1. Introduction
With the development of medical imaging technology, various imaging modals such as ultrasound (US) imaging, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET) and single-photon emission computed tomography (SPECT) are finding a range of applications in diagnosis and assessment of medical conditions that affect brain, breast, lungs, soft tissues, bones and so on [
1]. Owing to the difference in imaging mechanism and the high complexity of human histology, medical images of different modals provide a variety of complementary information about the human body. For example, CT is well-suited for imaging dense structures like non-metallic implants and bones with relatively less distortion. Likewise, MRI can visualize the pathological soft tissues better whereas PET can measure the amount of metabolic activity at a site in the body. Multimodal medical image fusion (MIF) aims to integrate complementary information from multimodal images into a single new image to improve the understanding of the clinical information in a new space. Thus, MIF plays an important role in diagnosis and treatment of diseases and has found wide clinical applications, such as US-MRI for prostate biopsy [
1], PET-CT in lung cancer [
2], MRI-PET in brain disease [
3] and SPECT-CT in breast cancer [
4].
Numerous image fusion algorithms have been proposed by working at pixel level, feature level or decision level. Among these methods, the pixel-level fusion scheme has been investigated most widely due to its advantage of containing the original measured quantities, easy implementation and computational efficiency [
5]. Existing pixel-level image fusion methods generally include substitution methods, multi-resolution fusion methods and neural network based methods. The substitution methods such as intensity hue saturation [
6,
7], principal component analysis [
8] based methods can be implemented with high efficiency but at the expense of reduced contrast and distortion of the spectral characteristics. Image fusion methods based on the multi-resolution decomposition techniques can preserve important image features better than substitution methods via the decomposition of images at a different scale to several components using pyramid (e.g., contrast pyramid [
9] and gradient pyramid [
10]), empirical mode decomposition [
11] or various transforms including wavelet transform [
12,
13,
14], curvelet transform [
15], ripplet transform [
16], contourlet transform [
17], non-subsampled contourlet transform (NSCT) [
18,
19,
20,
21,
22] and shift-invariant shearlet transform [
23,
24]. However, the transform based fusion methods involve much higher computational complexity than the substitution methods, and it is challenging to adaptively determine the involved parameters in these methods for the different medical images.
The various neural networks such as self-generating neural network [
25] and pulse coupled neural network (PCNN) have been used for image fusion. Different from some traditional neural networks, PCNN, as the third generation artificial neural network, has biological background and it is derived from the phenomena of synchronous pulse bursts in the visual cortex of mammals [
26,
27]. The PCNN based MIF method has gained much attention due to its great advantages of being very generic and requiring no training. The parallel image fusion method using multiple PCNNs has been proposed by Li et al. [
28]. The multi-channel PCNN (m-PCNN) based fusion method has been proposed by Wang et al. [
29], and it has been further improved by Zhao et al. [
30]. Recently, PCNN has been combined with multi-resolution decomposition methods such as the wavelet transform [
31], the NSCT [
32,
33,
34,
35,
36], the shearlet transform [
37,
38] and the empirical mode decomposition [
39]. These methods involve such disadvantages as high computational complexity, difficulty in adaptively determining PCNN parameters for various source images and image contrast reduction or loss of image details. In view of high computational complexity of PCNN, Zhan et al. [
40] have recently proposed a computationally more efficient spiking cortical model (SCM), a single-layer, local-connected and two-dimensional neural network. Wang et al. [
41] have presented a fusion method based on the firing times of the SCM (SCM-F). Despite the superiority of SCM over PCNN in computational efficiency, the SCM-F method will lead to loss of image details during fusion because it only utilizes the firing times of individual neurons in the SCM to establish the fusion rule, and employs a too simple fusion strategy.
To address the problem of unwanted image degradation during fusion for the above-mentioned fusion methods, we have proposed a distinctive SCM based weighting fusion method. In the proposed method, the weight is computed based on the multi-features of pulse outputs produced by SCM neurons in a neighborhood rather than the individual neurons. The multi-features include the entropy information of pulse outputs, which can characterize the gray-level information of source images, and the Weber local descriptor (WLD) feature [
42] of firing mapping images produced from pulse outputs, which can represent the local structural information of source images. Compared with the PCNN based fusion method, the proposed SCM based method using the multi-features of pulse outputs (SCM-M) has such advantages as higher computational efficiency, simpler parameter tuning as well as less contrast reduction and loss of image details. Meanwhile, the proposed SCM-M method can preserve the details of source images better than the SCM-F method. Extensive experiments on CT and MR images demonstrate the superiority of the proposed method over numerous state-of-the-art fusion methods.
