Spectral and Image Integrated Analysis of Hyperspectral Data for Waxy Corn Seed Variety Classification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Hyperspectral Imaging System
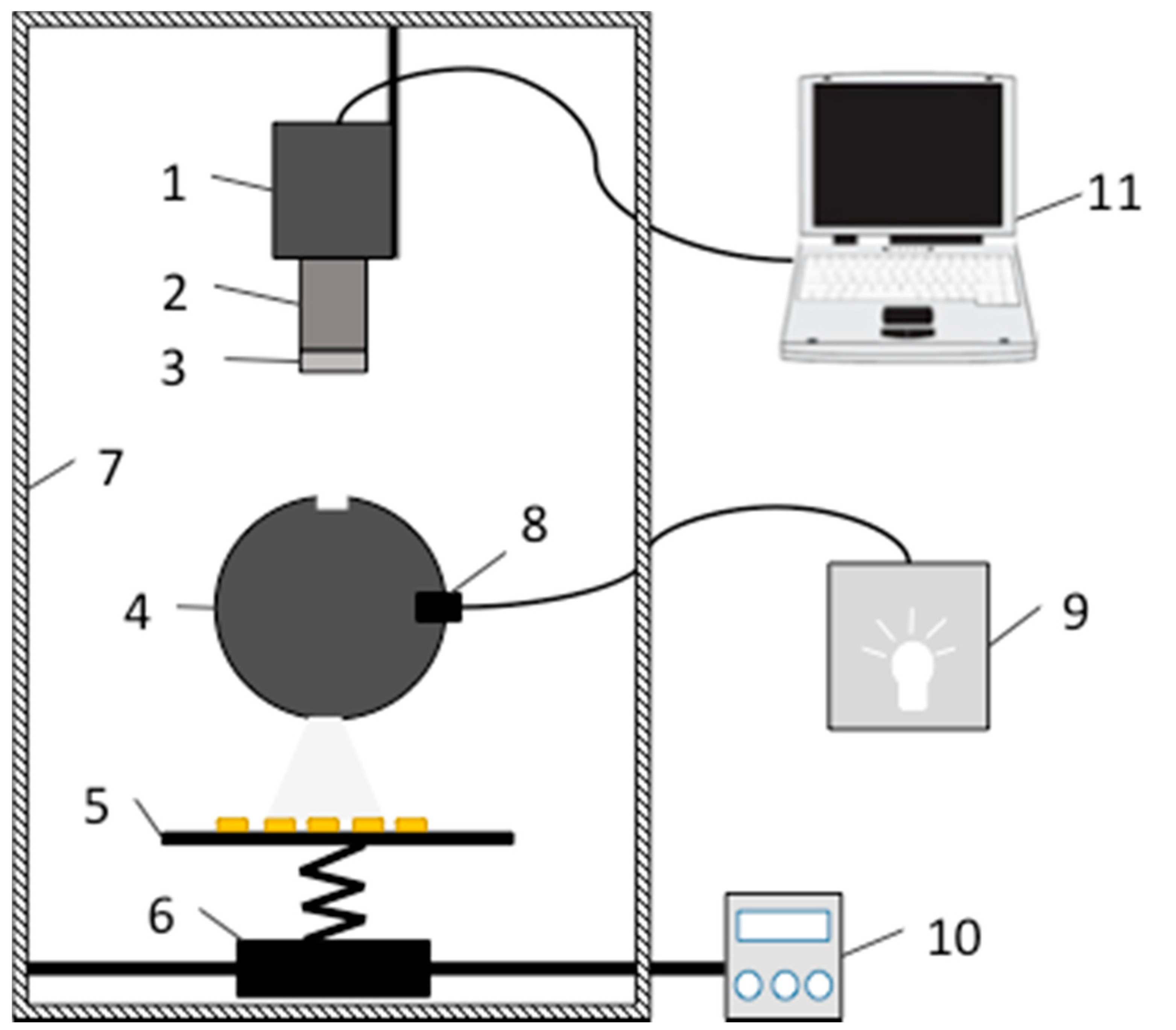
2.2. Seed Sample Selection and Preparation

2.3. Image Acquisition and Calibration
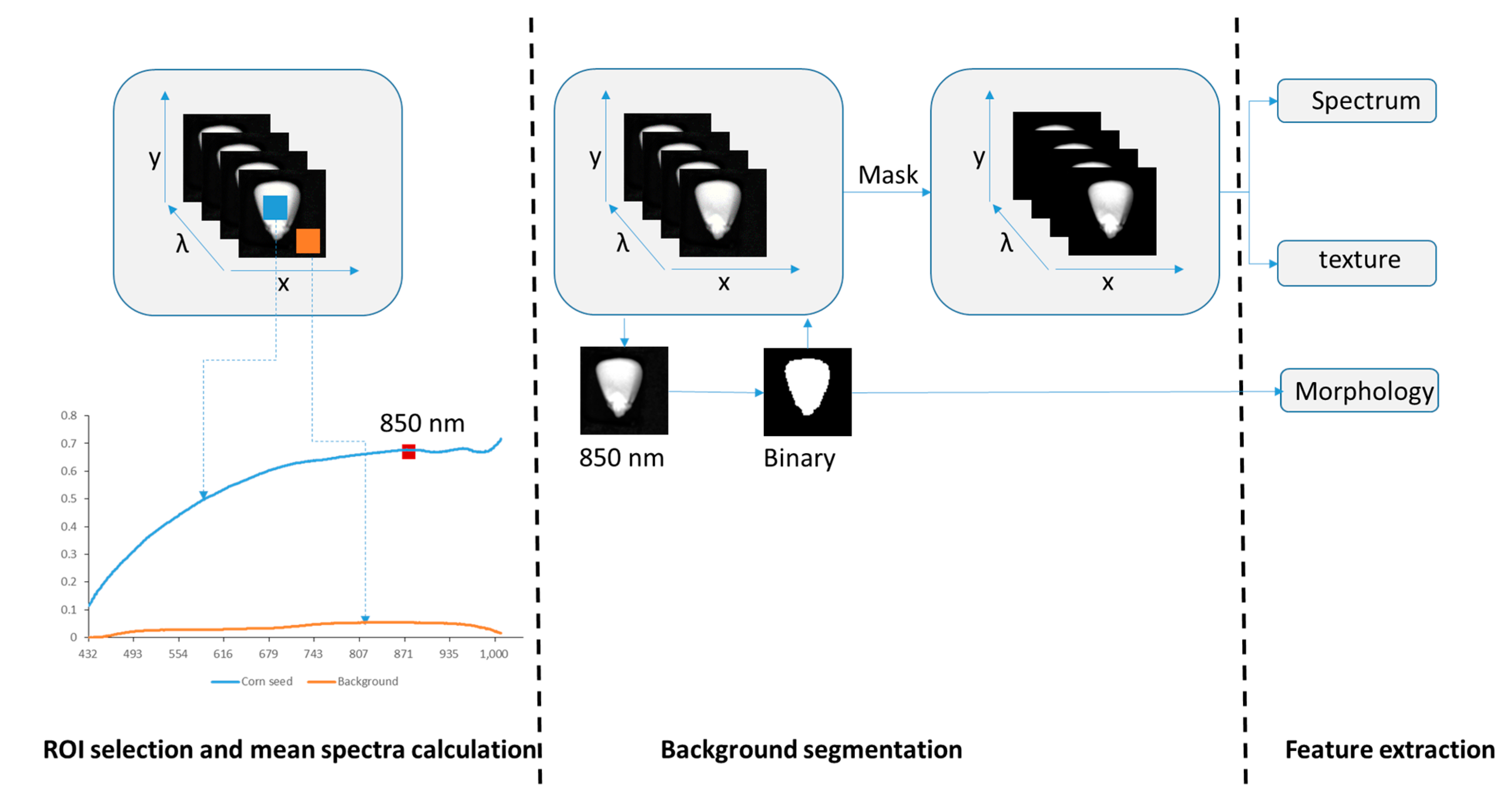
2.4. Spectral and Image Feature Extraction
2.4.1. Background Segmentation and Spectra Extraction
2.4.2. Spectra Preprocessing
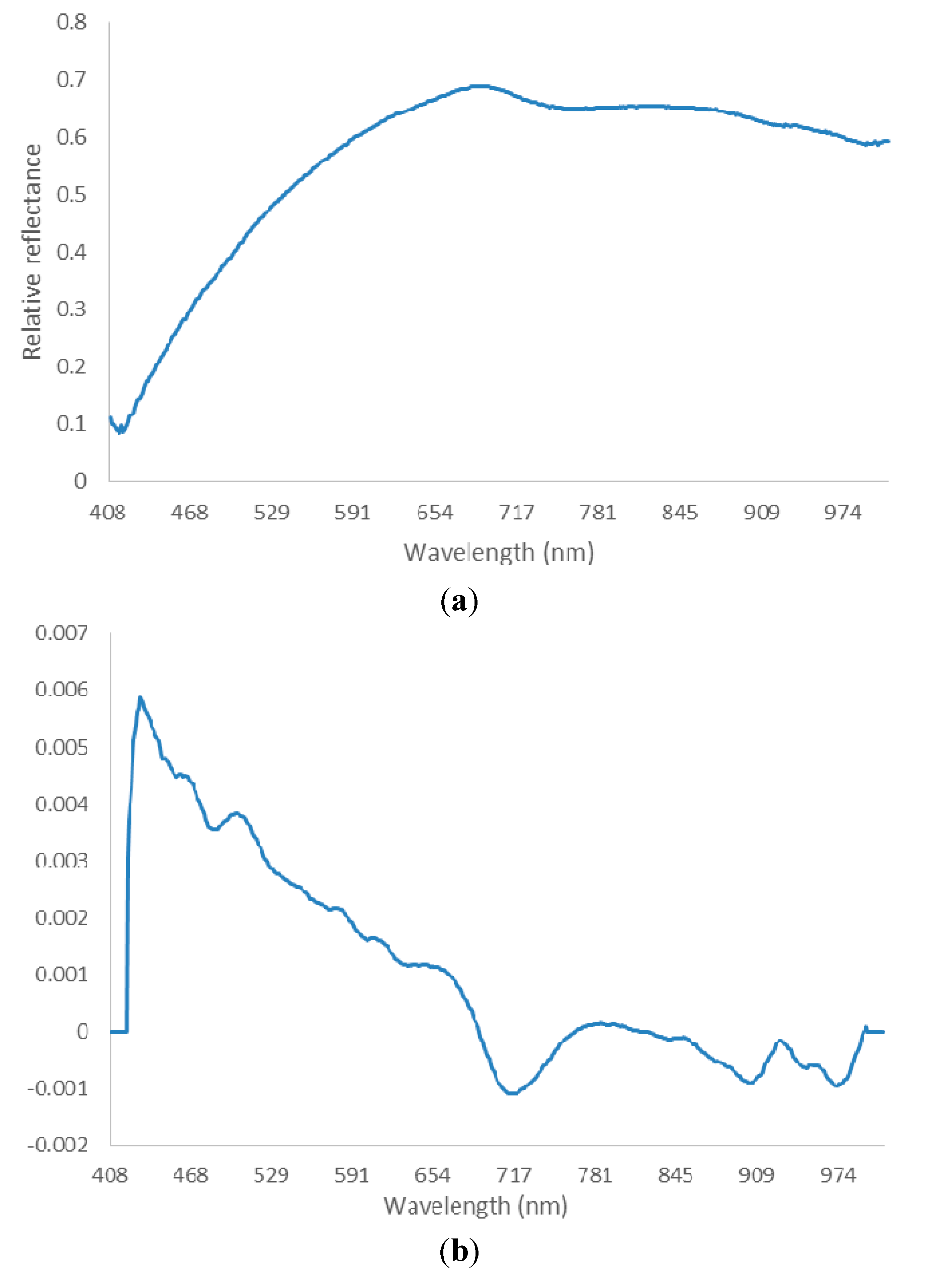
2.4.3. Optimal Wavelength Selection
2.4.4. Image Features Extraction
2.5. Development of Classification Models
3. Results and Discussion
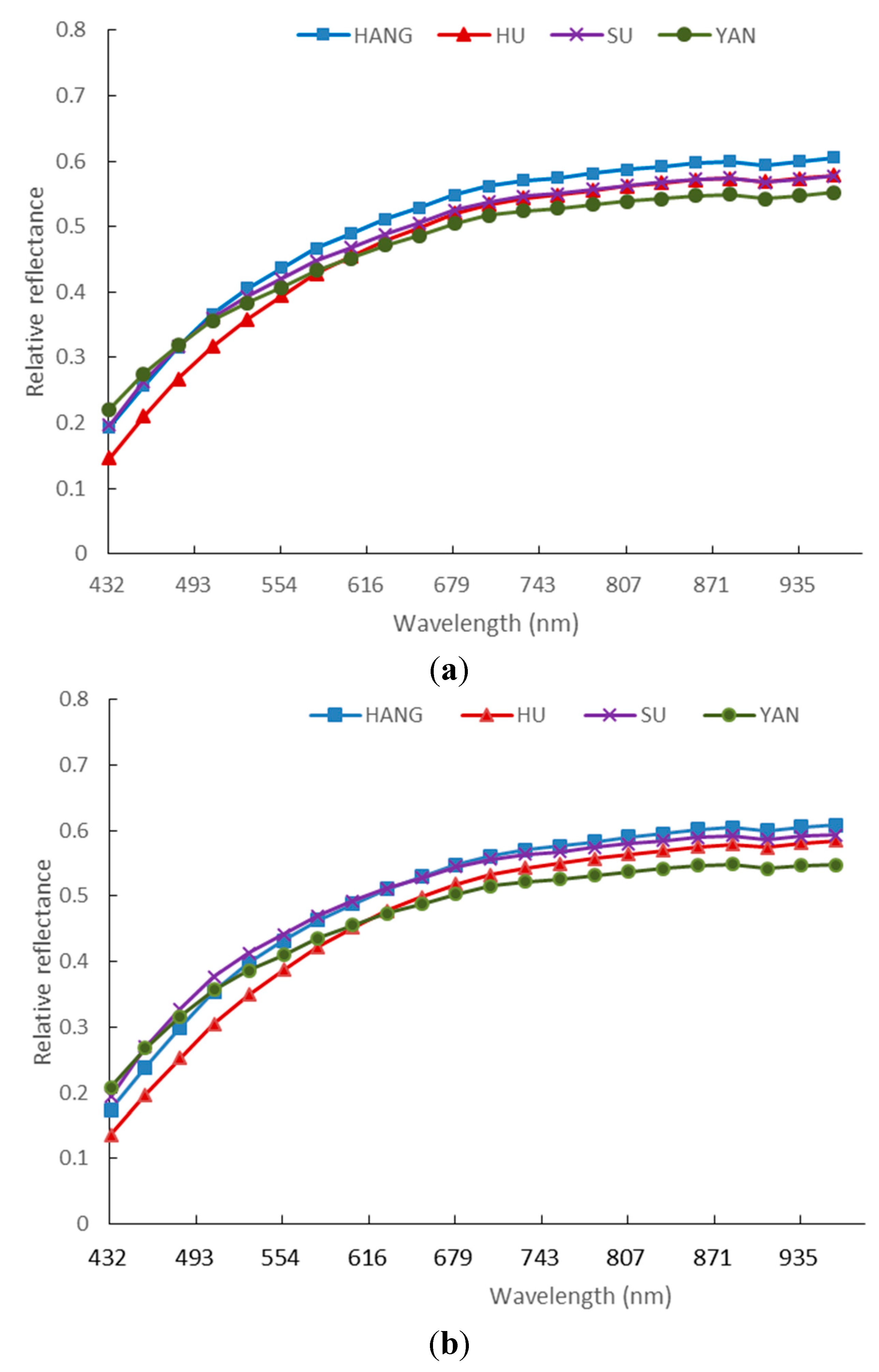
3.1. Spectra of Four Varieties of Maize Seeds
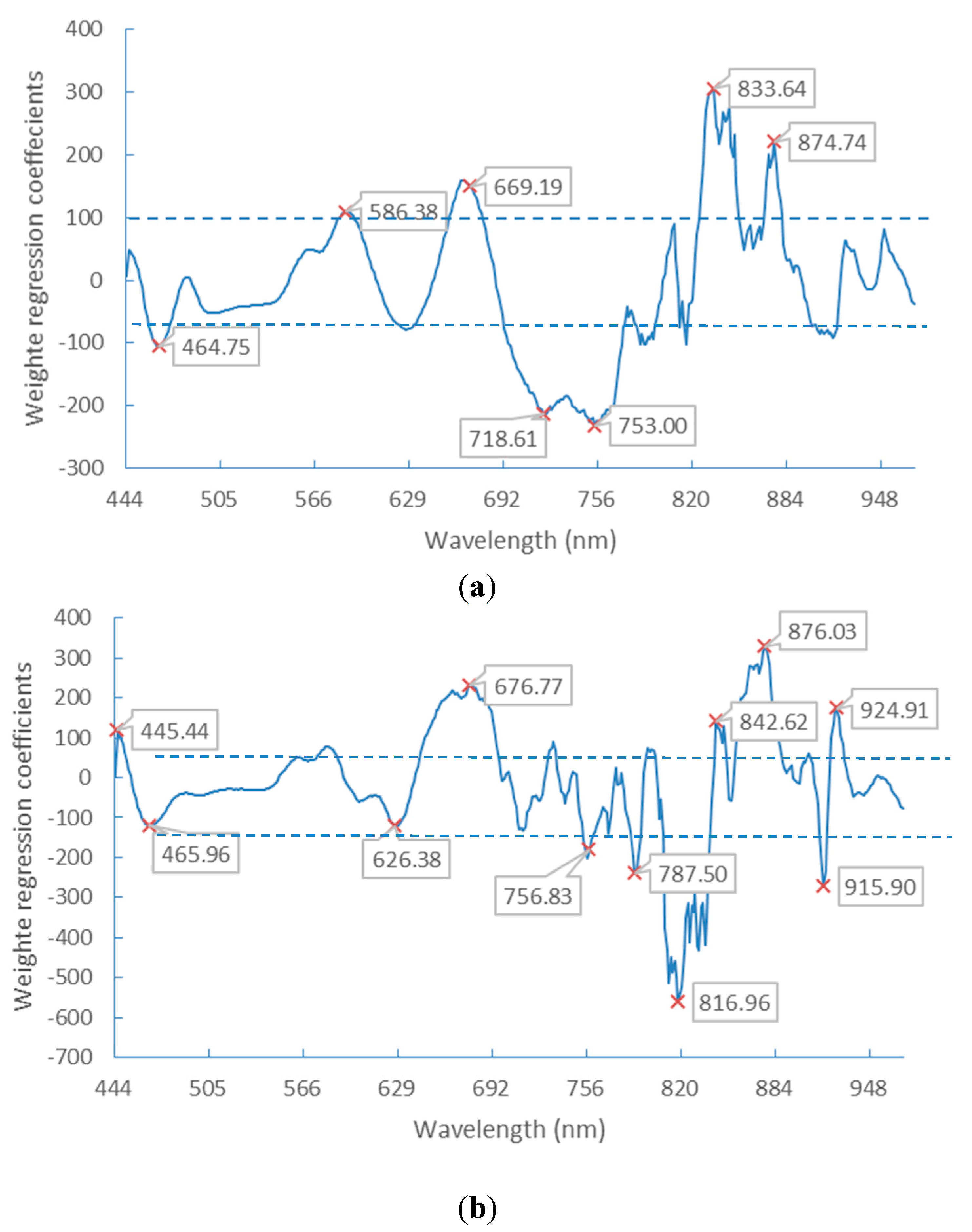
3.2. Optimal Spectral Wavebands

| Selected Wavelengths (nm) | |
|---|---|
| Germ-up images | 445.44, 447.85, 451.46, 453.87, 456.29, 458.70, 462.33, 469.59, 480.51, 491.46, 497.56, 506.11, 539.28, 581.40, 596.35, 626.38, 637.68, 891.46, 927.48, 935.20, 940.35, 945.50, 951.93, 959.65, 964.79, 968.65 |
| Germ-down images | 444.24, 462.33, 472.01, 481.73, 493.90, 506.11, 538.05, 561.53, 576.42, 588.87, 600.10, 616.35, 641.46, 661.61, 708.45, 801.58, 901.75, 960.94, 968.65 |
