xue, M.; Zhao, Y.; Meng, Z.; Wang, Q. Anti-diabetic drugs detection by raman spectrometry with molecular imprinted composite membrane. Chem. J. Chin. Univ. 2016, 37, 239–245. [Google Scholar]
- Yoshida, M.; Uezu, K.; Goto, M.; Furusaki, S. Metal ion imprinted microsphere prepared by surface molecular imprinting technique using water-in-oil-in-water emulsions. J. Appl. Polym. Sci. 1999, 73, 1223–1230. [Google Scholar] [CrossRef]
- Li, H.; Jiang, J.; Wang, Z.; Wang, X.; Liu, X.; Yan, Y.; Li, C. A high performance and highly-controllable core-shell imprinted sensor based on the surface-enhanced raman scattering for detection of r6g in water. J. Colloid Interface Sci. 2017, 501, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.Q.; Li, D.W.; Qu, L.L.; Long, Y.T. Surface-imprinted core-shell au nanoparticles for selective detection of bisphenol a based on surface-enhanced raman scattering. Anal. Chim. Acta 2013, 777, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Ki, C.D.; Oh, C.; Oh, S.-G.; Chang, J.Y. The use of a thermally reversible bond for molecular imprinting of silica spheres. J. Am. Chem. Soc. 2002, 124, 14838–14839. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, M.A.; Kust, P.R.; Deng, G.; Schoen, P.E.; Dordick, J.S.; Clark, D.S.; Gaber, B.P. Catalytic silica particles via template-directed molecular imprinting. Langmuir ACS J. Surfaces Colloids 2000, 16, 1759–1765. [Google Scholar] [CrossRef]
- Rao, M.S.; Dave, B.C. Selective intake and release of proteins by organically-modified silica sol−gels. J. Am. Chem. Soc. 1998, 120, 13270–13271. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Su, X.; Wang, T. Advancements of molecularly imprinted polymers in the food safety field. Analyst 2016, 141, 3540–3553. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, X.; Zhao, Y.; Chang, L.; Qi, J. High performance surface-enhanced raman scattering via dummy molecular imprinting onto silver microspheres. Chem. Commun. 2014, 50, 14331–14333. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Zhou, W.H.; Han, B.; Yang, H.H.; Chen, X.; Wang, X.R. Surface-imprinted core-shell nanoparticles for sorbent assays. Anal. Chem. 2007, 79, 5457–5461. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Long, Y.; Pan, J.; Li, K.; Liu, F. A method for coating colloidal particles with molecularly imprinted silica films. J. Mater. Chem. 2008, 18, 2849–2854. [Google Scholar] [CrossRef]
