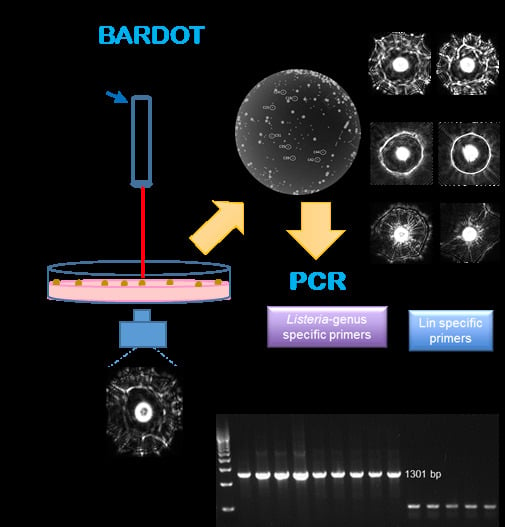
Novel PCR Assays Complement Laser Biosensor-Based Method and Facilitate Listeria Species Detection from Food
Abstract
:1. Introduction
2. Experimental Section
2.1. Bacterial Cultures, Growth and Ribotyping
2.2. Design of Lap Gene-Specific Primer Sets for Listeria Species
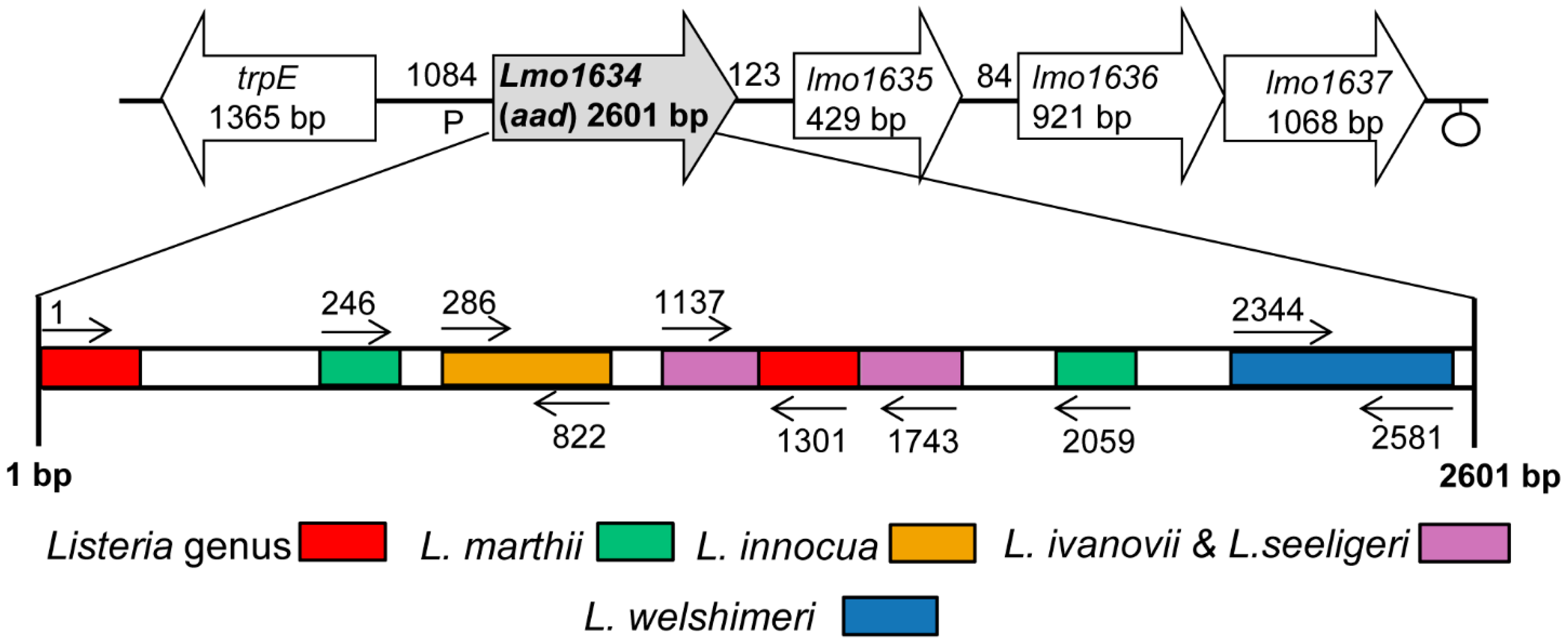
| Primer | Sequence a | Location in Lap Gene | Product Size (bp) | Specificity |
|---|---|---|---|---|
| ELAP-F1 | 5′CGGTCCCCGGGTACCATGGCAATTAAAGAAAATGCGGCC3′ | 1–1301 | 1301 | Listeria spp. (except L. grayi, L. rocourtiae) |
| LIS-R1 | 5′TTTGTGATACAGAGTTTTTACC3′ | |||
| Inn-F1 | 5′GGAGTTATTAACGAAGATACT3′ | 286–822 | 536 | L. innocua |
| Inn-R1 | 5′TTCTGCTTTTACTTCTTTAGCA3′ | |||
| IvaSee-F1 | 5′AAGCTGCAGTTATTCATTCC3′ | 1137–1743 | 606 | L. ivanovii,L. seeligeri |
| IvaSee-R1 | 5′ATCTAAGAATTTTTGTTTTAGT3′ | |||
| Wel-F1 | 5′TTCTCGTATTATCGGTTTACCA3′ | 2344–2581 | 237 | L. welshimeri |
| Wel-R1 | 5′GCTTCAAGATAGATTTCTTTCAA3′ | |||
| Mar-F1 | 5′AGAATATATTTGGAACAGCATC3′ | 246–2059 | 1813 | L. marthii |
| Mar-R1 | 5′GTTCGATTGCACGGATGGAAAG3′ |
2.3. PCR Conditions, Primers and DNA Extraction
2.4. Specificity and Sensitivity of Lap Gene Primers for Listeria Detection
2.5. Laser Optical Sensor and Scatter Image Analysis