The remainder of the paper is structured as follows.
Section 2 describes the spiking cortical model.
Section 3 presents the details of the proposed SCM-M method. The experimental results and discussions are presented in
Section 4. Conclusions and future research directions are given in
Section 5.
2. Spiking Cortical Model
The spiking cortical model (SCM) is derived from several other visual cortex models such as Eckhorn’s model [
26,
27]. The SCM has been specially designed for image processing applications. The structural model of the SCM is presented in
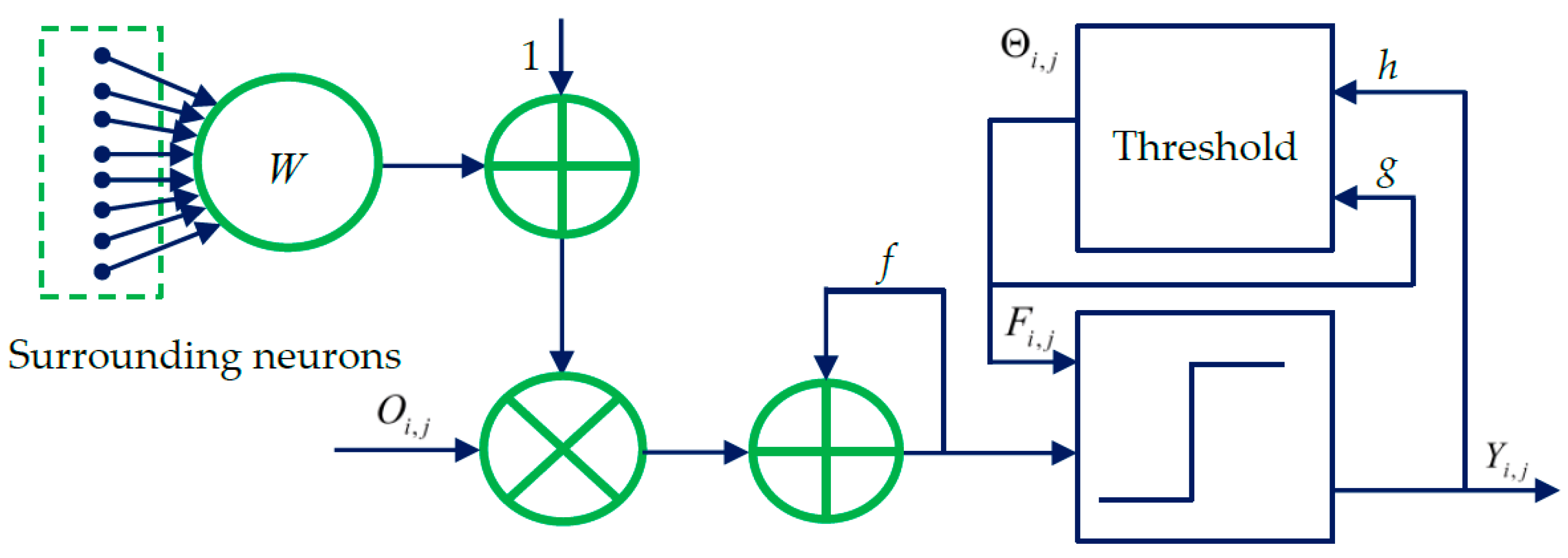
Figure 1. As shown in
Figure 1, each neuron
at (
i,
j) corresponds to one pixel in an input image, receiving its normalized intensity as feeding input