3.3. Classification by SVM and PLS-DA
| Image Type | Classification Method | Full Bands | Image Features | Selected Bands | Features Fusion | ||
|---|---|---|---|---|---|---|---|
| SPA | PLS-DA | SPA + Image Features | PLS-DA + Image Features | ||||
| Germ-Up Images | SVM | 94.6 | 86.6 | 96.2 | 66.1 | 98.2 | 77.4 |
| PLS-DA | 91 | 65.8 | 82 | 83.1 | 86.4 | 83.5 | |
| Germ-Down Images | SVM | 89.2 | 86.8 | 95 | 78.6 | 96.3 | 86.1 |
| PLS-DA | 88.4 | 67.4 | 86.5 | 86.8 | 91.6 | 86.8 | |
3.3.1. Classification Using Spectral Features
3.3.2. Classification Using Image Features
3.3.3. Classification Using both Spectral and Image Features
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Payne, R. Variety testing by official AOSA seed laboratories. J. Seed Technol. 1986, 10, 24–36. [Google Scholar]
- Chen, X.; Xun, Y.; Li, W.; Zhang, J. Combining discriminant analysis and neural networks for corn variety identification. Comput. Electron. Agric. 2010, 71, S48–S53. [Google Scholar] [CrossRef]
- Remund, K.M.; Dixon, D.A.; Wright, D.L.; Holden, L.R. Statistical considerations in seed purity testing for transgenic traits. Seed Sci. Res. 2001, 11, 101–120. [Google Scholar]
