Label-Free Aptasensors for the Detection of Mycotoxins
Abstract
:1. Introduction
2. Why Label-Free Detection?
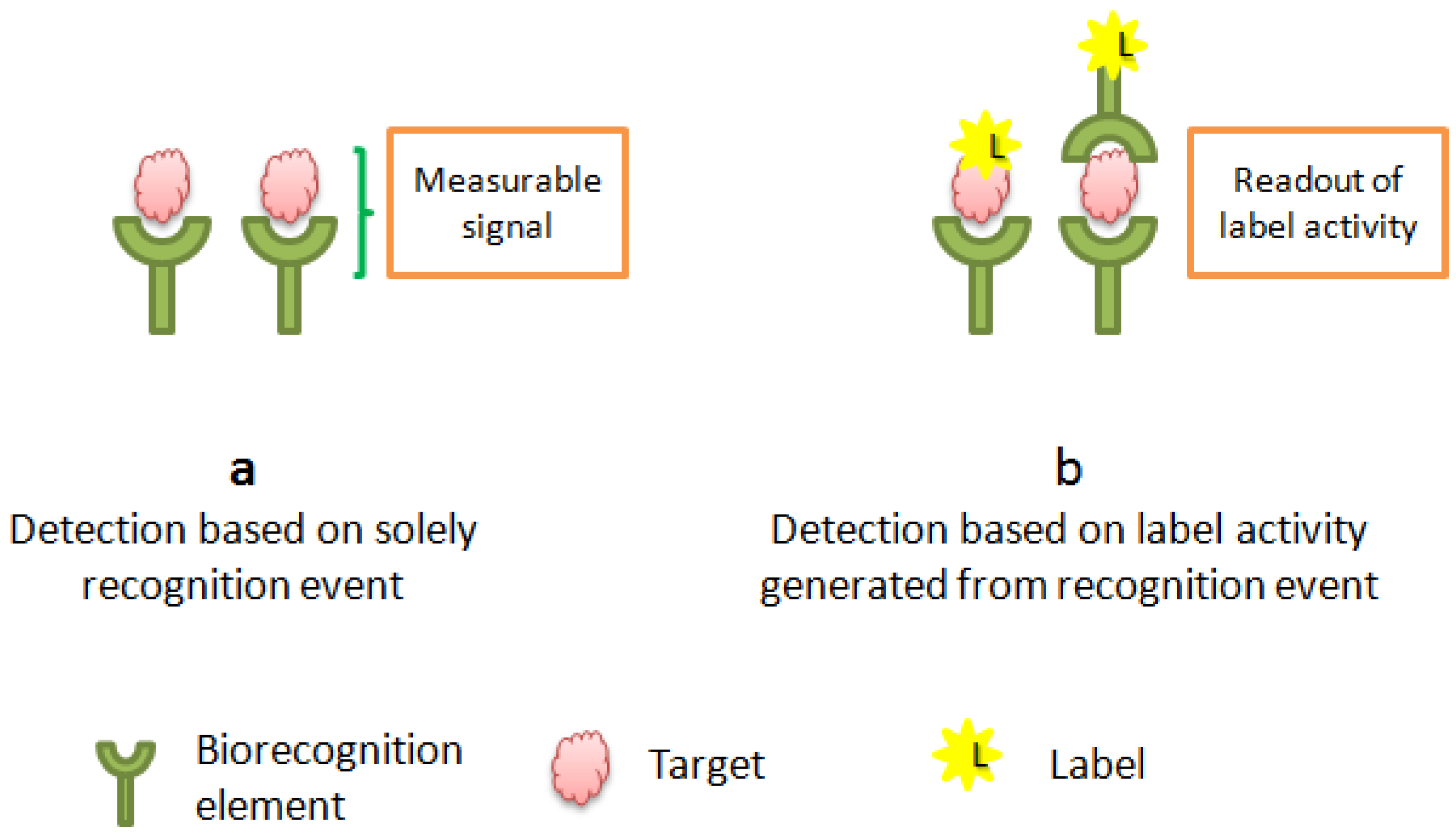
2.1. Labeled vs.Unlabeled Screening of Biomolecular Interactions
2.2. Label Free Detection Mechanism
3. Aptamers in Label-Free Biosensing
3.1. Label-Free Aptasensing Formats
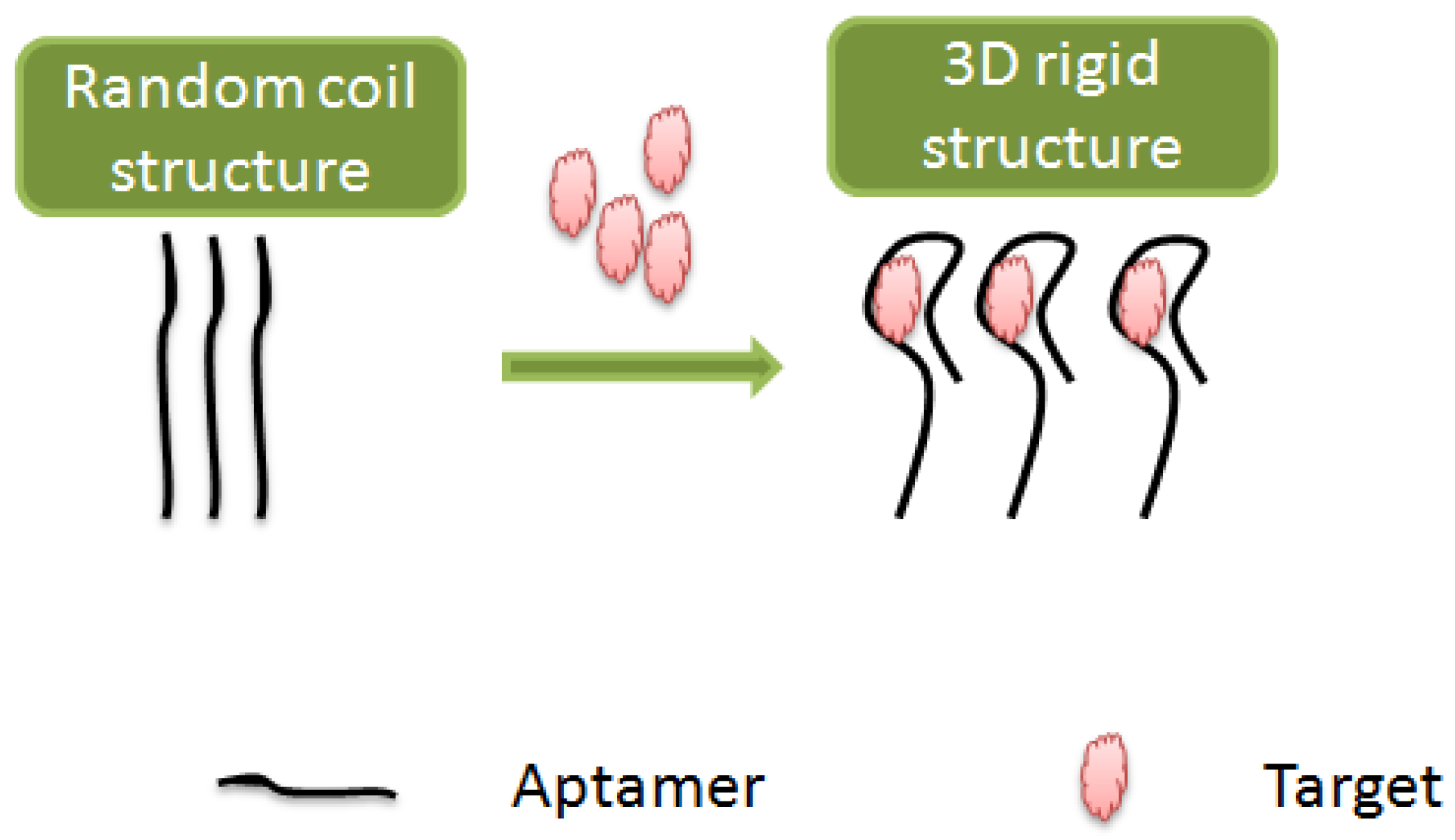
3.1.1. Structure Switchable Aptamer Assays
3.1.2. Aptamer Construct Assembly/Disassembly Based Assays
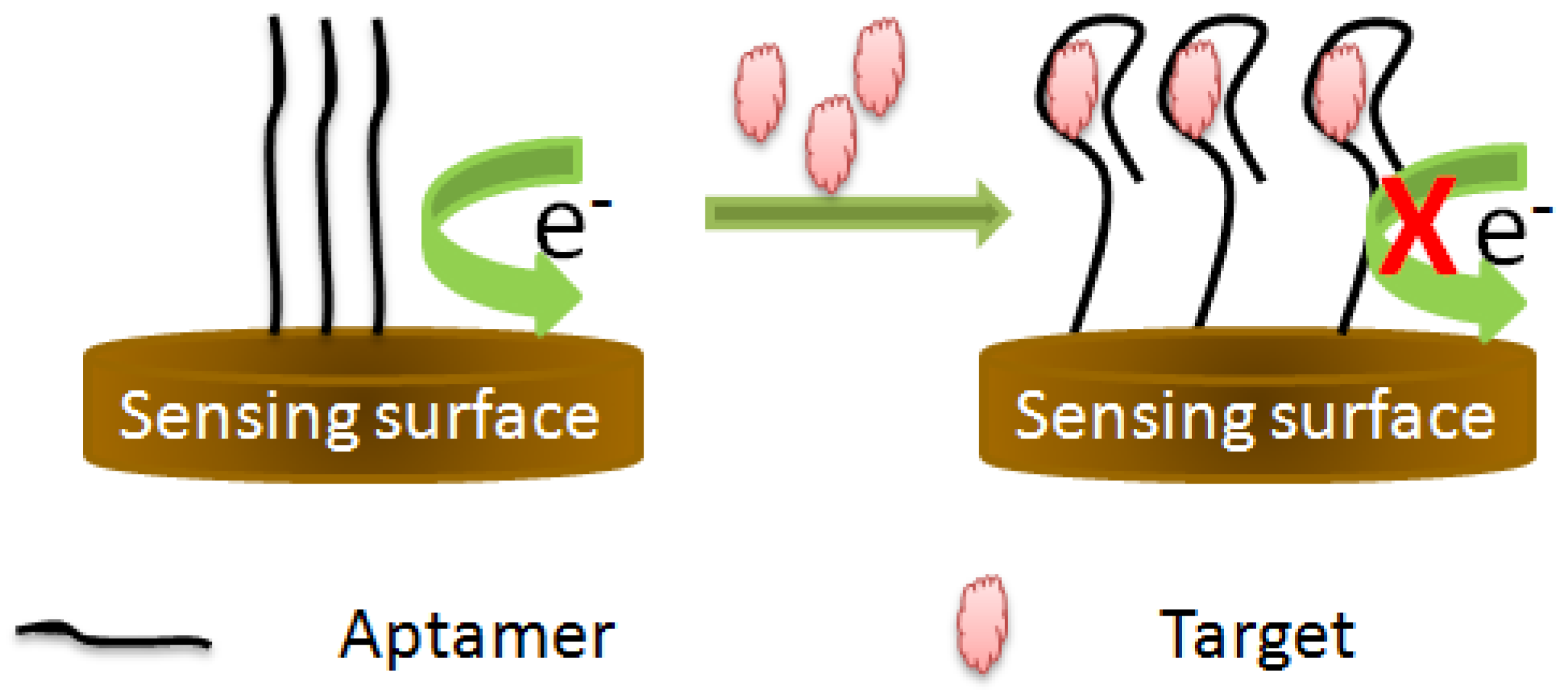
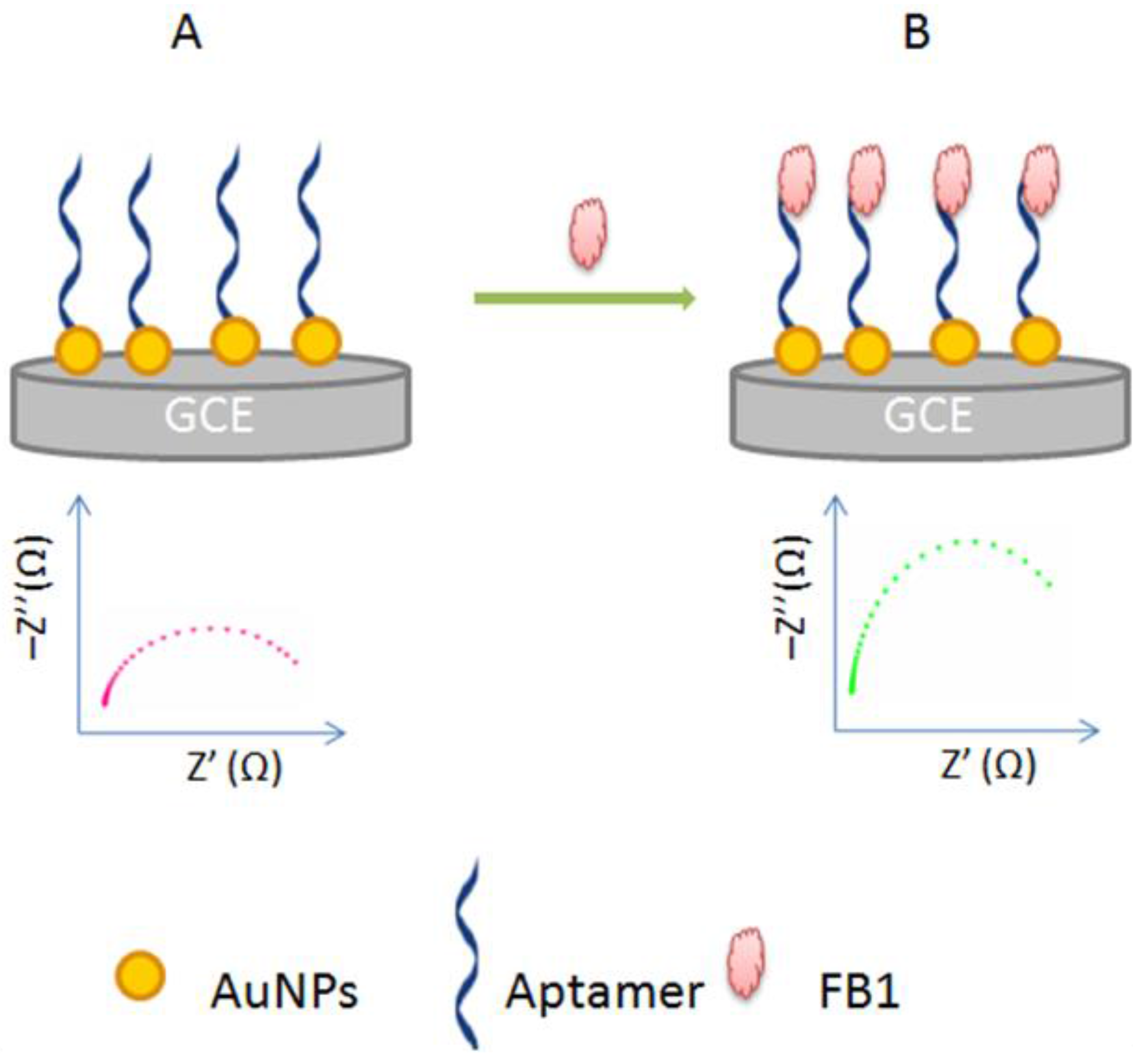
3.1.3. Target-Induced Variation in Charge Transfer Resistance
3.2. Aptamer Immobilization Techniques
3.3. Detection Methodologies in Label-Free Aptasensing
3.3.1. Optical Detection
Colorimetric Assays
Fluorescent Assays
3.3.2. Electrochemical Detection
3.3.3. Mechanical Detection
4. Label-Free Aptasensors for Myctotoxin Determination
4.1. Label-Free Aptasensors for OTA Detection
4.2. Label-Free Aptasensors for Aflatoxins Detection
4.3. Label-Free Aptasensors for Other Mycotoxins
5. Limitations and Challenges of Aptasensors
6. Conclusions and Future Prospects
Conflicts of Interest
References
- Mejri Omrani, N.; Hayat, A.; Korri-Youssoufi, H.; Marty, J.L. Electrochemical biosensors for food security: Mycotoxins detection. In Biosensors for Security and Bioterrorism Applications; Nikolelis, D.P., Nikoleli, G.-P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 469–490. [Google Scholar]
- Bazin, I.; Tria, S.A.; Hayat, A.; Marty, J.L. New biorecognition molecules in biosensors for the detection of toxins. Biosens. Bioelectron. 2016, 87, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Lehninger, A.L. Biochemistry, 2nd ed.; Worth Publishers: New York, NY, USA, 1977. [Google Scholar]
- Byfield, M.P.; Abuknesha, R.A. Biochemical aspects of biosensors. Biosens. Bioelectron. 1994, 9, 373–400. [Google Scholar] [CrossRef]
- Catanante, G.; Rhouati, A.; Hayat, A.; Marty, J.L. An overview of recent electrochemical immunosensing strategies for mycotoxins detection. Electroanalysis 2016, 28, 1750–1763. [Google Scholar] [CrossRef]
- Hammers, C.M.; Stanley, J.R. Antibody phage display: Technique and applications. J. Investig. Dermatol. 2014, 134, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Hayat, A.; Paniel, N.; Rhouati, A.; Marty, J.-L.; Barthelmebs, L. Recent advances in ochratoxin a-producing fungi detection based on pcr methods and ochratoxin a analysis in food matrices. Food Control 2012, 26, 401–415. [Google Scholar] [CrossRef]
- Guo, X.; Wen, F.; Zheng, N.; Luo, Q.; Wang, H.; Wang, H.; Li, S.; Wang, J. Development of an ultrasensitive aptasensor for the detection of aflatoxin B 1. Biosens. Bioelectron. 2014, 56, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.T.; Fung, C.W.; Cho, E.J.; Ellington, A.D. Real-Time rolling circle amplification for protein detection. Anal. Chem. 2007, 79, 3320–3329. [Google Scholar] [CrossRef] [PubMed]
- Weizmann, Y.; Beissenhirtz, M.K.; Cheglakov, Z.; Nowarski, R.; Kotler, M.; Willner, I. A virus spotlighted by an autonomous DNA machine. Angew. Chem. Int. Ed. Engl. 2006, 45, 7384–7388. [Google Scholar] [CrossRef] [PubMed]
