Simultaneous Imaging of Two Different Cancer Biomarkers Using Aptamer-Conjugated Quantum Dots
Abstract
:1. Introduction
2. Experimental Section
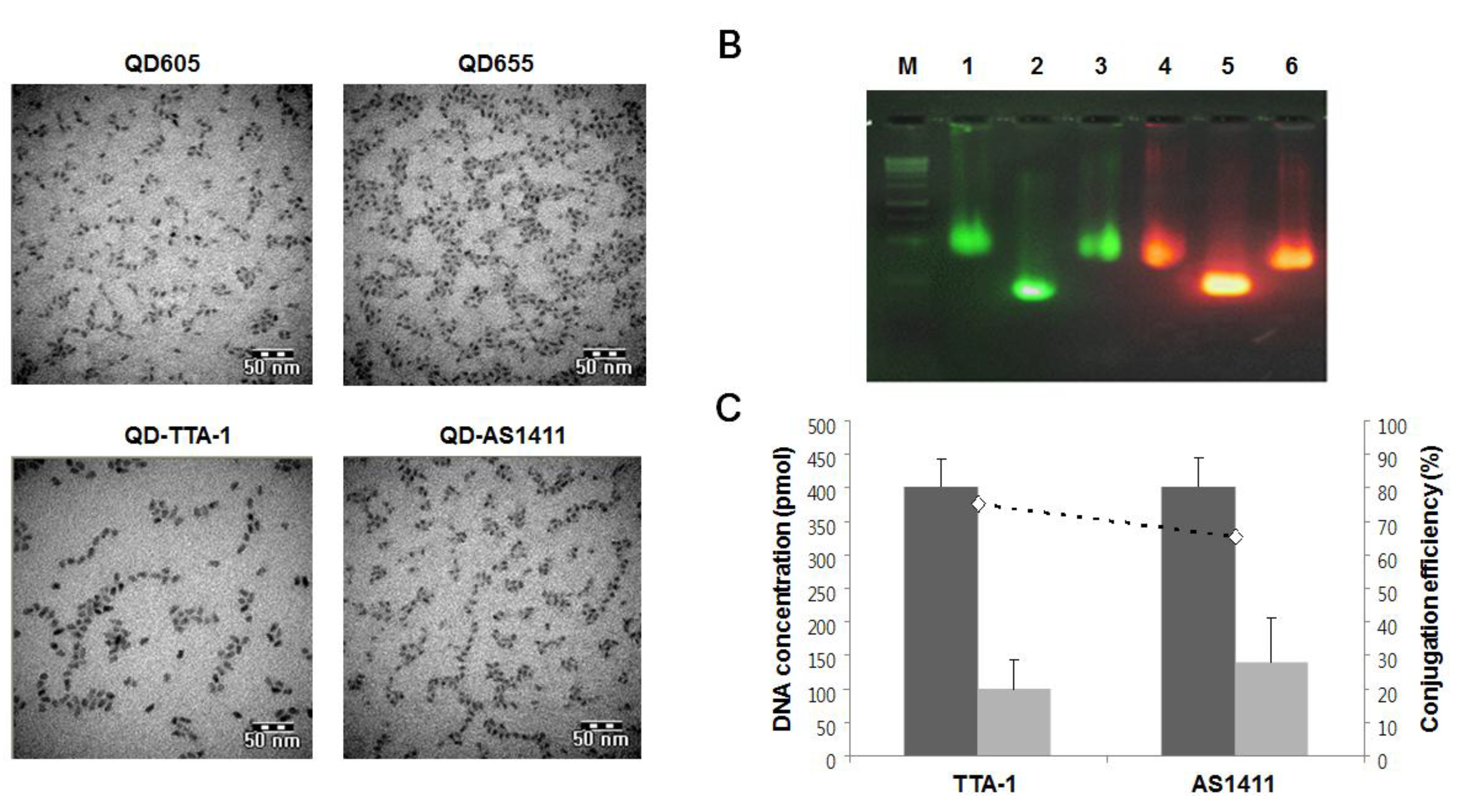
2.1. Preparation of QD-Aptamer Conjugates
2.2. Characterization of QD-Aptamer Conjugates
2.3. Cell Culture Conditions
2.4. Treatment with QD-Aptamer Conjugates and Measurement of Fluorescence Intensity
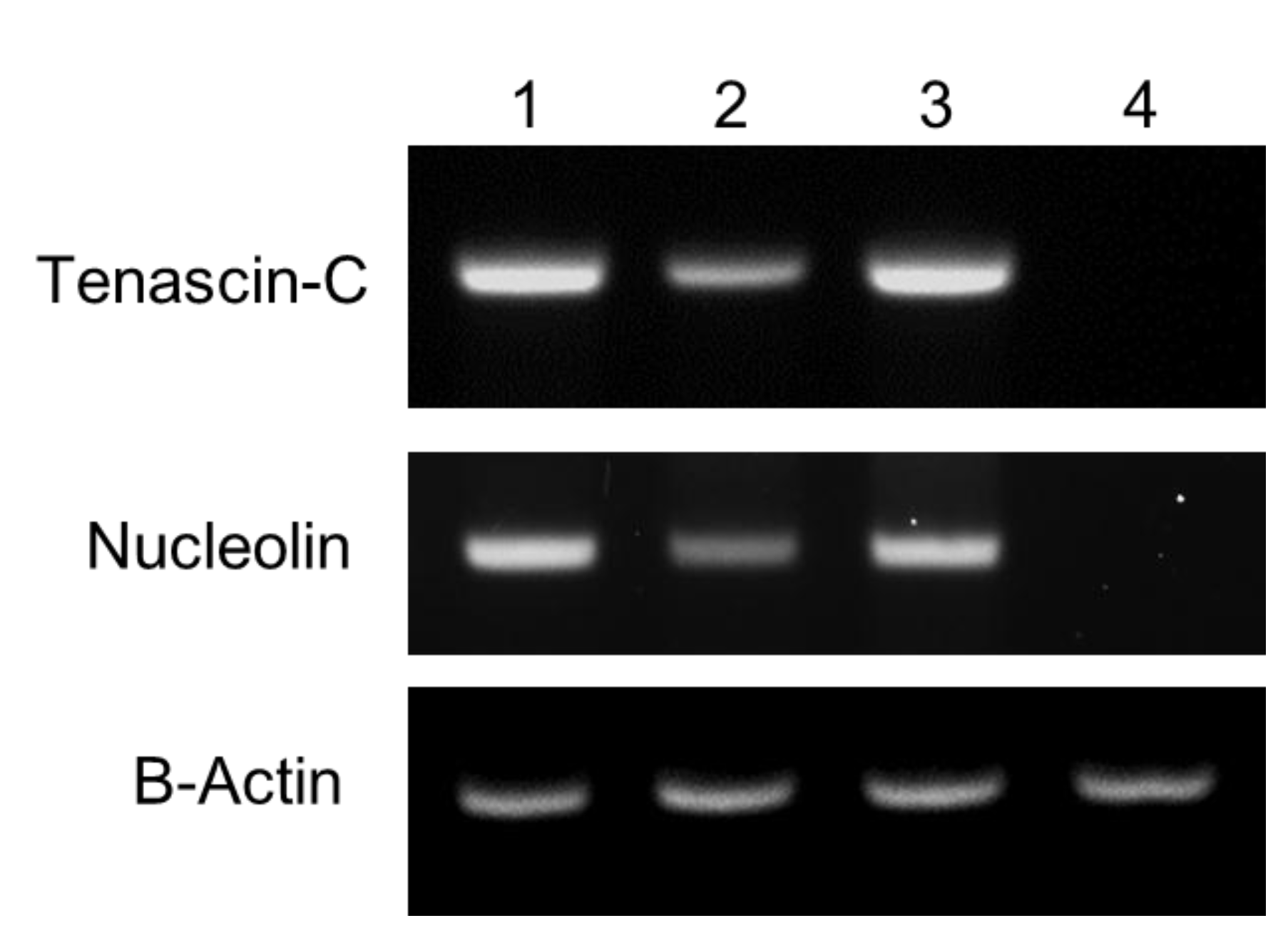
2.5. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
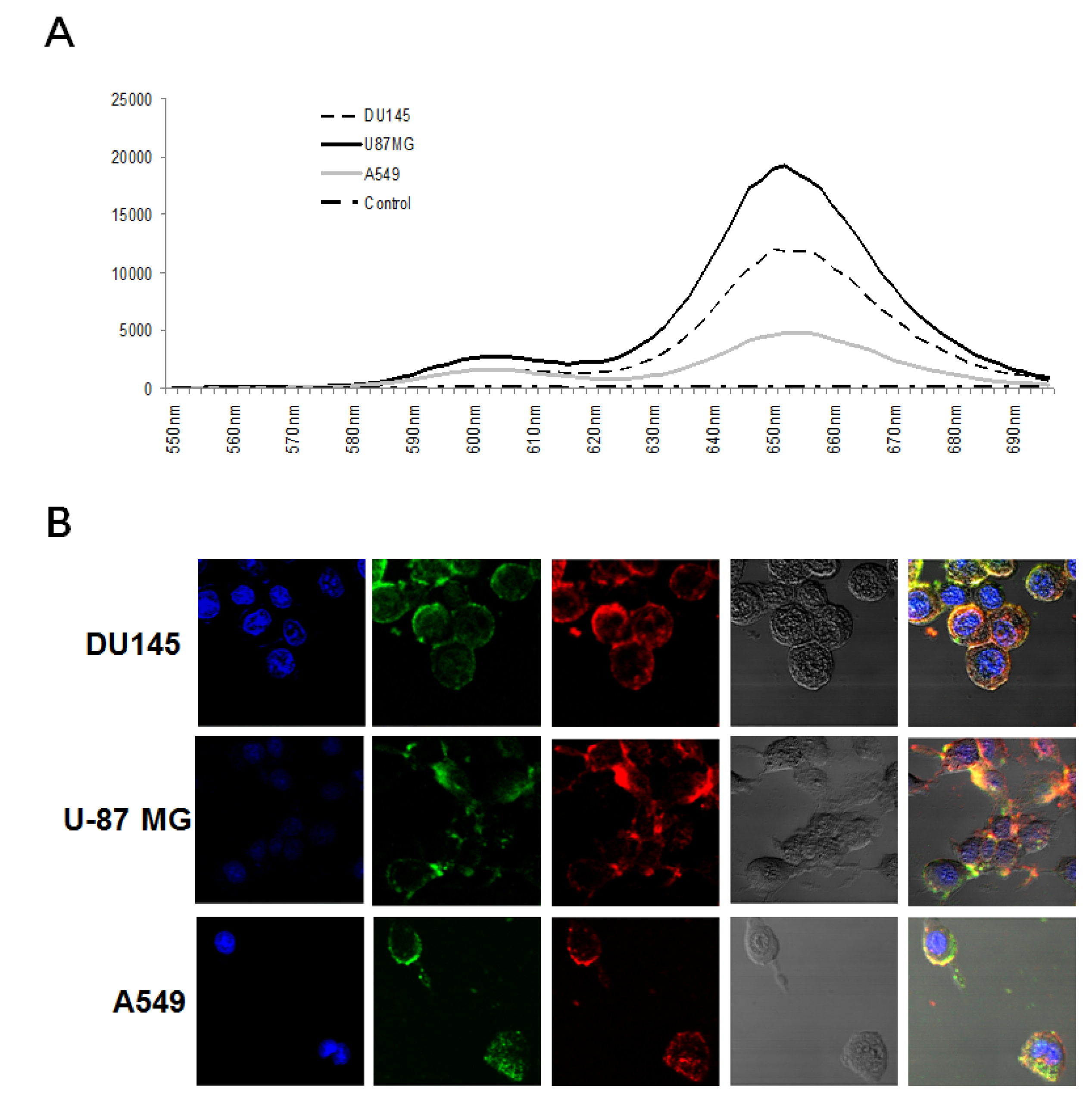
2.6. Confocal Microscopy
3. Results and Discussion
3.1. Characterization of Prepared QD-Aptamer Conjugates
3.2. Cancer-Targeting Specificity of QD-Aptamer Conjugates in Three Cancer Cell Lines
3.3. Simultaneous Imaging of Two Cancer Biomarkers Using QD-Aptamer Conjugates in Three Cancer Call Lines


4. Conclusions/Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Akerman, M.E.; Chan, W.C.; Laakkonen, P.; Bhatia, S.N.; Ruoslahti, E. Nanocrystal targeting in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 12617–12621. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Shin, D.W.; Chen, W.; Gheysens, O.; Cao, Q.; Wang, S.X.; Gambhir, S.S.; Chen, X. Peptide-labeled near-infrared quantum dots for imaging tumor