2.6. Detection and Identification of Listeria in Artificially Inoculated Food Samples
3. Results
3.1. Specificity and Sensitivity of Lap Gene Primers for Listeria Detection


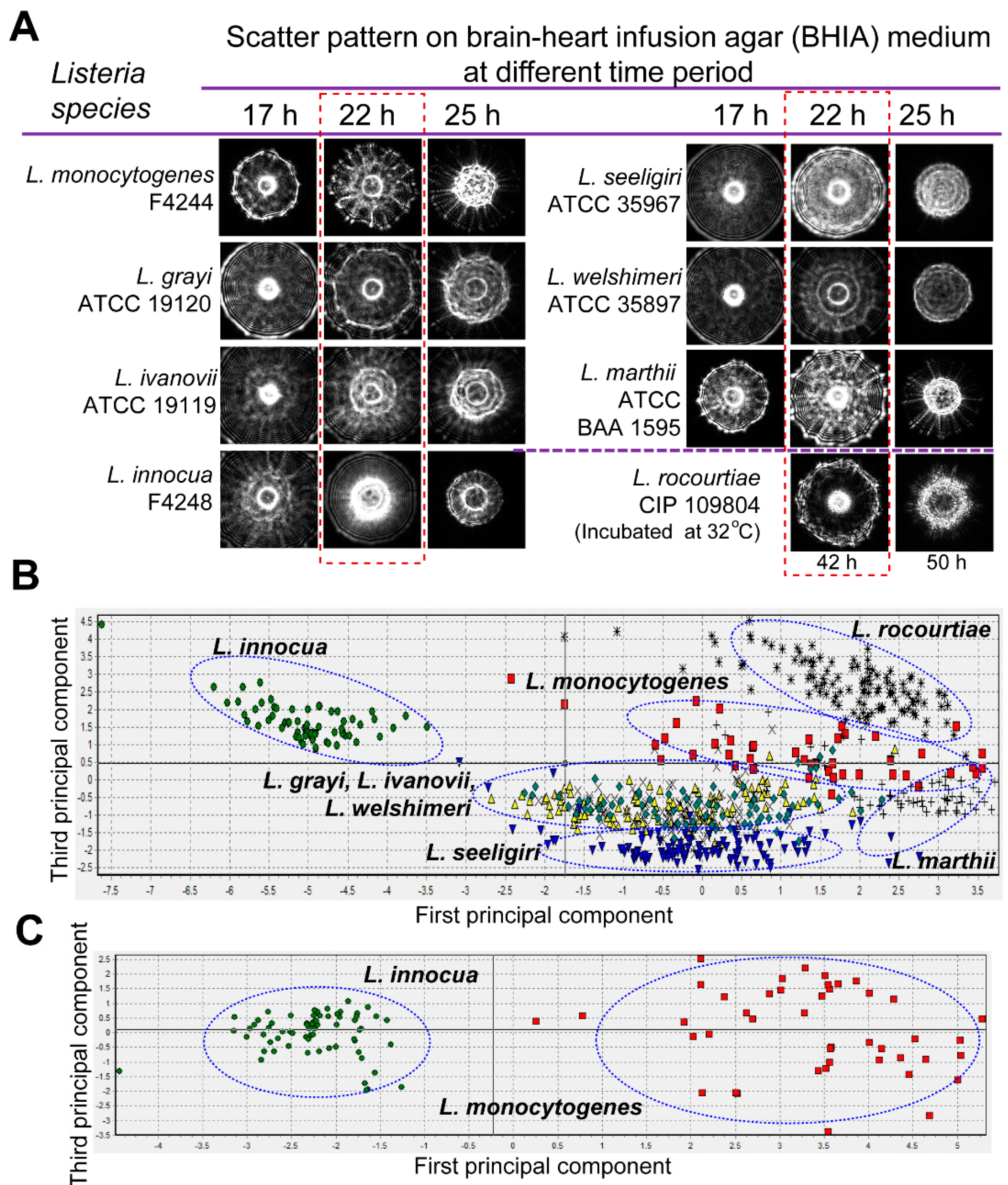
3.2. Scatter Image Library of Listeria Species and Serovars
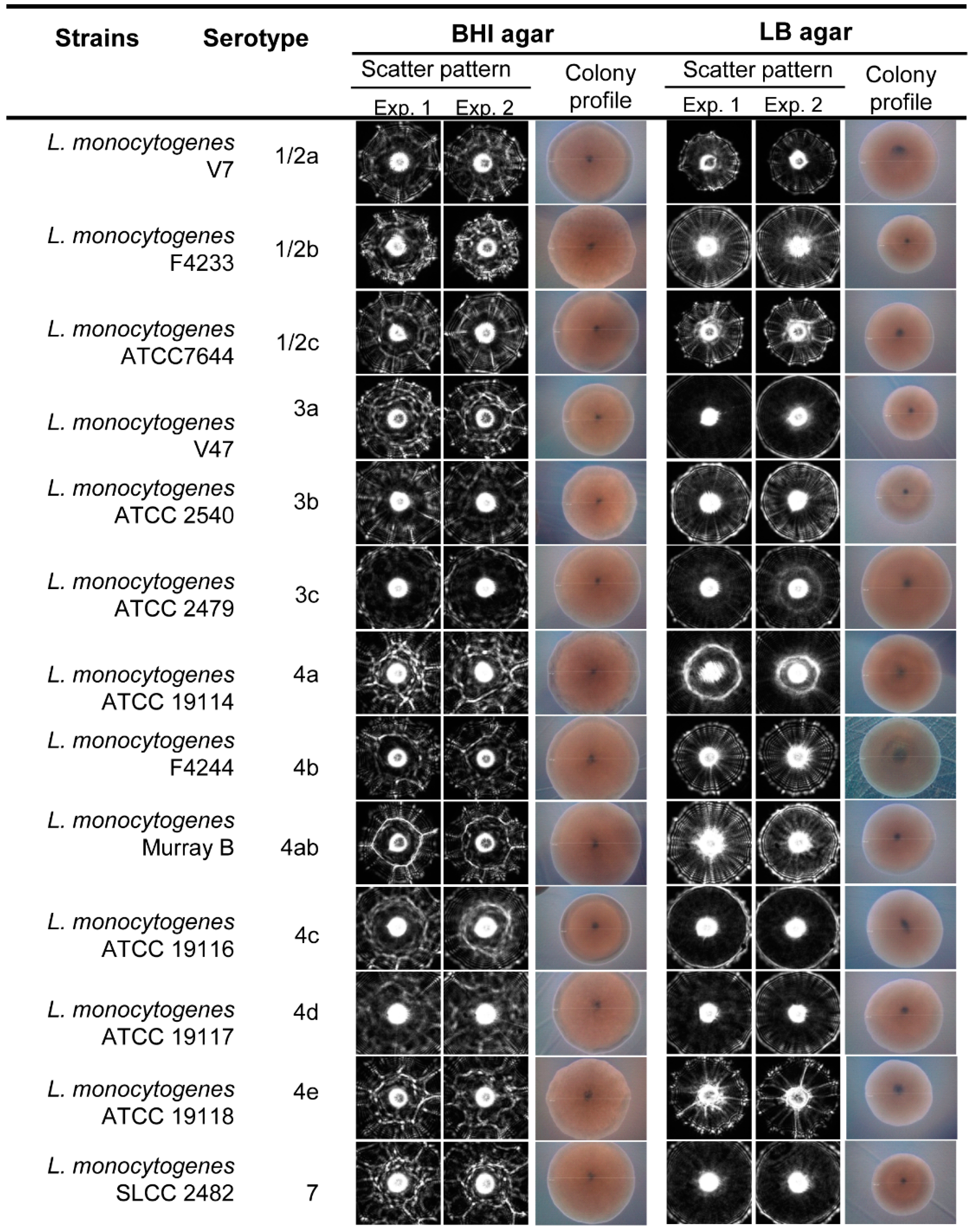
| Strains | Serotype | % Average Positive Predictive Value (PPV ± SD) | |
|---|---|---|---|
| BHI | LB | ||
| L. monocytogenes V7 | 1/2a | 81.8 ± 2.3 | 96.2 ± 1.9 |
| L. monocytogenes F4233 | 1/2b | 89.4 ± 1.1 | 96.8 ± 2.1 |
| L. monocytogenes ATCC7644 | 1/2c | 74.5 ± 3.2 | 81.3 ± 3.8 |
| L. monocytogenes V47 | 3a | 80.8 ± 1.8 | 77.6 ± 5.6 |
| L. monocytogenes ATCC 2540 | 3b | 74.2 ± 2.2 | 97.6 ± 1.7 |
| L. monocytogenes ATCC 2479 | 3c | 99.8 ± 0.9 | 85.2 ± 2.3 |
| L. monocytogenes ATCC 9114 | 4a | 93.2 ± 1.3 | 98.0 ± 1.8 |
| L. monocytogenes F4244 | 4b | 46.2 ± 3.5 | 88.8 ± 2.1 |
| L. monocytogenes Murray B | 4ab | 92.8 ± 2.9 | 85.4 ± 3.2 |
| L. monocytogenes ATCC 19116 | 4c | 98.8 ± 1.0 | 93.6 ± 2.8 |
| L. monocytogenes ATCC 19117 | 4d | 99.0 ± 0.5 | 88.2 ± 3.1 |
| L. monocytogenes ATCC 19118 | 4e | 65.4 ± 5.6 | 92.6 ± 1.7 |
| L. monocytogenes SLCC 2482 | 7 | 82.4 ± 2.4 | 90.2 ± 3.1 |
| Average precision rate | 82.9 ± 2.2 | 90.1 ± 2.7 | |
3.3. Detection and Verification of Listeria from Food Samples