- Patolsky, F.; Katz, E.; Willner, I. Amplified DNA detection by electrogenerated biochemiluminescence and by the catalyzed precipitation of an insoluble product on electrodes in the presence of the doxorubicin intercalator. Angew. Chem. Int. Ed. Engl. 2002, 41, 3398–3402. [Google Scholar] [CrossRef]
- Deng, C.Y.; Chen, J.H.; Nie, Z.; Wang, M.D.; Chu, X.C.; Chen, X.L.; Xiao, X.L.; Lei, C.Y.; Yao, S.Z. Impedimetric aptasensor with femtomolar sensitivity based on the enlargement of surface-charged gold nanoparticles. Anal. Chem. 2009, 81, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-Functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef] [PubMed]
- Schmitteckert, E.M.; Schlicht, H.-J. Detection of the human hepatitis b virus x-protein in transgenic mice after radioactive labelling at a newly introduced phosphorylation site. J. Gen. Virol. 1999, 80, 2501–2509. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.-J.; Wang, Z.-P. An aptamer-based fluorescence assay for ochratoxin A. Chin. J. Anal. Chem. 2011, 39, 300–304. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Y.; Marty, J.L.; Yang, X. Aptamer-Based colorimetric biosensing of ochratoxin a using unmodified gold nanoparticles indicator. Biosens. Bioelectron. 2011, 26, 2724–2727. [Google Scholar] [CrossRef] [PubMed]
- Rhouati, A.; Hayat, A.; Hernandez, D.B.; Meraihi, Z.; Munoz, R.; Marty, J.-L. Development of an automated flow-based electrochemical aptasensor for on-line detection of ochratoxin A. Sens. Actuators B Chem. 2013, 176, 1160–1166. [Google Scholar] [CrossRef]
- Daniels, J.S.; Pourmand, N. Label-Free impedance biosensors: Opportunities and challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dong, S.; Wang, E. Homogeneous analysis: Label-free and substrate-free aptasensors. Chem.–An Asian J. 2010, 5, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Hunt, H.K.; Armani, A.M. Label-Free biological and chemical sensors. Nanoscale 2010, 2, 1544–1559. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; Mayorga-Martinez, C.C.; Quesada-González, D.; Zamora-Gálvez, A.; de la Escosura-Muñiz, A.; Merkoçi, A. Label-Free impedimetric aptasensor for ochratoxin-a detection using iridium oxide nanoparticles. Anal. Chem. 2015, 87, 5167–5172. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Davis, J.J. Electrical biosensors and the label free detection of protein disease biomarkers. Chem. Soc. Rev. 2013, 42, 5944–5962. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhan, S.; Xu, L.; Shi, W.; Xi, T.; Zhan, X.; Zhou, P. A simple and label-free sensor for mercury (ii) detection in aqueous solution by malachite green based on a resonance scattering spectral assay. Chem. Commun. 2011, 47, 6027–6029. [Google Scholar] [CrossRef] [PubMed]
- Bülbül, G.; Hayat, A.; Andreescu, S. Ssdna functionalized nanoceria: A redox active aptaswitch for biomolecular recognition. Adv. Healthc. Mater. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D. In vitro selection of rna molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Rhouati, A.; Yang, C.; Hayat, A.; Marty, J.-L. Aptamers: A promising tool for ochratoxin a detection in food analysis. Toxins 2013, 5, 1988–2008. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, B.; Wang, E. “Fitting” makes “sensing” simple: Label-Free detection strategies based on nucleic acid aptamers. Acc. Chem. Res. 2012, 46, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Lakshmipriya, T.; Awazu, K. Colorimetric detection of controlled assembly and disassembly of aptamers on unmodified gold nanoparticles. Biosens. Bioelectron. 2014, 51, 115–123. [Google Scholar] [CrossRef] [PubMed]