- Manickavasagan, A.; Sathya, G.; Jayas, D.S.; White, N.D.G. Wheat class identification using monochrome images. J. Cereal Sci. 2008, 47, 518–527. [Google Scholar] [CrossRef]
- Manickavasagan, A.; Jayas, D.; White, N.; Paliwal, J. Wheat class identification using thermal imaging. Food Bioprocess Technol. 2010, 3, 450–460. [Google Scholar] [CrossRef]
- Yan, X.; Wang, J.; Liu, S.; Zhang, C. Purity identification of maize seed based on color characteristics. In Computer and Computing Technologies in Agriculture IV; Springer: Nanchang, China, 2011; pp. 620–628. [Google Scholar]
- Grillo, O.; Mattana, E.; Venora, G.; Bacchetta, G. Statistical seed classifiers of 10 plant families representative of the Mediterranean vascular flora. Seed Sci. Technol. 2010, 38, 455–476. [Google Scholar] [CrossRef]
- Mavi, K. The relationship between seed coat color and seed quality in watermelon Crimson sweet. Hortic. Sci. 2010, 37, 62–69. [Google Scholar]
- Wu, D.; Feng, L.; He, Y.; Bao, Y. Variety identification of Chinese cabbage seeds using visible and near-infrared spectroscopy. Trans. ASABE 2008, 51, 2193–2199. [Google Scholar] [CrossRef]