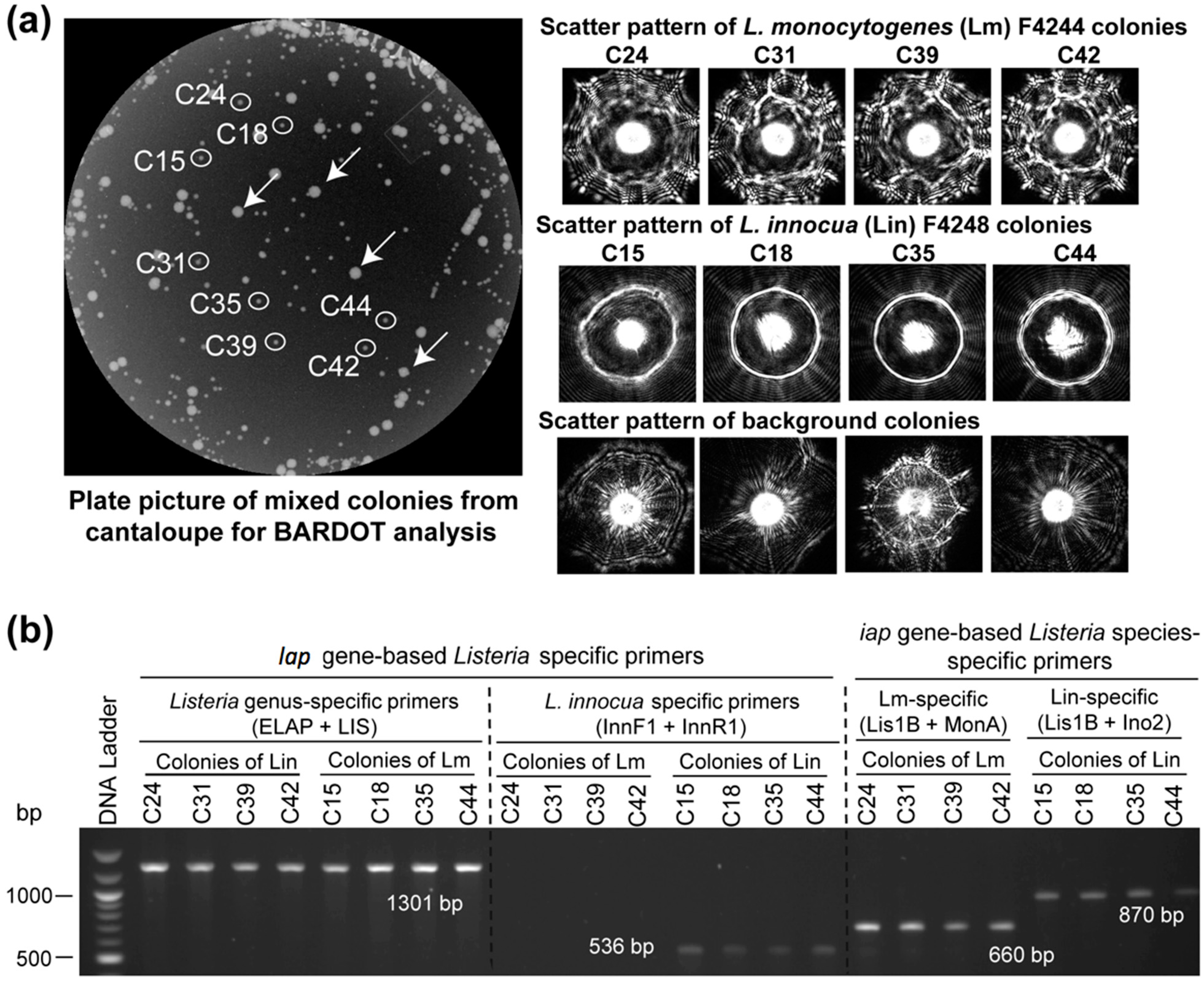
| Treatment a | Inoculation (CFU/25g) | Enrichment Time (h) in Fraser Broth at 37 °C | PCR b | |||
|---|---|---|---|---|---|---|
| Hotdog | Cantaloupe | |||||
| ELAP-F1/LIS-R1 | Inn-F1/Inn-R1 | ELAP-F1/LIS-R1 | Inn-F1/Inn-R1 | |||
| Uninoculated | 0 | 24 | − | − | − | − |
| L. innocua (Lin) | 100 | 24 | + | + | + | + |
| L. monocytogenes (Lm) | 100 | 24 | + | − | + | − |
| Lin and Lmc | 100 | 24 | + | + | + | + |
| Lb. casei | 100 | 24 | − | − | − | − |
| E. coli | 100 | 24 | − | − | − | − |
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jagadeesan, B.; Koo, O.K.; Kim, K.P.; Burkholder, K.M.; Mishra, K.K.; Aroonnual, A.; Bhunia, A.K. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 2010, 156, 2782–2795. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Fleishman Littlejohn, A.E.; Amalaradjou, M.A. R.; Singh, A.K.; Mishra, K.K.; La, D.; Kihara, D.; Bhunia, A.K. N-Terminal Gly224–Gly411 domain in Listeria adhesion protein interacts with host receptor Hsp60. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Burkholder, K.M.; Bhunia, A.K. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation, and induces expression of LAP receptor Hsp60. Infect. Immun. 2010, 78, 5062–5073. [Google Scholar] [CrossRef]