- De-los-Santos-Álvarez, N.; Lobo-Castañón, M.A.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Aptamers as recognition elements for label-free analytical devices. TrAC Trends Anal. Chem. 2008, 27, 437–446. [Google Scholar] [CrossRef]
- Citartan, M.; Gopinath, S.C.; Tominaga, J.; Tan, S.-C.; Tang, T.-H. Assays for aptamer-based platforms. Biosens. Bioelectron. 2012, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Neumann, O.; Zhang, D.; Tam, F.; Lal, S.; Wittung-Stafshede, P.; Halas, N.J. Direct optical detection of aptamer conformational changes induced by target molecules. Anal. Chem. 2009, 81, 10002–10006. [Google Scholar] [CrossRef] [PubMed]
- Hayat, A.; Yang, C.; Rhouati, A.; Marty, J.L. Recent advances and achievements in nanomaterial-based, and structure switchable aptasensing platforms for ochratoxin a detection. Sensors 2013, 13, 15187–15208. [Google Scholar] [CrossRef] [PubMed]
- Polsky, R.; Gill, R.; Kaganovsky, L.; Willner, I. Nucleic acid-functionalized pt nanoparticles: Catalytic labels for the amplified electrochemical detection of biomolecules. Anal. Chem. 2006, 78, 2268–2271. [Google Scholar] [CrossRef] [PubMed]
- Nutiu, R.; Li, Y. Structure-Switching signaling aptamers: Transducing molecular recognition into fluorescence signaling. Chem.–A Eur. J. 2004, 10, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.C.; Kawde, A.-N.; Wang, J. Aptamer biosensor for label-free impedance spectroscopy detection of proteins based on recognition-induced switching of the surface charge. Chem. Commun. 2005, 34, 4267–4269. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-J.; Nie, H.-G.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Electrochemical sensor for mercury (ii) based on conformational switch mediated by interstrand cooperative coordination. Anal. Chem. 2009, 81, 5724–5730. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.K.; Sen, D.; Yu, H.-Z. Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry 2009, 77, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.-A.; Leclerc, M. Optical sensors based on hybrid aptamer/conjugated polymer complexes. J. Am. Chem. Soc. 2004, 126, 1384–1387. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Sun, C.; Kang, Y.; Chen, J.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Label-Free electrochemical detection of nanomolar adenosine based on target-induced aptamer displacement. Electrochem. Commun. 2008, 10, 531–535. [Google Scholar] [CrossRef]
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heeger, A.J. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991. [Google Scholar] [CrossRef] [PubMed]