- Seregely, Z.; Deak, T.; Bisztray, G.D. Distinguishing melon genotypes using NIR spectroscopy. Chemometr. Intell. Lab. Syst. 2004, 72, 195–203. [Google Scholar] [CrossRef]
- Delwiche, S.; Graybosch, R.A. Identification of waxy wheat by near-infrared reflectance spectroscopy. J. Cereal Sci. 2002, 35, 29–38. [Google Scholar] [CrossRef]
- Agelet, L.E.; Hurburgh, C.R., Jr. Limitations and Current Applications of Near Infrared Spectroscopy for Single Seed Analysis. Talanta 2014, 121, 288–299. [Google Scholar] [CrossRef]
- Orman, B.A.; Schumann, R.A., Jr. Nondestructive single-kernel oil determination of maize by near-infrared transmission spectroscopy. J. Am. Oil Chem. Soc. 1992, 69, 1036–1038. [Google Scholar] [CrossRef]
- Delwiche, S.R. Single Wheat Kernel Analysis by Near-Infrared Transmittance: Protein-Content. Cereal Chem. 1995, 72, 11–16. [Google Scholar]
- Delwiche, S.R.; Graybosch, R.A.; Amand, P.S.; Bai, G. Starch Waxiness in Hexaploid Wheat (Triticum aestivum L.) by NIR Reflectance Spectroscopy. J. Agric. Food Chem. 2011, 59, 4002–4008. [Google Scholar] [CrossRef]
- Delwiche, S.R.; Graybosch, R.A.; Peterson, C.J. Predicting protein composition, biochemical properties, and dough-handling properties of hard red winter wheat flour by near-infrared reflectance. Cereal Chem. 1998, 75, 412–416. [Google Scholar] [CrossRef]