- Kim, K.P.; Jagadeesan, B.; Burkholder, K.M.; Jaradat, Z.W.; Wampler, J.L.; Lathrop, A.A.; Morgan, M.T.; Bhunia, A.K. Adhesion characteristics of Listeria adhesion protein (LAP)-expressing Escherichia coli to Caco-2 cells and of recombinant LAP to eukaryotic receptor Hsp60 as examined in a surface plasmon resonance sensor. FEMS Microbiol. Lett. 2006, 256, 324–332. [Google Scholar] [CrossRef]
- Henderson, B.; Martin, A. Bacterial moonlighting proteins and bacterial virulence. Curr. Top. Microbiol. Immunol. 2013, 358, 155–213. [Google Scholar]
- Burkholder, K.M.; Bhunia, A.K. Listeria monocytogenes and Host Hsp60—An invasive pairing. In Moonlighting Cell Stress Proteins in Microbial Infections, Heat Shok Proteins; Henderson, B., Ed.; Springer Science+Business Media: Dordrecht, Germany, 2013; pp. 267–282. [Google Scholar]
- Den Bakker, H.C.; Bundrant, B.N.; Fortes, E.D.; Orsi, R.H.; Wiedmann, M. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl. Environ. Microbiol. 2010, 76, 6085–6100. [Google Scholar] [CrossRef]
- Den Bakker, H.C.; Warchocki, S.; Wright, E.M.; Allred, A.F.; Ahlstrom, C.; Manuel, C.S.; Stasiewicz, M.J.; Burrell, A.; Roof, S.; Strawn, L.K.; et al. Listeria floridensis sp nov., Listeria aquatica sp nov., Listeria cornellensis sp nov., Listeria riparia sp nov and Listeria grandensis sp nov., from agricultural and natural environments. Int. J. Syst. Evol. Microbiol. 2014, 64, 1882–1889. [Google Scholar] [CrossRef]