- Jenkins, A.L.; Yin, R.; Jensen, J.L. Molecularly imprinted polymer sensors for pesticide and insecticide detection in water. Analyst 2001, 126, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yang, J.; Su, Q.; Cai, J.; Gao, Y. Selective solid-phase extraction using molecularly imprinted polymer for the analysis of polar organophosphorus pesticides in water and soil samples. J. Chromatogr. A 2005, 1092, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.P.; Kala, R.; Daniel, S. Metal ion-imprinted polymers—Novel materials for selective recognition of inorganics. Anal. Chim. Acta 2006, 578, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.M.; Narayanaswamy, R. Fluorescence sensor using a molecularly imprinted polymer as a recognition receptor for the detection of aluminium ions in aqueous media. Anal. Bioanal. Chem. 2006, 386, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Kugimiya, A.; Takeuchi, T. Molecularly imprinted polymer-coated quartz crystal microbalance for detection of biological hormone. Electroanalysis 1999, 11, 1158–1160. [Google Scholar] [CrossRef]
- Wackerlig, J.; Lieberzeit, P.A. Molecularly imprinted polymer nanoparticles in chemical sensing—Synthesis, characterisation and application. Sens. Actuators B Chem. 2015, 207, 144–157. [Google Scholar] [CrossRef]
- Lv, Y.; Tan, T.; Svec, F. Molecular imprinting of proteins in polymers attached to the surface of nanomaterials for selective recognition of biomacromolecules. Biotechnol. Adv. 2013, 31, 1172–1186. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Chen, Y.; Liu, Z. A boronate affinity sandwich assay: An appealing alternative to immunoassays for the determination of glycoproteins. Angew. Chem. Int. Ed. 2014, 53, 10386–10389. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Qin, Y.; Svec, F.; Tan, T. Molecularly imprinted plasmonic nanosensor for selective SERS detection of protein biomarkers. Biosens. Bioelectron. 2016, 80, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed]
- Jahn, I.J.; Zukovskaja, O.; Zheng, X.S.; Weber, K.; Bocklitz, T.W.; Cialla-May, D.; Popp, J. Surface-enhanced raman spectroscopy and microfluidic platforms: Challenges, solutions and potential applications. Analyst 2017, 142, 1022–1047. [Google Scholar] [CrossRef] [PubMed]
