Separation and Analysis of Adherent and Non-Adherent Cancer Cells Using a Single-Cell Microarray Chip
Abstract
:1. Introduction
2. Materials and Methods
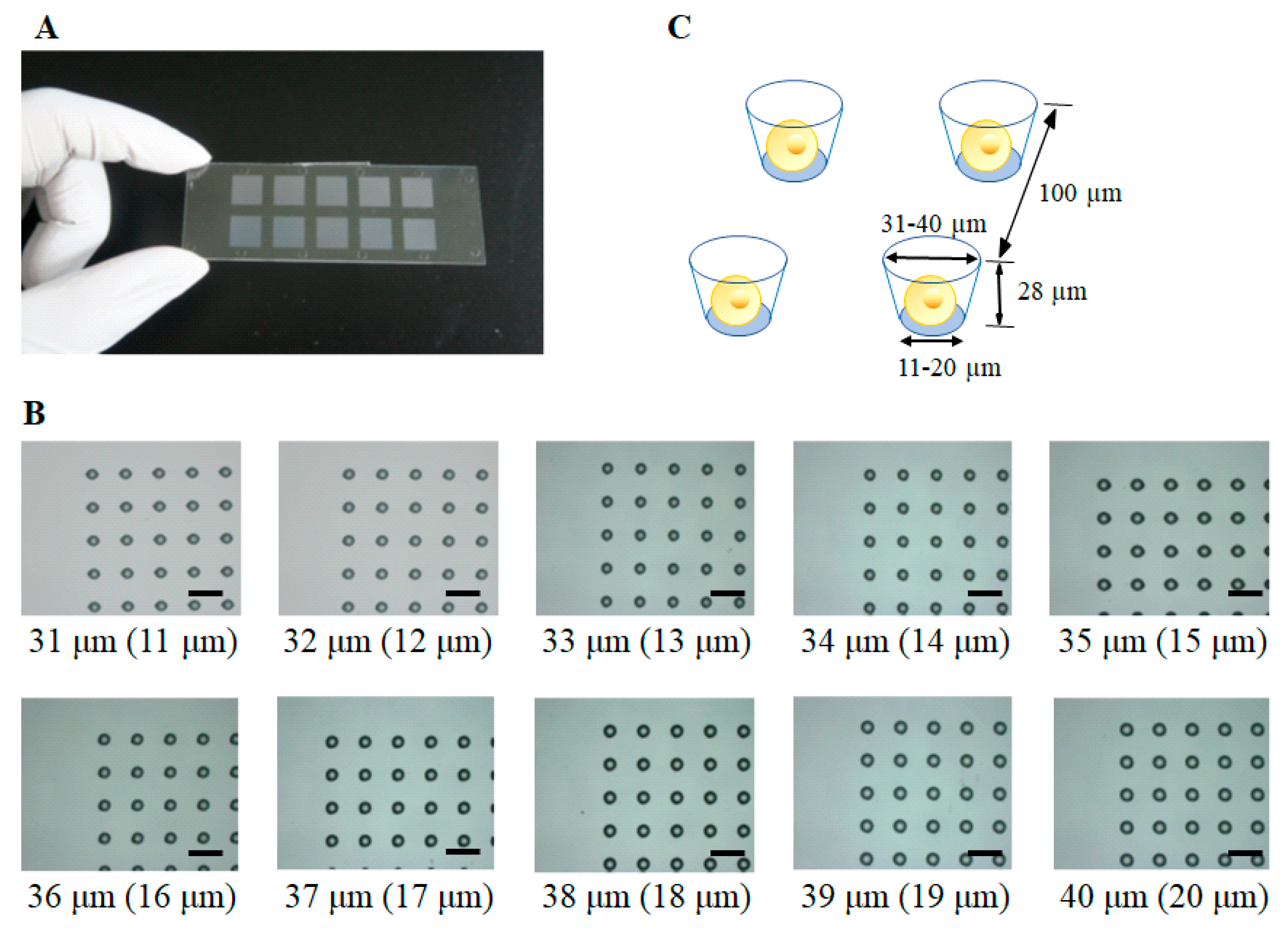
2.1. Construction of a Single-Cell Microarray Chip
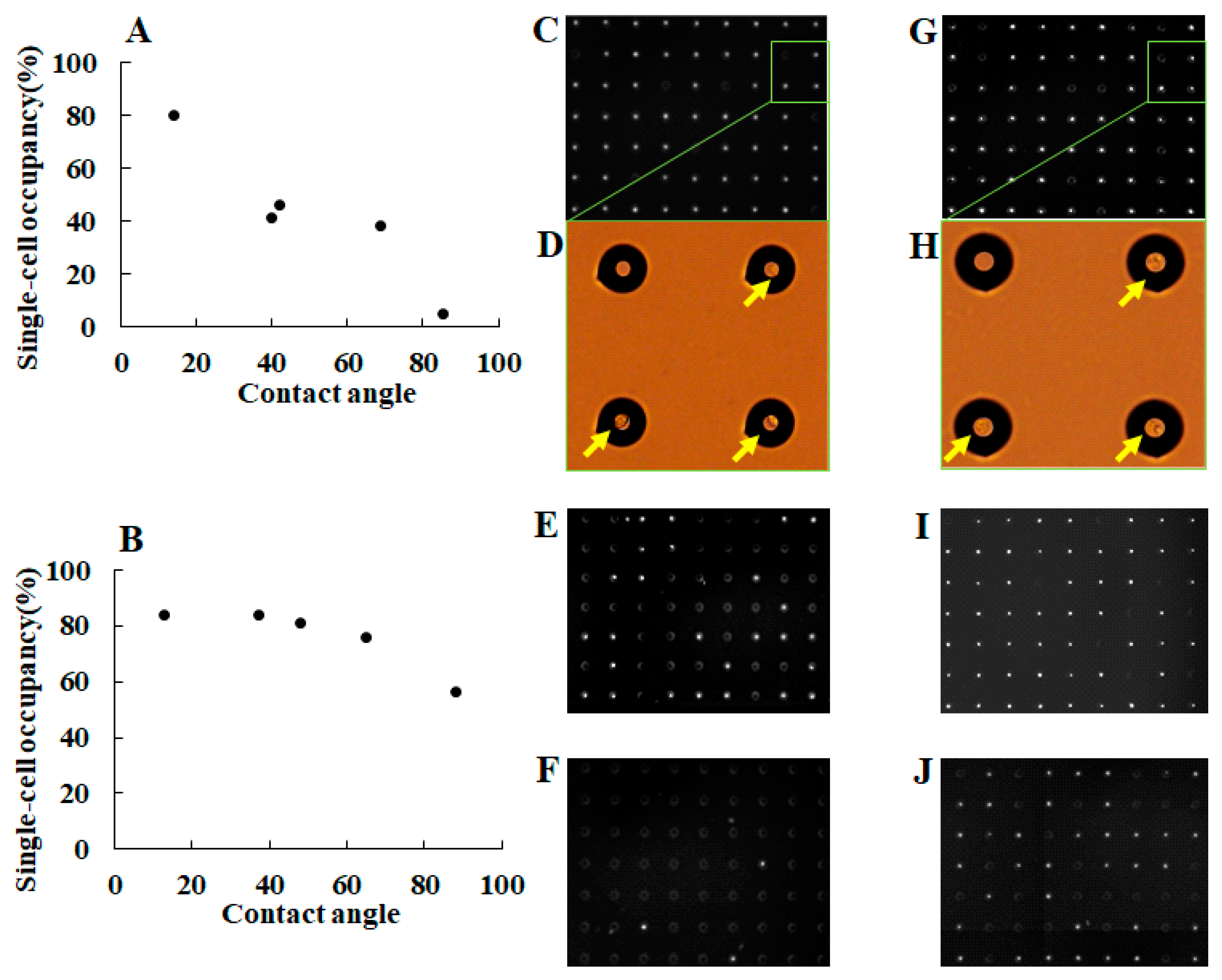
2.2. Chip Surface Treatment and Characterization
2.3. Cell Culture and Preparation
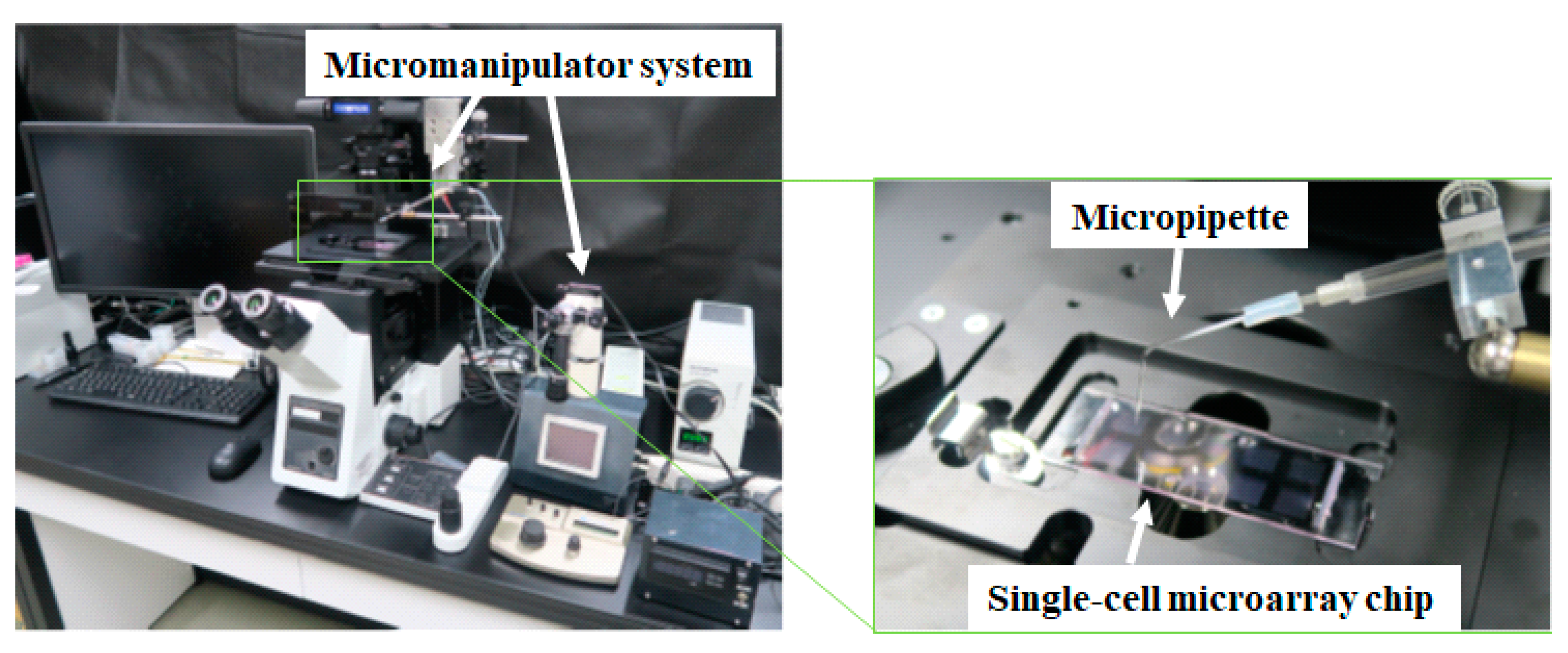
2.4. Single-Cell Separation and Analysis
2.5. Single-Cell Retrieval
3. Results and Discussion
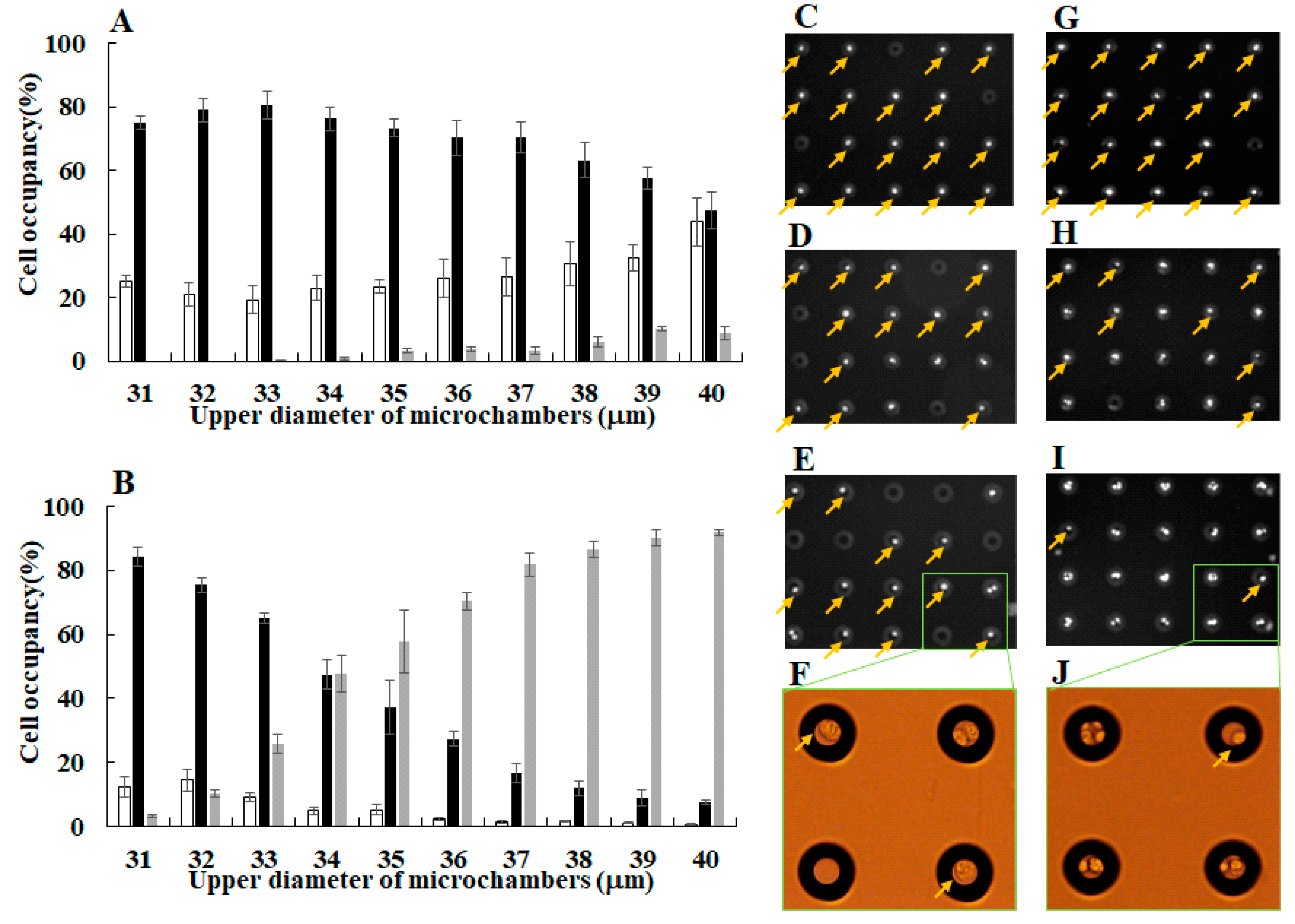
3.1. Separation of Different Types of Single Cancer Cells on a Single-Cell Microarray Chip
3.2. Identification of Different Types of Cancer Cells on a Single-Cell Microarray Chip
3.3. Retrieval of Different Types of Single Cancer Cells
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Carlo, D.D.; Lee, L.P. Dynamic single-cell analysis for quantitative biology. Anal. Chem. 2006, 78, 7918–7925. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.P.; Sklar, L.A. The emergence of flow cytometry for sensitive, real-time measurements of molecular interactions. Nat. Biotechnol. 1998, 16, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Krutzik, O.P.; Nolan, G.P. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat. Methods 2006, 3, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Cruz, I.; Ciudad, J.; Cruz, J.J.; Ramos, M.; Gómez-Alonso, A.; Adansa, J.C.; Rodríguez, C.; Orfao, A. Evaluation of multiparameter flow cytometry for the detection of breast cancer tumor cells in blood samples. Am. J. Clin. Pathol. 2005, 123, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Alunni-Fabbroni, M.; Sandri, M.T. Circulating tumour cells in clinical practice: Methods of detectionand possible characterization. Methods 2010, 50, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Krivacic, R.T.; Ladanyi, A.; Curry, D.N.; Hsieh, H.B.; Kuhn, P.; Bergsrud, D.E.; Kepros, J.F.; Barbera, T.; Ho, M.Y.; Chen, L.B.; et al. A rare-cell detector for cancer. Proc. Natl. Acad. Sci. USA 2004, 101, 10501–10504. [Google Scholar] [CrossRef] [PubMed]
- Kahn, H.J.; Presta, A.; Yang, L.Y.; Blondal, J.; Trudeau, M.; Lickley, L.; Holloway, C.; McCready, D.R.; Maclean, D.; Marks, A. Enumeration of circulating tumor cells in the blood of breast cancer patients after filtration enrichment: Correlation with disease stage. Breast Cancer Res. Treat. 