Bio-Optics Based Sensation Imaging for Breast Tumor Detection Using Tissue Characterization
Abstract
:1. Introduction
2. Sensor Design and Sensing Principle
2.1. Sensor Design
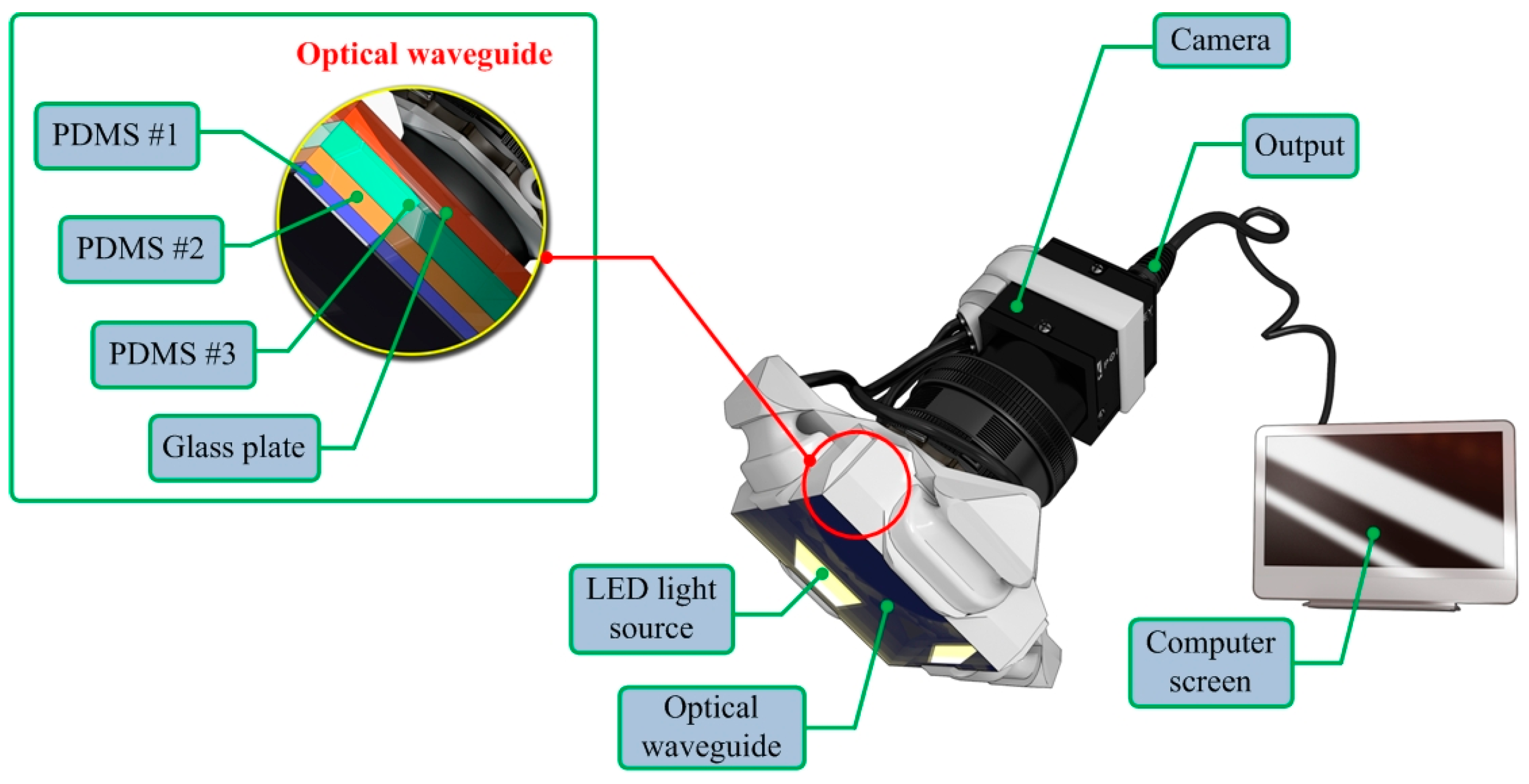
2.2. Sensing Principle

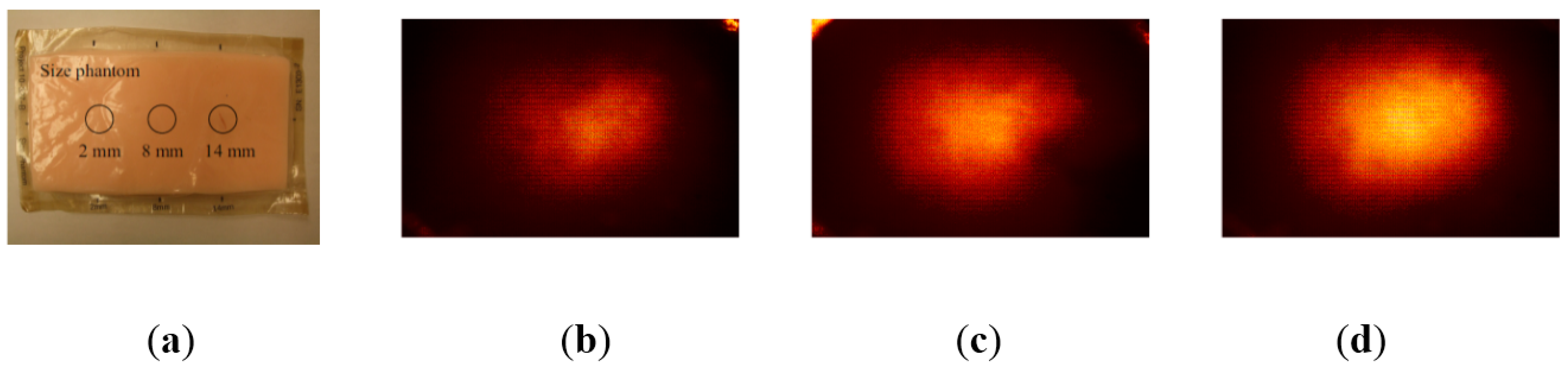
3. Tactile Data Processing Algorithm