- Hidi, I.J.; Jahn, M.; Weber, K.; Cialla-May, D.; Popp, J. Droplet based microfluidics: Spectroscopic characterization of levofloxacin and its SERS detection. Phys. Chem. Chem. Phys. PCCP 2015, 17, 21236–21242. [Google Scholar] [CrossRef] [PubMed]
- Strehle, K.R.; Cialla, D.; Rosch, P.; Henkel, T.; Kohler, M.; Popp, J. A reproducible surface-enhanced raman spectroscopy approach. Online SERS measurements in a segmented microfluidic system. Anal. Chem. 2007, 79, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Marz, A.; Ackermann, K.R.; Malsch, D.; Bocklitz, T.; Henkel, T.; Popp, J. Towards a quantitative SERS approach—Online monitoring of analytes in a microfluidic system with isotope-edited internal standards. J. Biophotonics 2009, 2, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Cheng, Z.; deMello, A.J.; Choo, J. Wash-free magnetic immunoassay of the PSA cancer marker using SERS and droplet microfluidics. Lab Chip 2016, 16, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.W.; Nieuwoudt, M.K.; Vargas, M.J.T.; Bodley, O.L.C.; Yohendiran, T.S.; Oosterbeek, R.N.; Williams, D.E.; Cather Simpson, M. Raman on a disc: High-quality raman spectroscopy in an open channel on a centrifugal microfluidic disc. Analyst 2017, 142, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kang, T.; Cho, H.; Choi, Y.; Lee, L.P. Additional amplifications of SERS via an optofluidic cd-based platform. Lab Chip 2009, 9, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Fang, Y.-C.; Lin, S.-Y.; Lin, Y.-J.; Yen, S.-Y.; Huang, C.-H.; Yang, C.-Y.; Chau, L.-K.; Wang, S.-C. Corona-induced micro-centrifugal flows for concentration of neisseria and salmonella bacteria prior to their quantitation using antibody-functionalized SERS-reporter nanobeads. Microchim. Acta 2017, 184, 1021–1028. [Google Scholar] [CrossRef]
- Yu, W.W.; White, I.M. Inkjet printed surface enhanced raman spectroscopy array on cellulose paper. Anal. Chem. 2010, 82, 9626–9630. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.W.; White, I.M. Inkjet-printed paper-based SERS dipsticks and swabs for trace chemical detection. Analyst 2013, 138, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Torul, H.; Ciftci, H.; Cetin, D.; Suludere, Z.; Boyaci, I.H.; Tamer, U. Paper membrane-based SERS platform for the determination of glucose in blood samples. Anal. Bioanal. Chem. 2015, 407, 8243–8251. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Si, J.; Li, Z. Fabrication techniques for microfluidic paper-based analytical devices and their applications for biological testing: A review. Biosens. Bioelectron. 2016, 77, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M. Three-dimensional microfluidic devices fabricated in layered paper and tape. Proc. Natl. Acad. Sci. USA 2008, 105, 19606–19611. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Crooks, R.M. Three-dimensional paper microfluidic devices assembled using the principles of origami. J. Am. Chem. Soc. 2011, 133, 17564–17566. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Gao, Z.; Luo, Y.; Liu, X.; Zhao, W.; Lin, B. Manual-slide-engaged paper chip for parallel SERS-immunoassay measurement of clenbuterol from swine hair. Electrophoresis 2016, 37, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Gao, Z.; Deng, Q.; Luo, Y.; Zou, L.; Lu, Y.; Zhao, W.; Lin, B. Picomolar detection of carcinoembryonic antigen in whole blood using microfluidics and surface-enhanced raman spectroscopy. Electrophoresis 2016, 37, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Z.; Zhang, Y.; Fei, J.; Chen, H.; Zong, S.; Cui, Y. In situ probing of cell–cell communications with surface-enhanced raman scattering (SERS) nanoprobes and microfluidic networks for screening of immunotherapeutic drugs. Nano Res. 2016, 10, 584–594. [Google Scholar] [CrossRef]
- Catala, C.; Mir-Simon, B.; Feng, X.; Cardozo, C.; Pazos-Perez, N.; Pazos, E.; Gómez-de Pedro, S.; Guerrini, L.; Soriano, A.; Vila, J.; et al. Online SERS quantification ofstaphylococcus aureusand the application to diagnostics in human fluids. Adv. Mater. Technol. 2016, 1, 1600163. [Google Scholar] [CrossRef]
- Fu, C.; Wang, Y.; Chen, G.; Yang, L.; Xu, S.; Xu, W. Aptamer-based surface-enhanced raman scattering-microfluidic sensor for sensitive and selective polychlorinated biphenyls detection. Anal. Chem. 2015, 87, 9555–9558. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Gao, R.; Ko, J.; Choi, N.; Lim, D.W.; Lee, E.K.; Chang, S.I.; Choo, J. Trace analysis of mercury(ii) ions using aptamer-modified Au/Ag core-shell nanoparticles and SERS spectroscopy in a microdroplet channel. Lab Chip 2013, 13, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Tetala, K.K.R.; Vijayalakshmi, M.A. A review on recent developments for biomolecule separation at analytical scale using microfluidic devices. Anal. Chim. Acta 2016, 906, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Abell, J.; Huang, Y.W.; Zhao, Y. On-chip ultra-thin layer chromatography and surface enhanced raman spectroscopy. Lab Chip 2012, 12, 3096–3102. [Google Scholar] [CrossRef] [PubMed]