- He, J.-L.; Wu, Z.-S.; Zhou, H.; Wang, H.-Q.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal. Chem. 2010, 82, 1358–1364. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, D.; Li, Y.; Qi, H.; Gao, Q.; Zhang, C. Label-Free and sensitive faradic impedance aptasensor for the determination of lysozyme based on target-induced aptamer displacement. Biosens. Bioelectron. 2009, 25, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Willner, I.; Zayats, M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. Engl. 2007, 46, 6408–6418. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-D.; Zhao, H.-T.; Chen, Z.-B.; Mu, X.-J.; Guo, L. Aptamer biosensor for label-free impedance spectroscopy detection of thrombin based on gold nanoparticles. Sens. Actuators B Chem. 2011, 157, 189–194. [Google Scholar] [CrossRef]
- Zhang, X.; Yadavalli, V.K. Surface immobilization of DNA aptamers for biosensing and protein interaction analysis. Biosens. Bioelectron. 2011, 26, 3142–3147. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, S.; Obubuafo, A.; Soper, S.A.; Spivak, D.A. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2008, 390, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Samanta, D.; Sarkar, A. Immobilization of bio-macromolecules on self-assembled monolayers: Methods and sensor applications. Chem. Soc. Rev. 2011, 40, 2567–2592. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y. Label-Free cell-based assays with optical biosensors in drug discovery. Assay Drug Dev. Technol. 2006, 4, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, J.; Zhao, S.; Shi, M.; Chen, Z.-F.; Liang, H. Label-Free colorimetric aptasensor based on nicking enzyme assisted signal amplification and dnazyme amplification for highly sensitive detection of protein. Anal. Chem. 2013, 85, 4423–4430. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Dong, S.; Wang, E. Label-Free colorimetric detection of aqueous mercury ion (Hg2+) using Hg2+-modulated g-quadruplex-based dnazymes. Anal. Chem. 2009, 81, 2144–2149. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lates, V.; Prieto-Simon, B.; Marty, J.L.; Yang, X. Aptamer-Dnazyme hairpins for biosensing of ochratoxin a. Biosens. Bioelectron. 2012, 32, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Kerman, K.; Tamiya, E. An overview of label-free electrochemical protein sensors. Sensors 2007, 7, 3442–3458. [Google Scholar] [CrossRef]
- Chang, B.-Y.; Park, S.-M. Electrochemical impedance spectroscopy. Ann. Rev. Anal. Chem. 2010, 3, 207–229. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, J.; Kosaka, P.M.; Ruz, J.J.; San Paulo, Á.; Calleja, M. Biosensors based on nanomechanical systems. Chem. Soc. Rev. 2013, 42, 1287–1311. [Google Scholar] [CrossRef] [PubMed]
- Raiteri, R.; Grattarola, M.; Butt, H.-J.; Skládal, P. Micromechanical cantilever-based biosensors. Sens. Actuators B Chem. 2001, 79, 115–126. [Google Scholar] [CrossRef]
- Janshoff, A.; Galla, H.J.; Steinem, C. Piezoelectric mass-sensing devices as biosensors—An alternative to optical biosensors? Angew. Chem. Int. Ed. Engl. 2000, 39, 4004–4032. [Google Scholar] [CrossRef]
- Zain, M.E. Impact of mycotoxins on humans and animals. J. Saudi Chem. Soc. 2011, 15, 129–144. [Google Scholar] [CrossRef]