- Weller, D.; Andrus, A.; Wiedmann, M.; den Bakker, H.C. Listeria booriae sp nov and Listeria newyorkensis sp nov., from food processing environments in the USA. Int. J. Syst. Evol. Microbiol. 2015, 65, 286–292. [Google Scholar] [CrossRef]
- Bertsch, D.; Rau, J.; Eugster, M.R.; Haug, M.C.; Lawson, P.A.; Lacroix, C.; Meile, L. Listeria fleischmannii sp nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 2013, 63, 526–532. [Google Scholar] [CrossRef]
- Leclercq, A.; Clermont, D.; Bizet, C.; Grimont, P.A.D.; le Fleche-Mateos, A.; Roche, S.M.; Buchrieser, C.; Cadet-Daniel, V.; le Monnier, A.; Lecuit, M.; et al. Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2210–2214. [Google Scholar] [CrossRef]
- Halter, E.L.; Neuhaus, K.; Scherer, S. Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int. J. Syst. Evol. Microbiol. 2013, 63, 641–647. [Google Scholar] [CrossRef]
- Silk, B.J.; Date, K.A.; Jackson, K.A.; Pouillot, R.; Holt, K.G.; Graves, L.M.; Ong, K.L.; Hurd, S.; Meyer, R.; Marcus, R.; et al. Invasive listeriosis in the foodborne diseases active surveillance network (FoodNet), 2004–2009: Further targeted prevention needed for higher-risk groups. Clin. Infect. Dis. 2012, 54, S396–S404. [Google Scholar] [CrossRef]