vasculature in living subjects. Nano Lett. 2006, 6, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Rao, J. Quantum dot bioconjugates for in vitro diagnostics & in vivo imaging. Cancer Biomark. 2008, 4, 307–319. [Google Scholar] [PubMed]
- Savla, R.; Taratula, O.; Garbuzenko, O.; Minko, T. Tumor targeted quantum dot-mucin 1aptamer-doxorubicin conjugate for imaging and treatment of cancer. J. Control. Release 2011, 153, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Tada, H.; Higuchi, H.; Wanatabe, T.M.; Ohuchi, N. In vivo real-time tracking of single quantum dots conjugated with monoclonal anti-HER2 antibody in tumors of mice. Cancer Res. 2007, 67, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.H.; Kim, S.; Kang, W.J.; Lee, J.H.; Kang, H.; Moon, S.H.; Hwang, D.W.; Ko, H.K.; Lee, D.S. In vitro derby imaging of cancer biomarkers using quantum dots. Small 2009, 5, 1207–1212. [Google Scholar] [PubMed]
- Kerbel, R.; Folkman, J. Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer 2002, 2, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.P.; Stehle, T.; Diefenbach, B.; Zhang, R.; Dunker, R.; Scott, D.L.; Joachimiak, A.; Goodman, S.L.; Arnaout, M.A. Crystal structure of the extracellular segment of integrin αVβ3. Science 2001, 294, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Hovanessian, A.G.; Soundaramourty, C.; Khoury, D.E.; Nondier, I.; Svab, J.; Krust, B. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLos ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Pollard, H.B. Molecular dissection of nucleolin’s role in growth and cell proliferation: New insights. FASEB J. 1999, 13, 1911–1922. [Google Scholar] [PubMed]
- Jordan, G. At the heart of the nucleolus. Nature 1987, 329, 489–490. [Google Scholar] [CrossRef] [PubMed]
- Hicke, B.J.; Stephens, A.W.; Gould, T.; Chang, Y.F.; Lynott, C.K.; Heil, J.; Borkowski, S.; Hilger, C.S.; Cook, G.; Warren, S.; et al. Tumor targeting by an aptamer. J. Nucl. Med. 2006, 47, 668–678. [Google Scholar] [PubMed]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef] [PubMed]
- Valizadeh, A.; Mikaeili, H.; Samiei, M.; Farkhani, S.M.; Zarghami, N.; Kouhi, M.; Akbarzadeh, A.; Davaran, S. Quantum dots: Synthesis, bioapplications, and toxicity. Nanoscale Res. Lett. 2012, 7. [Google Scholar] [CrossRef]
- Walling, M.A.; Novak, J.A.; Shepard, J.R.E. Quantum dots for live cell and in vivo imaging. Int. J. Mol. Sci. 2009, 10, 441–491. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, D.; Tempest, H.G.; Skinner, B.M.; Thornhill, A.R.; Ellis, M.; Griffin, D.K. Quantum dots as new-generation fluorochromes for FISH: An appraisal. Chromosome Res. 2009, 17, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geissler, D.; Charbonnière, L.J.; Ziessel, R.F.; Butlin, N.G.; Löhmannsröben, H.G.; Hildebrandt, N. Quantum dot biosensors for ultrasensitive multiplexed diagnostics. Angew. Chem. Int. Ed. 2010, 49, 1396–1401. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Kang, H.J.; Jang, H.; Lee, Y.J.; Lee, Y.S.; Ali, B.A.; Al-Khedhairy, A.A.; Kim, S. Simultaneous Imaging of Two Different Cancer Biomarkers Using Aptamer-Conjugated Quantum Dots. Sensors 2015, 15, 8595-8604. https://doi.org/10.3390/s150408595
Lee J, Kang HJ, Jang H, Lee YJ, Lee YS, Ali BA, Al-Khedhairy AA, Kim S. Simultaneous Imaging of Two Different Cancer Biomarkers Using Aptamer-Conjugated Quantum Dots. Sensors. 2015; 15(4):8595-8604. https://doi.org/10.3390/s150408595
Chicago/Turabian StyleLee, Jonghwan, Hyo Jin Kang, Hyeok Jang, Youn Jung Lee, Yong Seung Lee, Bahy A. Ali, Abdulaziz A. Al-Khedhairy, and Soonhag Kim. 2015. "Simultaneous Imaging of Two Different Cancer Biomarkers Using Aptamer-Conjugated Quantum Dots" Sensors 15, no. 4: 8595-8604. https://doi.org/10.3390/s150408595