- Rai, M.; Jogee, P.S.; Ingle, A.P. Emerging nanotechnology for detection of mycotoxins in food and feed. Int. J. Food Sci. Nutr. 2015, 66, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lates, V.; Prieto-Simón, B.; Marty, J.-L.; Yang, X. Rapid high-throughput analysis of ochratoxin a by the self-assembly of dnazyme–aptamer conjugates in wine. Talanta 2013, 116, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dong, X.; Liu, Q.; Wang, K. Label-Free colorimetric aptasensor for sensitive detection of ochratoxin a utilizing hybridization chain reaction. Anal. Chim. Acta 2015, 860, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Yang, G.; Chen, J.; Wang, S. A signal-on fluorescent aptasensor based on Tb3+ and structure-switching aptamer for label-free detection of ochratoxin a in wheat. Biosens. Bioelectron. 2013, 41, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Chen, A.; Liu, J.; Guan, Z.; Zhou, Y.; Xu, S.; Yang, S.; Li, C. A simple and sensitive approach for ochratoxin a detection using a label-free fluorescent aptasensor. PLoS ONE 2014, 9, e85968. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Velu, R.; Hill, K.; Bardóczy, V.; Mészáros, T.; DeRosa, M. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 2014, 6, 2435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Wang, M.; Liu, L.-J.; Leung, C.-H.; Ma, D.-L. Label-Free luminescent switch-on probe for ochratoxin a detection using a g-quadruplex-selective iridium (iii) complex. ACS Appl. Mater. Interfaces 2015, 7, 8313–8318. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Byun, J.-Y.; Mun, H.; Shim, W.-B.; Shin, Y.-B.; Li, T.; Kim, M.-G. A regeneratable, label-free, localized surface plasmon resonance (lspr) aptasensor for the detection of ochratoxin A. Biosens. Bioelectron. 2014, 59, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.; Matharu, Z.; Malhotra, B.D. Polyaniline langmuir-blodgett film based aptasensor for ochratoxin a detection. Biosens. Bioelectron. 2011, 26, 4006–4011. [Google Scholar] [CrossRef] [PubMed]
- Castillo, G.; Lamberti, I.; Mosiello, L.; Hianik, T. Impedimetric DNA aptasensor for sensitive detection of ochratoxin a in food. Electroanalysis 2012, 24, 512–520. [Google Scholar] [CrossRef]
- Hayat, A.; Sassolas, A.; Marty, J.-L.; Radi, A.-E. Highly sensitive ochratoxin a impedimetric aptasensor based on the immobilization of azido-aptamer onto electrografted binary film via click chemistry. Talanta 2013, 103, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Hayat, A.; Catanante, G.; Ocaña, C.; Marty, J.-L. A label free aptasensor for ochratoxin A detection in cocoa beans: An application to chocolate industries. Anal. Chim. Acta 2015, 889, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Hayat, A.; Andreescu, S.; Marty, J.-L. Design of peg-aptamer two piece macromolecules as convenient and integrated sensing platform: Application to the label free detection of small size molecules. Biosens. Bioelectron. 2013, 45, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Hayat, A.; Haider, W.; Rolland, M.; Marty, J.-L. Electrochemical grafting of long spacer arms of hexamethyldiamine on a screen printed carbon electrode surface: Application in target induced ochratoxin a electrochemical aptasensor. Analyst 2013, 138, 2951–2957. [Google Scholar] [CrossRef] [PubMed]