and the local stimuli from its neighboring neurons as the linking input. The feeding input and the liking input are combined together as the internal activity
of
. The neuron
will fire and a pulse output
will be generated if
exceeds a dynamic threshold
. The above process can be expressed by [
40]:
where
f and
g are decay constants less than 1;
is the scalar of large value;
n denotes the number of iterations (
,
is the maximum iteration times); and
is the synaptic weight between
and its linking neuron
and it is defined as:
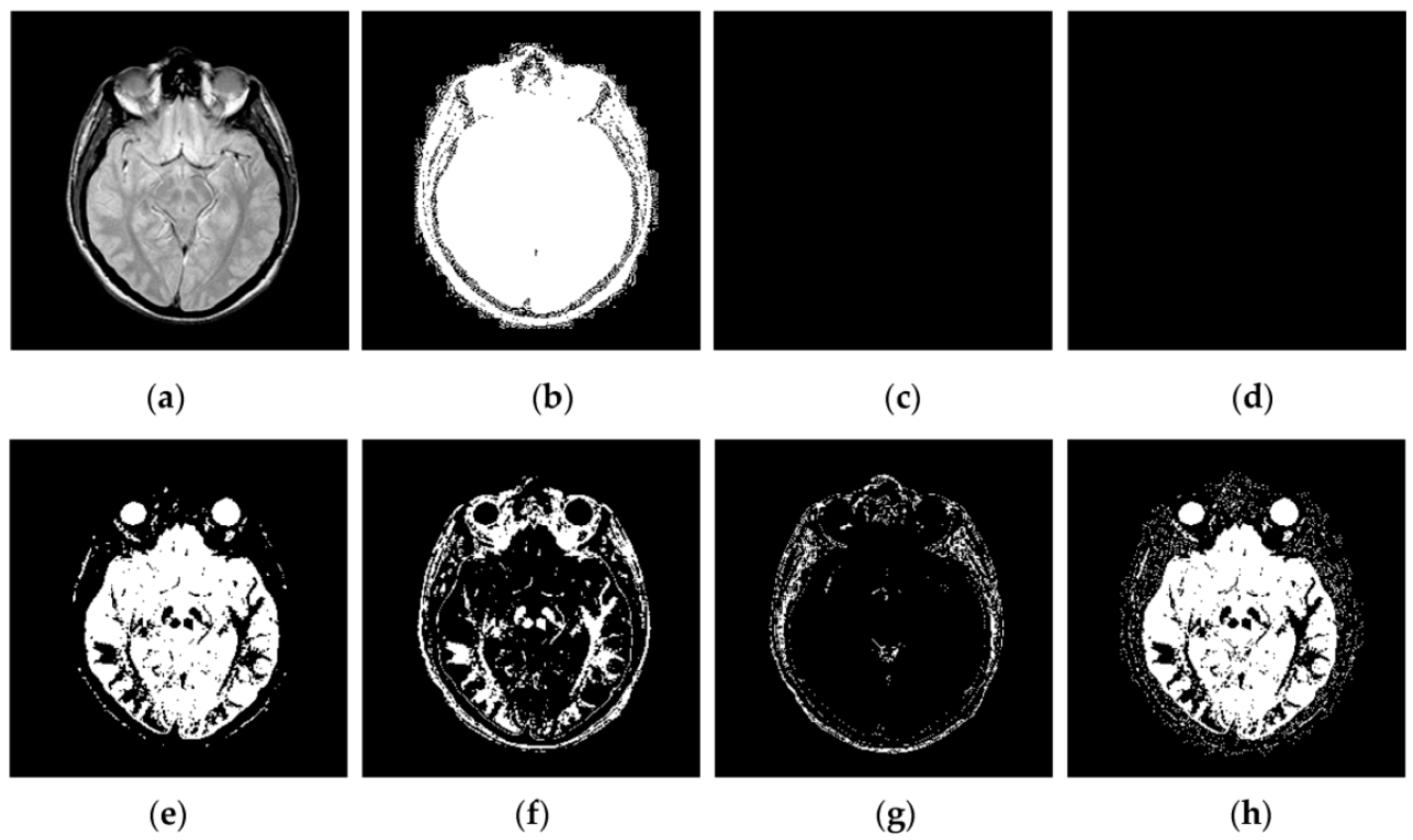
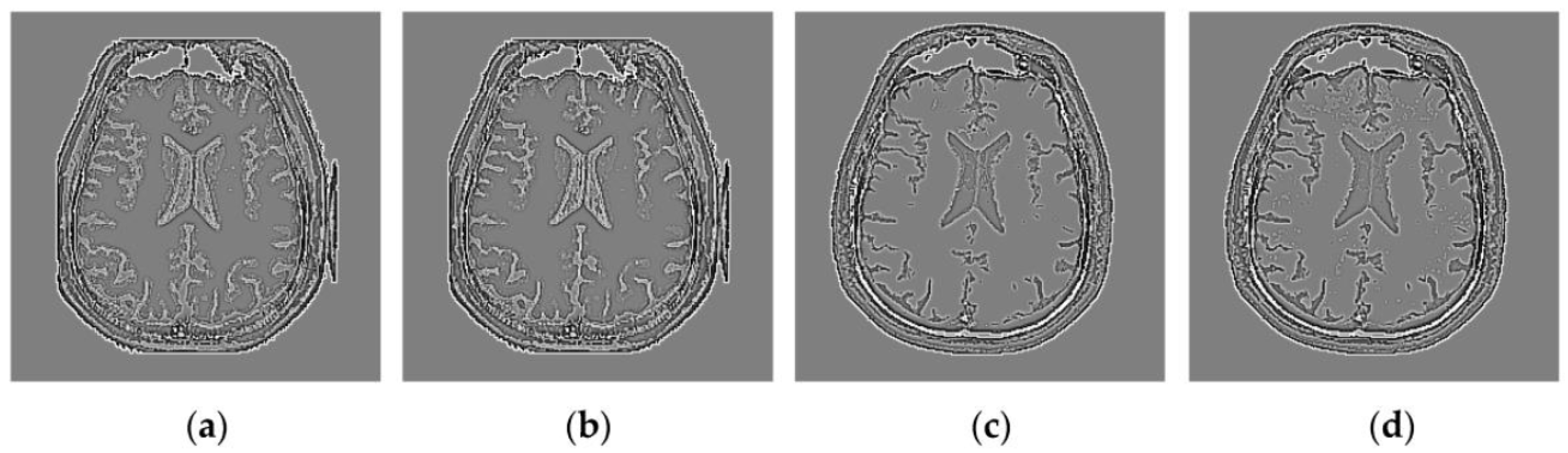