- (1)
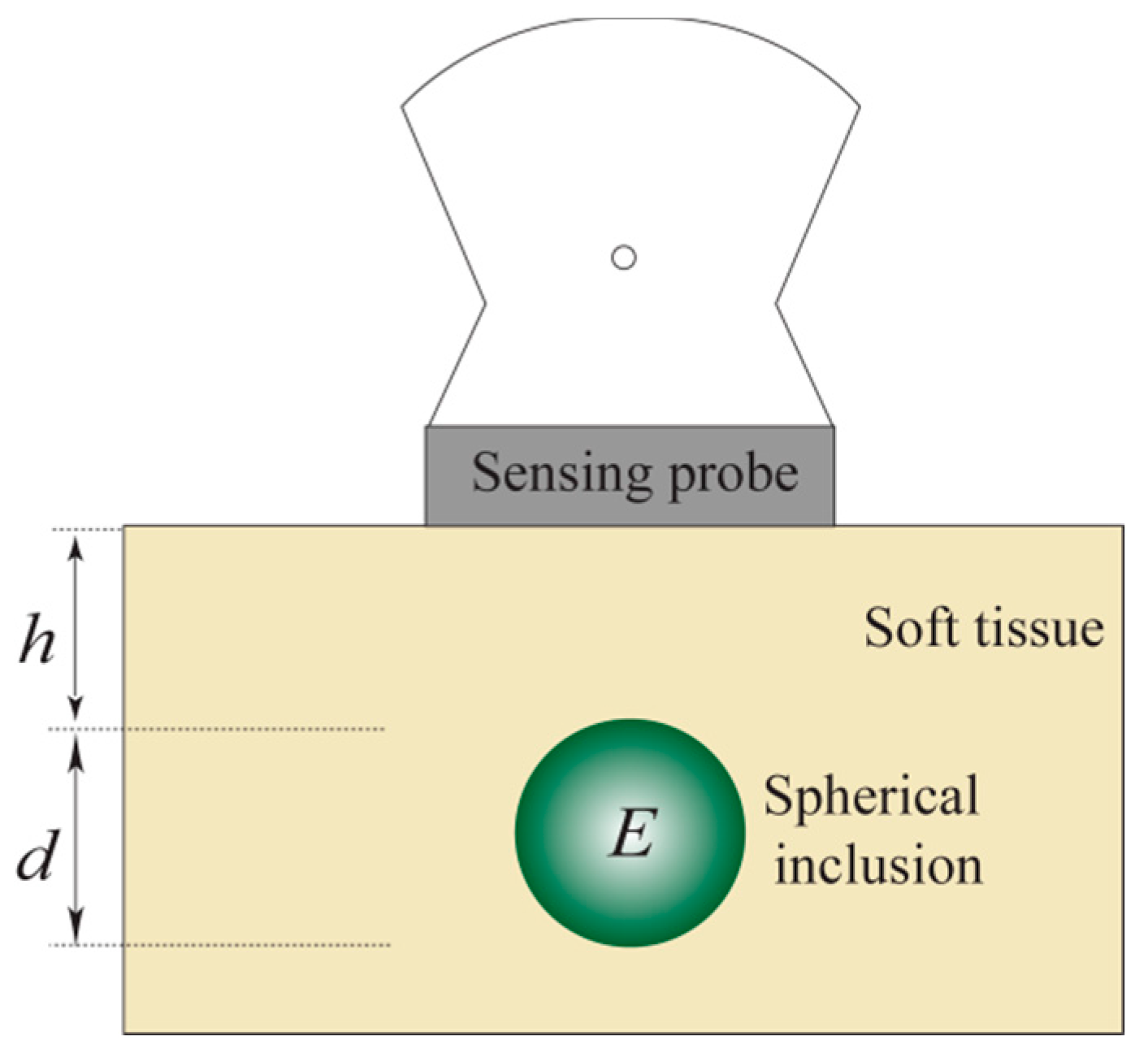
- Most breast tumors are found in the upper outer quadrant of the breast where the tissue is relatively thin and flat [18]. Therefore, in our model, the tissue is approximated as a slab of material of constant thickness that is fixed to a flat, incompressible chest wall.