- Evtugyn, G.; Porfireva, A.; Stepanova, V.; Kutyreva, M.; Gataulina, A.; Ulakhovich, N.; Evtugyn, V.; Hianik, T. Impedimetric aptasensor for ochratoxin a determination based on au nanoparticles stabilized with hyper-branched polymer. Sensors 2013, 13, 16129–16145. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Hayat, A.; Andreescu, S. Biomolecular detection at ssdna-conjugated nanoparticles by nano-impact electrochemistry. Biosens. Bioelectron. 2017, 87, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Chen, J.; Xie, G.; Li, C.; Ping, H.; Ma, Z.; Lu, A. Visual and microplate detection of aflatoxin b2 based on nacl-induced aggregation of aptamer-modified gold nanoparticles. Microchim. Acta 2015, 182, 995–1001. [Google Scholar] [CrossRef]
- Luan, Y.; Chen, Z.; Xie, G.; Chen, J.; Lu, A.; Li, C.; Fu, H.; Ma, Z.; Wang, J. Rapid visual detection of aflatoxin b1 by label-free aptasensor using unmodified gold nanoparticles. J. Nanosci. Nanotechnol. 2015, 15, 1357–1361. [Google Scholar] [CrossRef] [PubMed]
- Seok, Y.; Byun, J.-Y.; Shim, W.-B.; Kim, M.-G. A structure-switchable aptasensor for aflatoxin b1 detection based on assembly of an aptamer/split dnazyme. Anal. Chim. Acta 2015, 886, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Shim, W.-B.; Mun, H.; Joung, H.-A.; Ofori, J.A.; Chung, D.-H.; Kim, M.-G. Chemiluminescence competitive aptamer assay for the detection of aflatoxin b1 in corn samples. Food Control 2014, 36, 30–35. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, X.; Cai, S.; Wu, D.; Chen, M.; Wang, S.; Zhang, J. A fluorescent aptasensor based on DNA-scaffolded silver-nanocluster for ochratoxin a detection. Biosens. Bioelectron. 2014, 57, 226–231. [Google Scholar] [CrossRef] [PubMed]
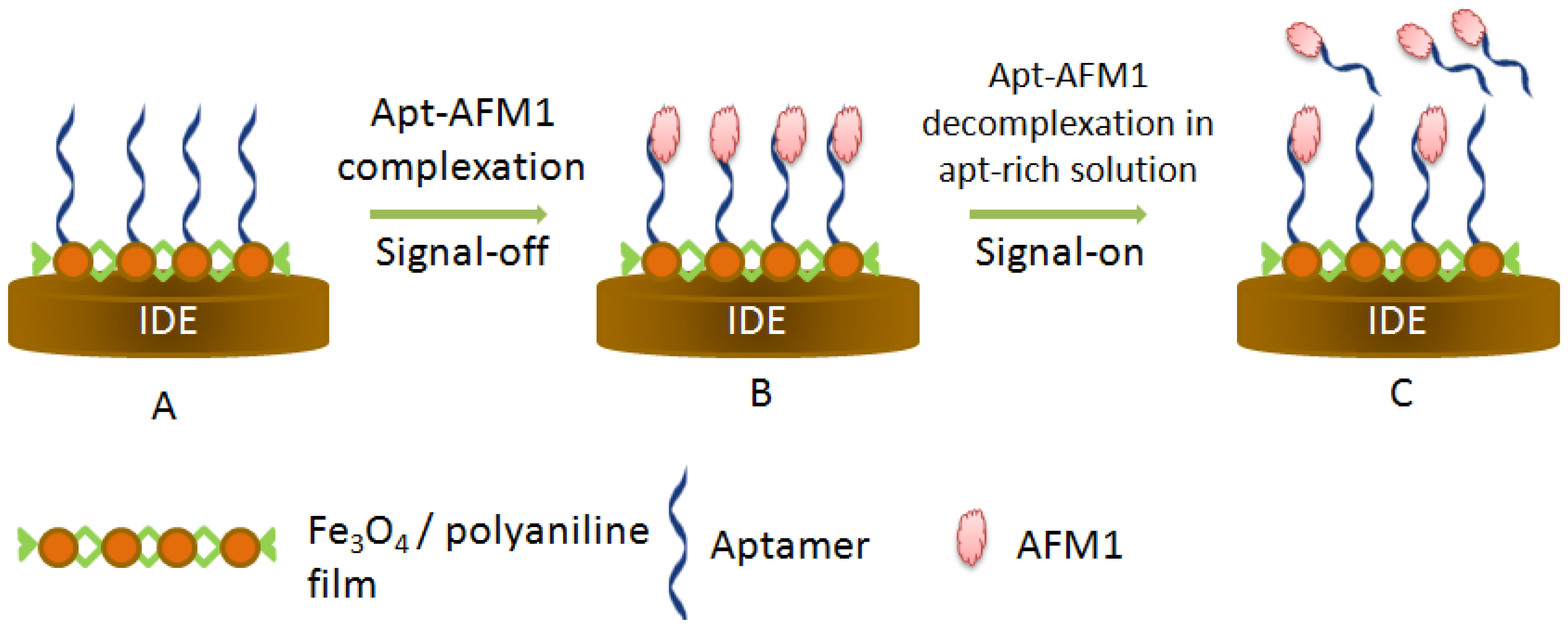
- Nguyen, B.H.; Dai Tran, L.; Do, Q.P.; Le Nguyen, H.; Tran, N.H.; Nguyen, P.X. Label-Free detection of aflatoxin m1 with electrochemical Fe3O4/polyaniline-based aptasensor. Mater. Sci. Eng. C 2013, 33, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Istamboulié, G.; Paniel, N.; Zara, L.; Granados, L.R.; Barthelmebs, L.; Noguer, T. Development of an impedimetric aptasensor for the determination of aflatoxin m1 in milk. Talanta 2016, 146, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Goud, K.Y.; Catanante, G.; Hayat, A.; Satyanarayana, M.; Gobi, K.V.; Marty, J.L. Disposable and portable electrochemical aptasensor for label free detection of aflatoxin b1 in alcoholic beverages. Sens. Actuators B Chem. 2016, 235, 466–473. [Google Scholar] [CrossRef]
- Castillo, G.; Spinella, K.; Poturnayová, A.; Šnejdárková, M.; Mosiello, L.; Hianik, T. Detection of aflatoxin b1 by aptamer-based biosensor using pamam dendrimers as immobilization platform. Food Control 2015, 52, 9–18. [Google Scholar] [CrossRef]
- Chen, X.; Huang, Y.; Ma, X.; Jia, F.; Guo, X.; Wang, Z. Impedimetric aptamer-based determination of the mold toxin fumonisin b1. Microchim. Acta 2015, 182, 1709–1714. [Google Scholar] [CrossRef]
- Chen, X.; Bai, X.; Li, H.; Zhang, B. Aptamer-Based microcantilever array biosensor for detection of fumonisin b-1. RSC Adv. 2015, 5, 35448–35452. [Google Scholar] [CrossRef]
- Amaya-González, S.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.; Lobo-Castañón, M. Aptamer-Based analysis: A promising alternative for food safety control. Sensors 2013, 13, 16292–16311. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin a with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef] [PubMed]