- Spielbauer, G.; Armstrong, P.; Baier, J.W.; Allen, W.B.; Richardson, K.; Shen, B.; Settles, A.M. High-throughput near-infrared reflectance spectroscopy for predicting quantitative and qualitative composition phenotypes of individual maize kernels. Cereal Chem. 2009, 86, 556–564. [Google Scholar] [CrossRef]
- Cogdill, R.P.; Hurburgh, C.R.; Rippke, G.R.; Bajic, S.J.; Jones, R.W.; McClelland, J.F.; Jensen, T.C.; Liu, J. Single-kernel maize analysis by near-infrared hyperspectral imaging. Trans. ASAE 2004, 47, 311–320. [Google Scholar] [CrossRef]
- Casasent, D.; Chen, X.W. Aflatoxin detection in whole corn kernels using hyperspectral methods. Proc. SPIE 2004, 5271, 275–284. [Google Scholar]
- Bauriegel, E.; Giebel, A.; Herppich, W.B. Hyperspectral and Chlorophyll Fluorescence Imaging to Analyse the Impact of Fusarium culmorum on the Photosynthetic Integrity of Infected Wheat Ears. Sensors 2011, 11, 3765–3779. [Google Scholar] [CrossRef]
- Yao, H.B.; Hruska, Z.; Kincaid, R.; Brown, R.L.; Bhatnagar, D.; Cleveland, T.E. Detecting maize inoculated with toxigenic and atoxigenic fungal strains with fluorescence hyperspectral imagery. Biosyst. Eng. 2013, 115, 125–135. [Google Scholar] [CrossRef]