- Anonymous. 2008 Listeriosis Outbreak in Ontario Epidemiologic Summary. Available online: http://www.health.gov.on.ca/en/public/publications/disease/docs/listeriosis_outbreak_epi_sum.pdf (accessed on 26 May 2015).
- Fretz, R.; Pichler, J.; Sagel, U.; Much, P.; Ruppitsch, W.; Pietzka, A.T.; Stöger, A.; Huhulescu, S.; Heuberger, S.; Appl, G.; et al. Update: Multinational listeriosis outbreak due to “Quargel”, a sour milk curd cheese, caused by two different L. monocytogenes serotype 1/2a strains, 2009–2010. Eurosurveillance 2010, 15, 1–2. [Google Scholar]
- CDC. Multistate outbreak of listeriosis associated with Jensen Farms cantaloupe—United States, August–September 2011. MMWR. Morb. Mortal. Weekly Rep. 2011, 60, 1357–1358. [Google Scholar]
- Anonymous. Multistate Outbreak of Listeriosis Linked to Commercially Produced, Prepackaged Caramel Apples Made from Bidart Bros. Apples (Final Update). Available online: http://www.cdc.gov/listeria/outbreaks/caramel-apples-12-14/ (accessed on 2 September 2015).
- Anonymous. Multistate Outbreak of Listeriosis Linked to Blue Bell Creameries Products (Final Update). Available online: http://www.cdc.gov/listeria/outbreaks/ice-cream-03-15/ (accessed on 2 September 2015).