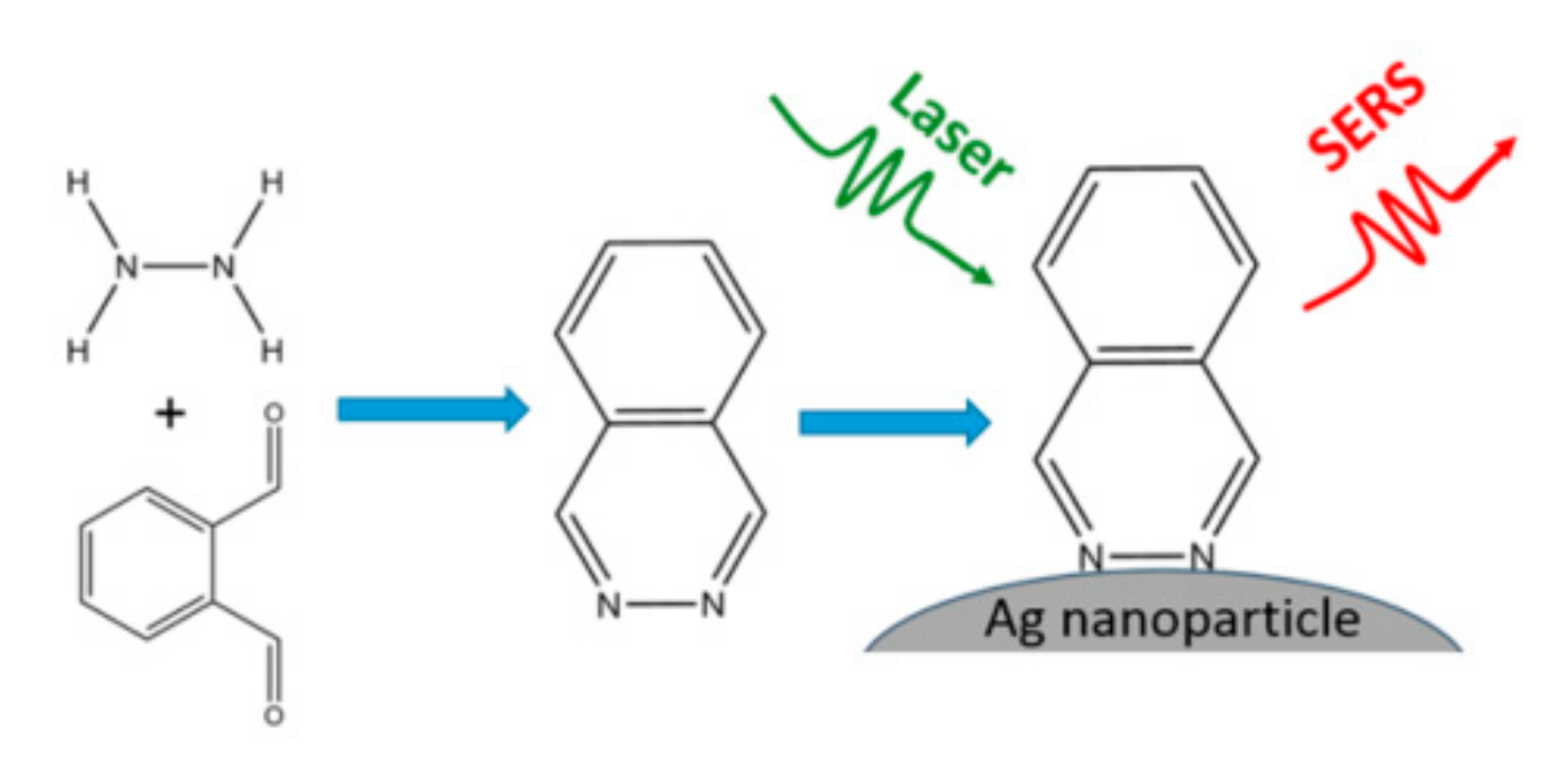
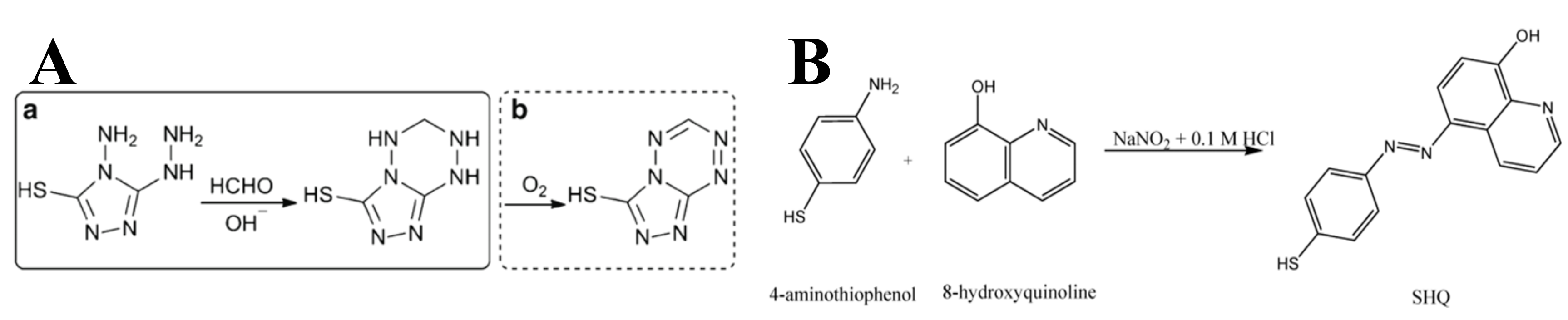
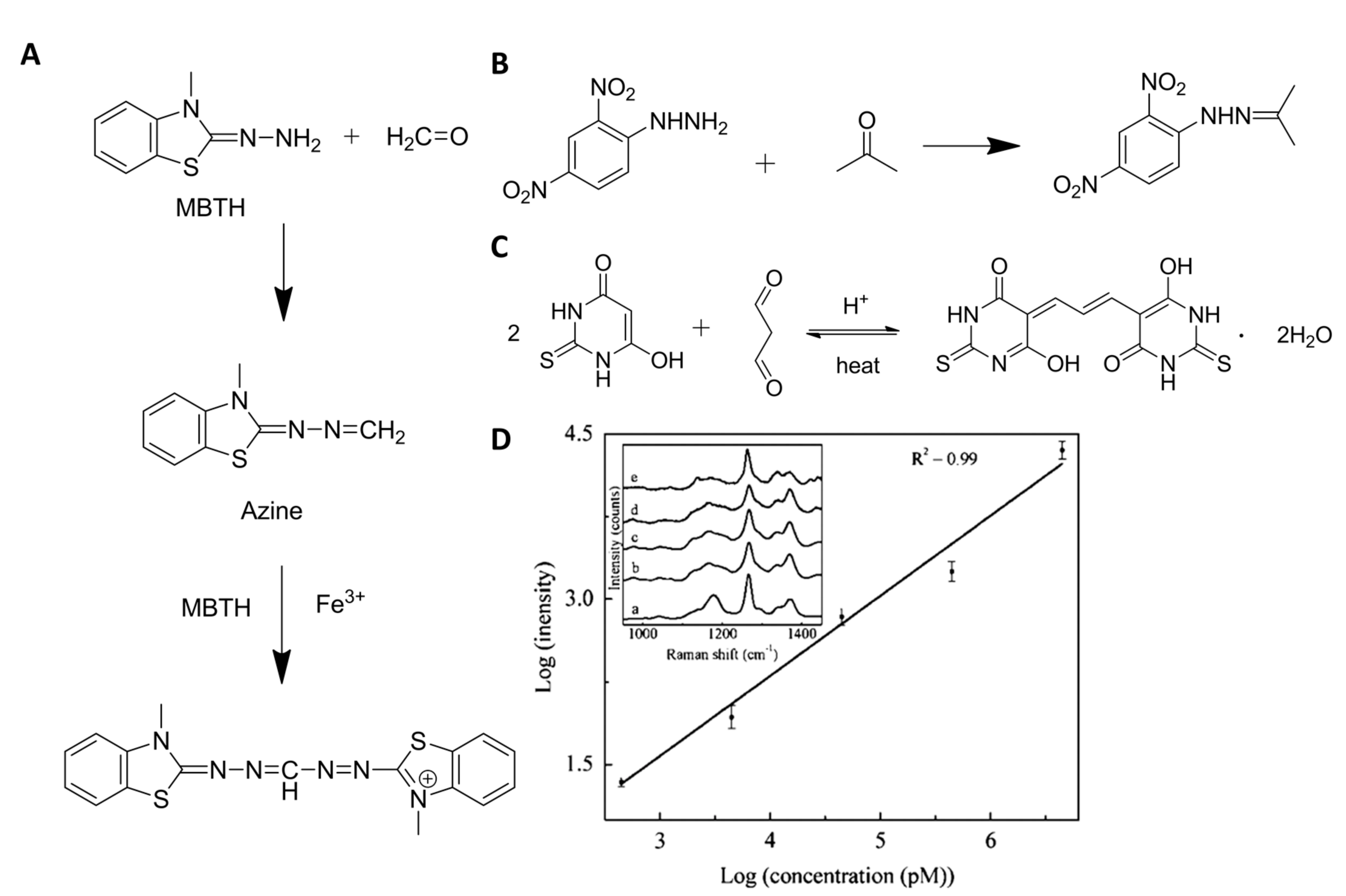



| Analytes | Reagents | Substrates | Matrix | LOD 1 | Linear Range 2 | Ref. |
|---|---|---|---|---|---|---|
| hydrazine | ortho-phthaldialdehyde | AgNPs | water | 8.5 × 10−11 M | 10−5–10−10 M | [6] |
| ethylene and SO2 | bromine-thiourea and OPA-NH4+, respectively | AuNPs | fruits | 1.7 μg/L and 12.0 μg/L, respectively | 14.7–21.1 μg/L and 35.1–35.9 μg/L | [7] |
| formaldehyde | 4-amino-5-hydrazino-3-mercapto-1,2,4-triazole | AgNPs | water and food | 0.15 μg/L | 1–1000 μg/L | [18] |
| formaldehyde | 3-Methyl-2-benzothiazolinone hydrazone | Au/SiO2 NPs | aquatic products | 0.17μg/L | 0.40–4.8 μg/L | [20] |
| acetone | 2,4-dinitrophenylhydrazine | iodide modified Ag nanoparticles (Ag IMNPs) | urine | 0.09 mM | 9.6–96 mg/L | [17] |
| malondialdehyde | thiobarbituric acid | AgNPs | 0.45 nM | 0.45–4.5 μM | [21] | |
| sodium dithionite | bis[4,4′-dithiodiphenyl azo-phenol] | A single cabbage-like Au microparticle | industry | 0.08 μM | 0.5–20 μM | [22] |
| acetone | 4-(methylthio)benzaldehyde | Ag@AuNCs | [24] | |||
| HNO3 and H2SO4 | ammonium hydroxide | Klarite substrate | 100 ppb | [25] | ||
| NO | o-phenylenediamine | AuNPs | living cells | 10−7 M | [31] |
| Target Analyte | Matrix | LOD | Ref. | |
|---|---|---|---|---|
| metal ion | Pb2+ | Water | 20 nM | [62] |
| As3+ | Water | 59 ppt | [63] | |
| As3+ | Lake water | 0.1 ppb | [64] | |
| Hg2+ | Water | 5 nM | [65] | |
| Hg2+ | Lake water | 100 fM | [66] | |
| Hg2+ | River water | 2.5 nM | [67] | |
| Small molecules | profenofos | Apple juice | 5 ppm | [69] |
| phorate | Apple juice | 0.1 ppm | [69] | |
| phorate | Apple skin | 0.05 mg/L | [85] | |
| omethoate | Apple juice | 5 ppm | [69] | |
| isocarbophos | Apple juice | 1 ppm | [69] | |
| ATP | Water | 10 μM | [70] | |
| AFB1 | Peanut milk | 0.48 pg/mL | [72] | |
| AFB1 | Peanut | 0.4 fg/mL | [73] | |
| biomacromolecules | PSA | Serum | 5 pg/mL | [74] |
| PSA | Serum | 3.2 × 10−20 M | [75] | |
| Thrombin | Serum | 5.7 × 10−17 M | [75] | |