Through iterative computation, the SCM neurons output the temporal series of binary pulse images. The temporal series contain much useful information of input images. To explain this point better,
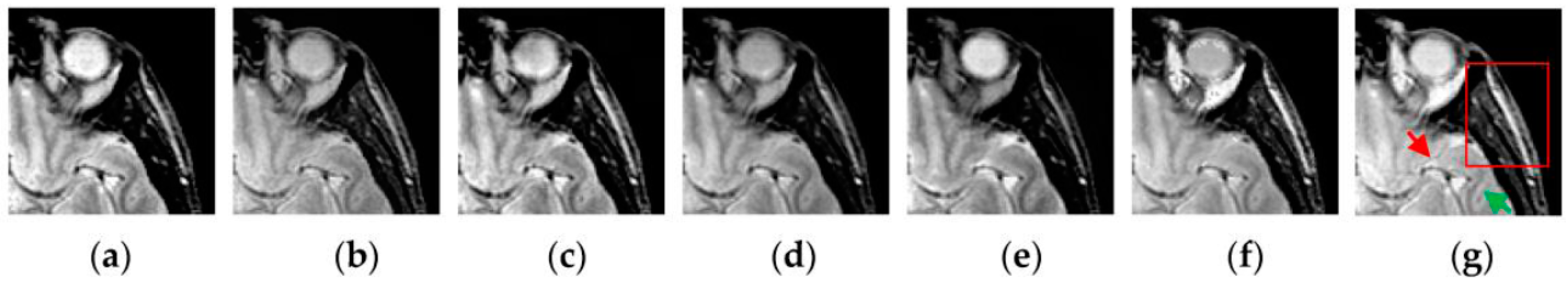
Figure 2 shows the temporal series produced by the SCM with
f = 0.9,
g = 0.3,
h = 20 and
Nmax = 7 operating on an input MR image shown in
Figure 2a. In
Figure 2, we can see that during the various iterations, the output binary images contain different image information and the outputs of the SCM typically represent such important information as the segments and edges of the input image. The observation from
Figure 2 indicates that the SCM can describe human visual perception. Therefore, the pulse outputs of the SCM can be utilized for image fusion.