2004, 86, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia1, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 11, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.A.; Okagbare, P.I.; Feng, J.; Hupert, M.L.; Patterson, D.; Göttert, J.; McCarley, R.L.; Nikitopoulos, D.; Murphy, M.C.; Soper, S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M.; Hayata, T.; Fukuda, Y.; Arakaki, A.; Yoshino, T.; Tanaka, T.; Matsunaga, T. Size-selective microcavity array for rapid and efficient detection of circulating tumor cells. Anal. Chem. 2010, 82, 6629–6635. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.G.; Levenberg, S.; Langer, R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004, 22, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Akagi, Y.; Sathuluri, R.R.; Morita, Y.; Tamiya, E. Optimization of fluorescent cell-based assays for high-throughput analysis using microchamber array chip formats. Sci. Technol. Adv. Mater. 2004, 5, 343–349. [Google Scholar] [CrossRef]
- Yamahira, S.; Yamaguchi, S.; Kawahara, M.; Nagamune, T. Collagen surfaces modified with photo-cleavable polyethylene glycol-lipid support versatile single-cell arrays of both non-adherent and adherent cells. Macromol. Biosci. 2014, 14, 1670–1676. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Kishi, H.; Tokimitsu, Y.; Kondo, S.; Honda, R.; Sathuluri, R.R.; Omori, M.; Tamiya, E.; Muraguchi, A. Single-cell microarra.y chip for analyzing cellular responses. Anal. Chem. 2005, 77, 8050–8056. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.; Ozawa, T.; Tajiri, K.; Obata, T.; Kondo, S.; Kinoshita, K.; Kadowaki, S.; Takahashi, K.; Sugiyama, T.; Kishi, H.; Muraguchi, A. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat. Med. 2009, 15, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, A.R.; Throndset, W.R.; Whelan, R.J.; Leach, A.M.; Zare, R.N.; Liaor, Y.H.; Farrell, K.; Manger, I.D.; Daridon, A. Microfluidic device for single-cell analysis. Anal. Chem. 2003, 75, 3581–3586. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Cheng, W.; Zhang, Z.; Pang, D.W.; Cheng, J.K.; Cui, D.F. Transport, location, and quantal release monitoring of single cells on a microfluidic device. Anal. Chem. 2004, 76, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.Y.; Li, P.C.H. A three-dimensional flow control concept for single-cell experiments on a microchip. 1. Cell selection, cell retention, cell culture, cell balancing, and cell scanning. Anal. Chem. 2004, 76, 5273–5281. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, P.C. Microfluidic selection and retention of a single cardiac myocyte, on-chip dye loading, cell contraction by chemical stimulation, and quantitative fluorescent analysis of intracellular calcium. Anal. Chem. 2005, 77, 4315–4322. [Google Scholar] [CrossRef] [PubMed]
- Ino, K.; Okochi, M.; Honda, H. Application of Magnetic Force-Based Cell Patterning for Controlling Cell-Cell Interactions in Angiogenesis. Biotechnol. Bioeng. 2009, 102, 882–890. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamura, S.; Yamada, E.; Kimura, F.; Miyajima, K.; Shigeto, H. Separation and Analysis of Adherent and Non-Adherent Cancer Cells Using a Single-Cell Microarray Chip. Sensors 2017, 17, 2410. https://doi.org/10.3390/s17102410
Yamamura S, Yamada E, Kimura F, Miyajima K, Shigeto H. Separation and Analysis of Adherent and Non-Adherent Cancer Cells Using a Single-Cell Microarray Chip. Sensors. 2017; 17(10):2410. https://doi.org/10.3390/s17102410
Chicago/Turabian StyleYamamura, Shohei, Eriko Yamada, Fukiko Kimura, Kumiko Miyajima, and Hajime Shigeto. 2017. "Separation and Analysis of Adherent and Non-Adherent Cancer Cells Using a Single-Cell Microarray Chip" Sensors 17, no. 10: 2410. https://doi.org/10.3390/s17102410