| DNA | Serum | 100 fM | [78] | |
| MiRNA | Serum | 10 fM | [78] | |
| Mucin-1 | Serum | 4.3 aM | [77] | |
| Alpha fetoprotein | Serum | 0.097 aM | [76] | |
| Lysozyme | [52] | |||
| living organisms | S. typhimurium | Pork | 15 cfu/mL | [79] |
| S. aureus | Pork | 35 cfu/mL | [79] | |
| Hela cells | [81] | |||
| CTC | Blood | 1 cell/mL | [83] | |
| CTC | Blood | 20 cells/mL | [84] | |
| mapping | ATP | intracellular ATP molecules | [70] | |
| Lysozyme | different substrates | [52] | ||
| membrane protein | Hela cells | [58] | ||
| membrane protein | HepG2 cells | [81] | ||
| Target Detector | MIPs-Sensor Type | Functional Monomer | Matrix | LOD | Ref. |
|---|---|---|---|---|---|
| Chlorpyrifos | MISPE-SERS | Methacrylic acid (MAA) | Apple juice | 0.01 mg/L | [91] |
| Sudan dyes | MISPE-SERS | 3-(trimethoxysilyl)propyl methacrylate (TPM) | chili powder | 1.5 ng/g | [92] |
| Melamine | MISPE-SERS | MAA | Milk | 0.005 mM | [93] |
| Chloramphenicol | MISPE-SERS | Acrylamide (AM) | Honey and milk | 0.1 ppm | [94] |
| Alpha-tocopherol | MISPE-SERS | MAA | Vegetable oils | 10 ppb | [95] |
| Sudan IV | Au NPs doped MIPs | MAA | Sudan derivatives | [89] | |
| Theophylline | Ag NPs doped MIPs | MAA | Green tea drinks | 3.5 μM | [97] |
| Metformin | MIM-SERS | MAA | Hypoglycemic agents | 5 mg/mL | [104] |
| R6G | Core-shell | AM | Water | 10−12 M | [106] |
| Bisphenol A | Core-shell | 3-(triethoxysilyl)propyl isocyanate (TEPIC) | River water | 0.1 mg/L | [107] |
| Transferrin | Core-shell | Polydopamine (PDA) | human serum | 10−8 M | [123] |
| Glycoproteins | Sandwich structure | Boronic acids | human serum | 13.8 ± 3.3 ng/mL | [122] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Cao, S.; Yan, R.; Wang, Z.; Wang, D.; Yang, H. Selectivity/Specificity Improvement Strategies in Surface-Enhanced Raman Spectroscopy Analysis. Sensors 2017, 17, 2689. https://doi.org/10.3390/s17112689
Wang F, Cao S, Yan R, Wang Z, Wang D, Yang H. Selectivity/Specificity Improvement Strategies in Surface-Enhanced Raman Spectroscopy Analysis. Sensors. 2017; 17(11):2689. https://doi.org/10.3390/s17112689
Chicago/Turabian StyleWang, Feng, Shiyu Cao, Ruxia Yan, Zewei Wang, Dan Wang, and Haifeng Yang. 2017. "Selectivity/Specificity Improvement Strategies in Surface-Enhanced Raman Spectroscopy Analysis" Sensors 17, no. 11: 2689. https://doi.org/10.3390/s17112689