- Wang, S.; Orsi, R.H. Listeria. In Foodborne Infections and Intoxications, 4th ed.; Morris, J.G.J., Potter, M.E., Eds.; Elsevier: New York, NY, USA, 2013; pp. 199–216. [Google Scholar]
- FDA. Guidance for industry: Control of Listeria monocytogenes in refrigerated or frozen ready-to-eat foods; Draft guidance. In US Department of Health and Human Services: Center for Food Safety and Applied Nutrition; Rockville, MD, USA, 2008. [Google Scholar]
- Gil, R.; Silva, F.J.; Pereto, J.; Moya, A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004, 68, 518–537. [Google Scholar] [CrossRef]
- Bae, E.; Banada, P.P.; Huff, K.; Bhunia, A.K.; Robinson, J.P.; Hirleman, E.D. Biophysical modeling of forward scattering from bacterial colonies using scalar diffraction theory. Appl. Opt. 2007, 46, 3639–3648. [Google Scholar] [CrossRef]
- Banada, P.P.; Guo, S.; Bayraktar, B.; Bae, E.; Rajwa, B.; Robinson, J.P.; Hirleman, E.D.; Bhunia, A.K. Optical forward-scattering for detection of Listeria monocytogenes and other Listeria species. Biosens. Bioelectron. 2007, 22, 1664–1671. [Google Scholar] [CrossRef]
- Singh, A.K.; Bettasso, A.M.; Bae, E.; Rajwa, B.; Dundar, M.M.; Forster, M.D.; Liu, L.; Barrett, B.; Lovchik, J.; Robinson, J.P.; et al. Laser optical sensor, a label-free on-plate Salmonella enterica colony detection tool. mBio 2014, 5. [Google Scholar] [CrossRef]
- Ahmed, W.M.; Bayraktar, B.; Bhunia, A.K.; Hirleman, E.D.; Robinson, J.P.; Rajwa, B. Classification of bacterial contamination using image processing and distributed computing. IEEE J. Biomed. Health Inform. 2013, 17, 232–239. [Google Scholar] [CrossRef]
- Lathrop, A.A.; Jaradat, Z.W.; Haley, T.; Bhunia, A.K. Characterization and application of a Listeria monocytogenes reactive monoclonal antibody C11E9 in a resonant mirror biosensor. J. Immunol. Methods 2003, 281, 119–128. [Google Scholar] [CrossRef]
- Gray, K.M.; Bhunia, A.K. Specific detection of cytopathogenic Listeria monocytogenes using a two-step method of immunoseparation and cytotoxicity analysis. J. Microbiol. Methods 2005, 60, 259–268. [Google Scholar] [CrossRef]
- Den Bakker, H.C.; Cummings, C.A.; Ferreira, V.; Vatta, P.; Orsi, R.H.; Degoricija, L.; Barker, M.; Petrauskene, O.; Furtado, M.R.; Wiedmann, M. Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 2010, 11. [Google Scholar] [CrossRef]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Gouin, E.; Mengaud, J.; Cossart, P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 1994, 62, 3550–3553. [Google Scholar]
- Bubert, A.; Hein, I.; Rauch, M.; Lehner, A.; Yoon, B.; Goebel, W.; Wagner, M. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 1999, 65, 4688–4692. [Google Scholar]
- Longhi, C.; Maffeo, A.; Penta, M.; Petrone, G.; Seganti, L.; Conte, M.P. Detection of Listeria monocytogenes in Italian-style soft cheeses. J. Appl. Microbiol. 2003, 94, 879–885. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Method 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Tang, Y.; Kim, H.; Singh, A.K.; Aroonnual, A.; Bae, E.; Rajwa, B.; Fratamico, P.M.; Bhunia, A.K. Light scattering sensor for direct identification of colonies of Escherichia coli serogroups O26, O45, O103, O111, O121, O145 and O157. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Isolation and Identification of Listeria monocytogenes from Red Meat, Poultry and Egg Products, and Environmental Samples; USDA-FSIS: Washington, DC, USA, 2013; Volume MLG 8.09, p. 21.
- Banada, P.P.; Huff, K.; Bae, E.; Rajwa, B.; Aroonnual, A.; Bayraktar, B.; Adil, A.; Robinson, J.P.; Hirleman, E.D.; Bhunia, A.K. Label-free detection of multiple bacterial pathogens using light-scattering sensor. Biosens. Bioelectron. 2009, 24, 1685–1692. [Google Scholar] [CrossRef]
- Curiale, M.S.; Sons, T.; Fanning, L.; Lepper, W.; McIver, D.; Garramone, S.; Mozola, M. Deoxyribonucleic acid hybridization method for the detection of Listeria in dairy products, seafoods, and meats: Collaborative study. J. AOAC Int. 1994, 77, 602–617. [Google Scholar]
- Rocourt, J.; Cossart, P. Listeria Monocytogenes; ASM Press: Washington, DC, USA, 1997. [Google Scholar]