- Le, L.C.; Cruz-Aguado, J.A.; Penner, G.A. Dna ligands for aflatoxin and zearalenone. U.S. Patent 13/391,426, 6 September 2012. [Google Scholar]
- Ma, X.; Wang, W.; Chen, X.; Xia, Y.; Duan, N.; Wu, S.; Wang, Z. Selection, characterization and application of aptamers targeted to aflatoxin b2. Food Control 2015, 47, 545–551. [Google Scholar] [CrossRef]
- Malhotra, S.; Pandey, A.; Rajput, Y.; Sharma, R. Selection of aptamers for aflatoxin m1 and their characterization. Jo. Mol. Recognit. 2014, 27, 493–500. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Bradley, C.R.; Girolamo, A.D.; Visconti, A.; Miller, J.D.; DeRosa, M.C. Screening and initial binding assessment of fumonisin b1 aptamers. Int. J. Mol. Sci. 2010, 11, 4864–4881. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Xia, Y.; Ma, X.; Zhu, C.; Jiang, Y.; Ding, Z.; Wang, Z. Selection and characterization of single stranded DNA aptamers recognizing fumonisin b1. Microchim. Acta 2014, 181, 1317–1324. [Google Scholar] [CrossRef]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Ma, X.; Xia, Y.; Zhu, C.; Jiang, Y.; Wang, Z. Selection and identification of ssdna aptamers recognizing zearalenone. Anal. Bioanal. Chem. 2013, 405, 6573–6581. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; Smith, M.; Goudreau, D.N.; DeRosa, M.C. Current status and future prospects for aptamer-based mycotoxin detection. J. AOAC Int. 2016, 99, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2016. [Google Scholar] [CrossRef] [PubMed]
- Mascini, M. Aptamers in Bioanalysis; Wiley: Florence, Italy, 2009. [Google Scholar]
- Eaton, B.E.; Gold, L.; Hicke, B.J.; Janjié, N.; Jucker, F.M.; Sebesta, D.P.; Tarasow, T.M.; Willis, M.C.; Zichi, D.A. Post-selex combinatorial optimization of aptamers. Bioorgan. Med. Chem. 1997, 5, 1087–1096. [Google Scholar] [CrossRef]


| Technique | Principle | Advantages | Limitations | Example of Bioconjugation |
|---|---|---|---|---|
| Physical adsorption | Electrostatic forces Van der Waals interactions | Simple and rapid | Weak attachment Random orientation of aptamers | Direct attachment on metals surfaces and surfaces coated with hydrophobic polymers |
| Covalent attachment | Interactions between the surface functional groups and aptamer’s chemical groups | Wide range of functional groups Flexibility | Multiple conjugation steps Non specific binding | NHS ester chemistry Click chemistry |
| SAMs | Amphiphilic molecules: hydrophilic and hydrophobic groups with respective affinity to the transducer and the aptamer | Stability Oriented recognition | More suitable with silicon and gold surfaces | Thiols and alkyne disulfides on gold Alcohols on glass |
| Mycotoxin | Detection | Assay Principle | Linear Range (µg/L) | LOD (µg/L) | Sample | Ref |
|---|---|---|---|---|---|---|
| OTA | Colorimetric | HRP mimicking DNAzyme | 3.6–120 | 12 | Wine | [62] |
| Colorimetric | HRP mimicking DNAzyme, Hybridization chain reaction | 0.004–0.96 | 0.004 | Yellow rice, wine, wheat flour | [63] | |
| Colorimetric | Structure switching aptamer Nanoceria | 0.08–12 | 0.06 | Milk | [25] | |
| Fluorescence | Structure switching aptamer Tb3+, magnetic sepatation | 0.1–1 | 0.02 | Wheat | [64] | |