- Delwiche, S.R.; Souza, E.J.; Kim, M.S. Limitations of single kernel near-infrared hyperspectral. imaging of soft wheat for milling quality. Biosyst. Eng. 2013, 115, 260–273. [Google Scholar] [CrossRef]
- Williams, P.; Geladi, P.; Fox, G.; Manley, M. Maize kernel hardness classification by near infrared (NIR) hyperspectral imaging and multivariate data analysis. Anal. Chim. Acta 2009, 653, 121–130. [Google Scholar] [CrossRef]
- Mahesh, S.; Manickavasagan, A.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Feasibility of near-infrared hyperspectral imaging to differentiate Canadian wheat classes. Biosyst. Eng. 2008, 101, 50–57. [Google Scholar] [CrossRef]
- Serranti, S.; Cesare, D.; Marini, F.; Bonifazi, G. Classification of oat and groat kernels using NIR hyperspectral imaging. Talanta 2013, 103, 276–284. [Google Scholar] [CrossRef]
- Zhang, X.L.; Liu, F.; He, Y.; Li, X.L. Application of Hyperspectral Imaging and Chemometric Calibrations for Variety Discrimination of Maize Seeds. Sensors 2012, 12, 17234–17246. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Pu, H.; Sun, D.-W.; Gao, W.; Xiong, Z. Use of Hyperspectral Imaging to Discriminate the Variety and Quality of Rice. Food Anal. Methods 2015, 8, 515–523. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemometr. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Galvão, R.K.H.; Araujo, M.C.U.; José, G.E.; Pontes, M.J.C.; Silva, E.C.; Saldanha, T.C.B. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef]
- Galvao, R.K.H.; Araujo, M.C.U.; Fragoso, W.D.; Silva, E.C.; Jose, G.E.; Soares, S.F.C.; Paiva, H.M. A variable elimination method to improve the parsimony of MLR models using the successive projections algorithm. Chemometr. Intell. Lab. Syst. 2008, 92, 83–91. [Google Scholar] [CrossRef]
- Liu, D.; Sun, D.-W.; Zeng, X.-A. Recent advances in wavelength selection techniques for hyperspectral image processing in the food industry. Food Bioprocess Technol. 2014, 7, 307–323. [Google Scholar] [CrossRef]
- Paliwal, J.; Visen, N.; Jayas, D. Evaluation of neural network architectures for cereal grain classification using morphological features. J. Agric. Eng. Res. 2001, 79, 361–370. [Google Scholar] [CrossRef]
- Liu, Z.-Y.; Cheng, F.; Ying, Y.-B.; Rao, X.-Q. Identification of rice seed varieties using neural network. J. Zhejiang Univ. Sci. B 2005, 6, 1095–1100. [Google Scholar] [CrossRef]