- Besse, N.G.; Barre, L.; Buhariwalla, C.; Vignaud, M.L.; Khamissi, E.; Decourseulles, E.; Nirsimloo, M.; Chelly, M.; Kalmokoff, M. The overgrowth of Listeria monocytogenes by other Listeria spp. in food samples undergoing enrichment cultivation has a nutritional basis. Int. J. Food Microbiol. 2010, 136, 345–351. [Google Scholar] [CrossRef]
- Ryser, E.T. Foodborne listeriosis. In Listeria, Listeriosis, and Food Safety; Ryser, E.T., Marth, E.H., Eds.; Marcel Decker: New York, NY, USA, 1999; pp. 299–358. [Google Scholar]
- Graves, L.M.; Helsel, L.O.; Steigerwalt, A.G.; Morey, R.E.; Daneshvar, M.I.; Roof, S.E.; Orsi, R.H.; Fortes, E.D.; Milillo, S.R.; den Bakker, H.C.; et al. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 2010, 60, 1280–1288. [Google Scholar] [CrossRef]
- Den Bakker, H.C.; Manuel, C.S.; Fortes, E.D.; Wiedmann, M.; Nightingale, K.K. Genome sequencing identifies Listeria fleischmannii subsp coloradonensis subsp nov., isolated from a ranch. Int. J. Syst. Evol. Microbiol. 2013, 63, 3257–3268. [Google Scholar] [CrossRef]
- Koo, O.K.; Aroonnual, A.; Bhunia, A.K. Human heat-shock protein 60 receptor-coated paramagnetic beads show improved capture of Listeria monocytogenes in the presence of other Listeria in food. J. Appl. Microbiol. 2011, 111, 93–104. [Google Scholar] [CrossRef]
- Kim, H.; Bhunia, A.K. Secreted Listeria adhesion protein (Lap) influences Lap-mediated Listeria monocytogenes paracellular translocation through epithelial barrier. Gut Pathog. 2013, 5. [Google Scholar] [CrossRef]
- Winters, D.K.; Maloney, T.P.; Johnson, M.G. Rapid detection of Listeria monocytogenes by a PCR assay specific for an aminopeptidase. Mol. Cell. Probes 1999, 13, 127–131. [Google Scholar] [CrossRef]
- Gilot, P.; Content, J. Specific identification of Listeria welshimeri and Listeria monocytogenes by PCR assays targeting a gene encoding a fibronectin-binding protein. J. Clin. Microbiol. 2002, 40, 698–703. [Google Scholar] [CrossRef]
- Bhunia, A.K.; Bae, E.; Rajwa, B.; Robinson, J.P.; Hirleman, E.D. Utilization of optical forward scatter image biological database: Foodborne pathogen colony differentiation and detection. In Omics, Microbial Modeling and Technologies for Foodborne Pathogens; Yan, X., Juneja, V.K., Fratamico, P.M., Smith, J.L, Eds.; Lancaster, PA, USA, 2012; pp. 553–578. [Google Scholar]
- Singh, A.K.; Sun, X.; Bai, X.; Kim, H.; Abdalhaseib, M.U.; Bae, E.; Bhunia, A.K. Label-free, non-invasive light scattering sensor for rapid screening of Bacillus colonies. J. Microbiol. Methods 2015, 109, 56–66. [Google Scholar] [CrossRef]
- He, Y.; Reed, S.; Bhunia, A.K.; Gehring, A.; Nguyen, L.H.; Irwin, P.L. Rapid identification and classification of Campylobacter spp. using laser optical scattering technology. Food Microbiol. 2015, 47, 28–35. [Google Scholar] [CrossRef]
- Huff, K.; Aroonnual, A.; Littlejohn, A.E.F.; Rajwa, B.; Bae, E.; Banada, P.P.; Patsekin, V.; Hirleman, E.D.; Robinson, J.P.; Richards, G.P.; et al. Light-scattering sensor for real-time identification of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae colonies on solid agar plate. Microb. Biotechnol. 2012, 5, 607–620. [Google Scholar] [CrossRef]
- Liu, D. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 2006, 55, 645–659. [Google Scholar] [CrossRef]
- Nelson, K.E.; Fouts, D.E.; Mongodin, E.F.; Ravel, J.; de Boy, R.T.; Kolonay, J.F.; Rasko, D.A.; Angiuoli, S.V.; Gill, S.R.; Paulsen, I.T.; et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004, 32, 2386–2395. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-P.; Singh, A.K.; Bai, X.; Leprun, L.; Bhunia, A.K. Novel PCR Assays Complement Laser Biosensor-Based Method and Facilitate Listeria Species Detection from Food. Sensors 2015, 15, 22672-22691. https://doi.org/10.3390/s150922672
Kim K-P, Singh AK, Bai X, Leprun L, Bhunia AK. Novel PCR Assays Complement Laser Biosensor-Based Method and Facilitate Listeria Species Detection from Food. Sensors. 2015; 15(9):22672-22691. https://doi.org/10.3390/s150922672
Chicago/Turabian StyleKim, Kwang-Pyo, Atul K. Singh, Xingjian Bai, Lena Leprun, and Arun K. Bhunia. 2015. "Novel PCR Assays Complement Laser Biosensor-Based Method and Facilitate Listeria Species Detection from Food" Sensors 15, no. 9: 22672-22691. https://doi.org/10.3390/s150922672