| fluorescence | Structure switching aptamer Pico green dye | 1–100,000 | 1 | Beer | [65] | |
| Fluorescence | SYBR green dye | 3.6–40 | 3.6 | ------- | [66] | |
| Luminescence | Structure switching aptamer Iridium(III) | 2–60 | 2 | ------- | [67] | |
| LSPR | Structure switching aptamer Red shift of LSPR band | 0.4–400 | 0.4 | Ground corn | [68] | |
| EIS | OTA-induced change in Rct | 0.1–10 | 0.1 | -------- | [69] | |
| EIS | OTA-induced change in Rct [Fe(CN)6]−3/−4 | 0.04–40 | 0.048 | Coffee, flour, wine | [70] | |
| EIS | OTA-induced change in Rct [Fe(CN)6]−3/−4 | 1.25 ×10−3–0.5 | 0.25 × 10−3 | Beer | [71] | |
| EIS | OTA-induced change in Rct [Fe(CN)6]−3/−4 | 0.15–2.5 | 0.15 | Cocoa | [72] | |
| EIS | Structure switching aptamer OTA-induced change in Rct | 0.12 ×10−3–5.5 ×10−3 | 0.12 × 10−3 | Beer | [73] | |
| CV | Structure switching aptamer OTA-induced change in Rct | 0.12–8.5 | 0.1 | Beer | [74] | |
| EIS | Structure switching aptamer OTA-induced change in Rct | 0.04–40 | 0.008 | Beer | [75] | |
| EIS | Structure switching aptamer OTA-induced change in Rct | 0.004–40 | 0.0056 | Wine | [22] | |
| Nano-impact electrochemistry | Structure switching aptamer OTA-induced collision frequency changes | 0.028–4 | 0.02 | ------ | [76] | |
| AFB2 | Colorimetric | Structure switching aptamer AuNPs aggregation | 0.025–10 | 0.025 | Beer | [77] |
| AFB1 | Colorimetric | Structure switching aptamer AuNPs aggregation | 0.025–100 | 0.025 | ----- | [78] |
| AFB1 | Colorimetric | HRP mimicking DNAzyme | 0.1–1.0 × 104 | 0.054 | Ground corn | [79] |
| AFB1 | Chemiluminescence | HRP mimicking DNAzyme | 0.1–10 | 0.11 | Corn | [80] |
| OTA and AFB1 | Fluorescence | Target-induced strand displacement DNA-scaffolded silver nanoculsters | 0.001–0.05 | 0.0002 and 0.0003 | Rice, corn, wheat | [81] |
| AFM1 | CV, SWV | Target-induced blocking of chargetransfer to the electrode surface | 0.006–0.06 | 0.00198 | -------- | [82] |
| AFM1 | EIS | Target-induced change in Rct | 0.002–0.15 | 0.00115 | Milk | [83] |
| AFB1 | EIS | Target-induced change in Rct | 0.125–16 | 0.12 | Beer and wine | [84] |
| AFB1 | CV, EIS | Target-induced blocking of chargetransfer to the electrode surface | 0.03–3.125 | 0.125 | Peanuts | [85] |
| AFB1 | RT-qPCR | Target-induced strand displacement | 5 × 10−5–5 | 0.000025 | Chinese wildrye hay and infant rice cereal samples | [9] |
| FB1 | EIS | target-induced change in Rct | 72–720 × 103 | 1.44 | Maize samples | [86] |
| Microcantilever | Target-induced change in surface stress | 33 | 100–40,000 | ---------------- | [87] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rhouati, A.; Catanante, G.; Nunes, G.; Hayat, A.; Marty, J.-L. Label-Free Aptasensors for the Detection of Mycotoxins. Sensors 2016, 16, 2178. https://doi.org/10.3390/s16122178
Rhouati A, Catanante G, Nunes G, Hayat A, Marty J-L. Label-Free Aptasensors for the Detection of Mycotoxins. Sensors. 2016; 16(12):2178. https://doi.org/10.3390/s16122178
Chicago/Turabian StyleRhouati, Amina, Gaelle Catanante, Gilvanda Nunes, Akhtar Hayat, and Jean-Louis Marty. 2016. "Label-Free Aptasensors for the Detection of Mycotoxins" Sensors 16, no. 12: 2178. https://doi.org/10.3390/s16122178