- Pourreza, A.; Pourreza, H.; Abbaspour-Fard, M.-H.; Sadrnia, H. Identification of nine Iranian wheat seed varieties by textural analysis with image processing. Comput. Electron. Agric. 2012, 83, 102–108. [Google Scholar] [CrossRef]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2. [Google Scholar] [CrossRef]
- Choudhary, R.; Mahesh, S.; Paliwal, J.; Jayas, D.S. Identification of wheat classes using wavelet features from near infrared hyperspectral images of bulk samples. Biosyst. Eng. 2009, 102, 115–127. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Hong, H.; You, Z.; Cheng, F. Spectral and Image Integrated Analysis of Hyperspectral Data for Waxy Corn Seed Variety Classification. Sensors 2015, 15, 15578-15594. https://doi.org/10.3390/s150715578
Yang X, Hong H, You Z, Cheng F. Spectral and Image Integrated Analysis of Hyperspectral Data for Waxy Corn Seed Variety Classification. Sensors. 2015; 15(7):15578-15594. https://doi.org/10.3390/s150715578
Chicago/Turabian StyleYang, Xiaoling, Hanmei Hong, Zhaohong You, and Fang Cheng. 2015. "Spectral and Image Integrated Analysis of Hyperspectral Data for Waxy Corn Seed Variety Classification" Sensors 15, no. 7: 15578-15594. https://doi.org/10.3390/s150715578