- (2)
- (3)
- We assume that both the tissue and the inclusion are linear and isotropic. Glandular and adipose tissues, which account for most of the breast tissue, are well modeled by isotropic materials [21].
- (4)
- In this model, the indentation is made by the sensing probe of the TSIS with finite length. The interaction between the sensing probe and the tissue is assumed to be frictionless.
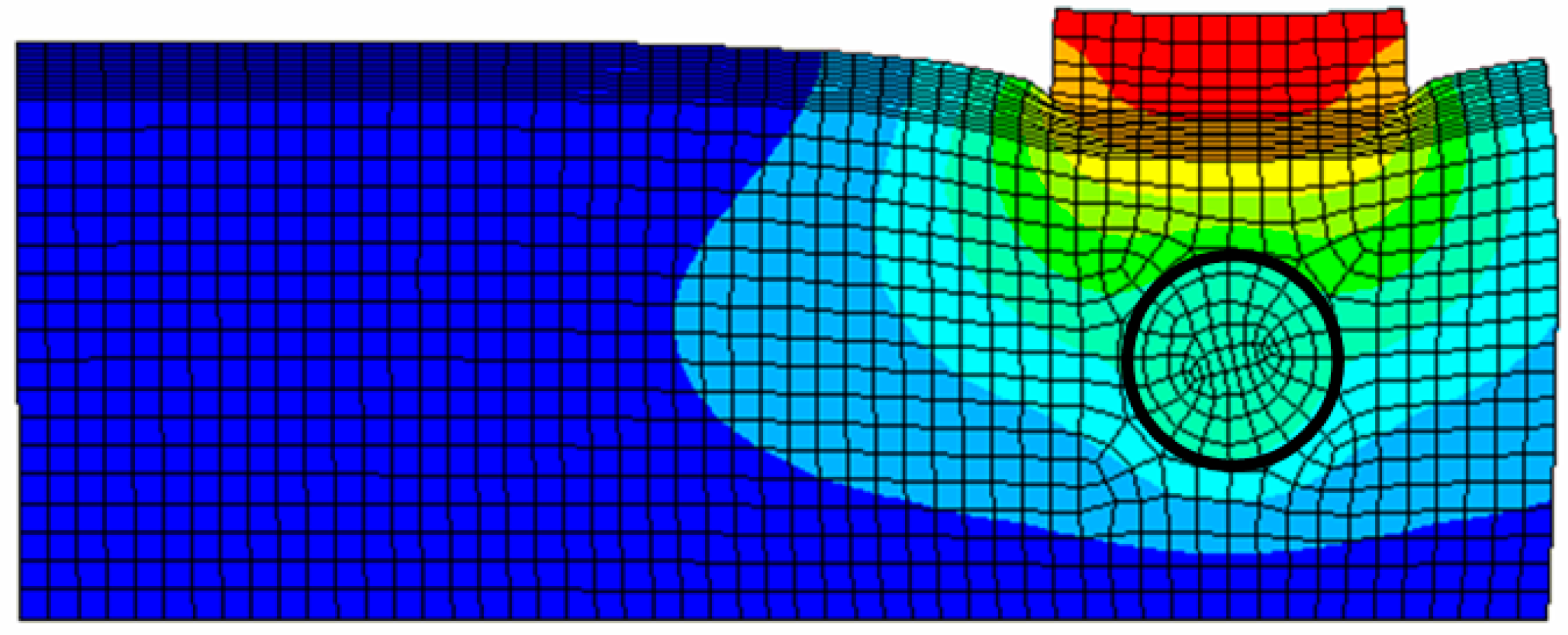
3.1. Forward Modeling–Finite Element Method
- (1)
- The biological tissue and inclusion are elastic and isotropic. It means the properties of a material are identical in all directions.
- (2)
- The Poisson’s ratio of each material is set to 0.49 because the breast can be considered an incompressible material. The incompressible material means the material incapable of or resistant to compression.
- (3)
- The biological tissue is assumed to be sitting on non-deformable hard surfaces like bones.
- (4)
- The tissue cross-section is a square with dimensions 120 mm 120 mm. The sensing probe of the TSIS is a square shape with dimensions of 25 mm 25 mm, which corresponds to the sensing probe size in the laboratory design discussed in Section 2.
3.2. Tactile Data from FEM
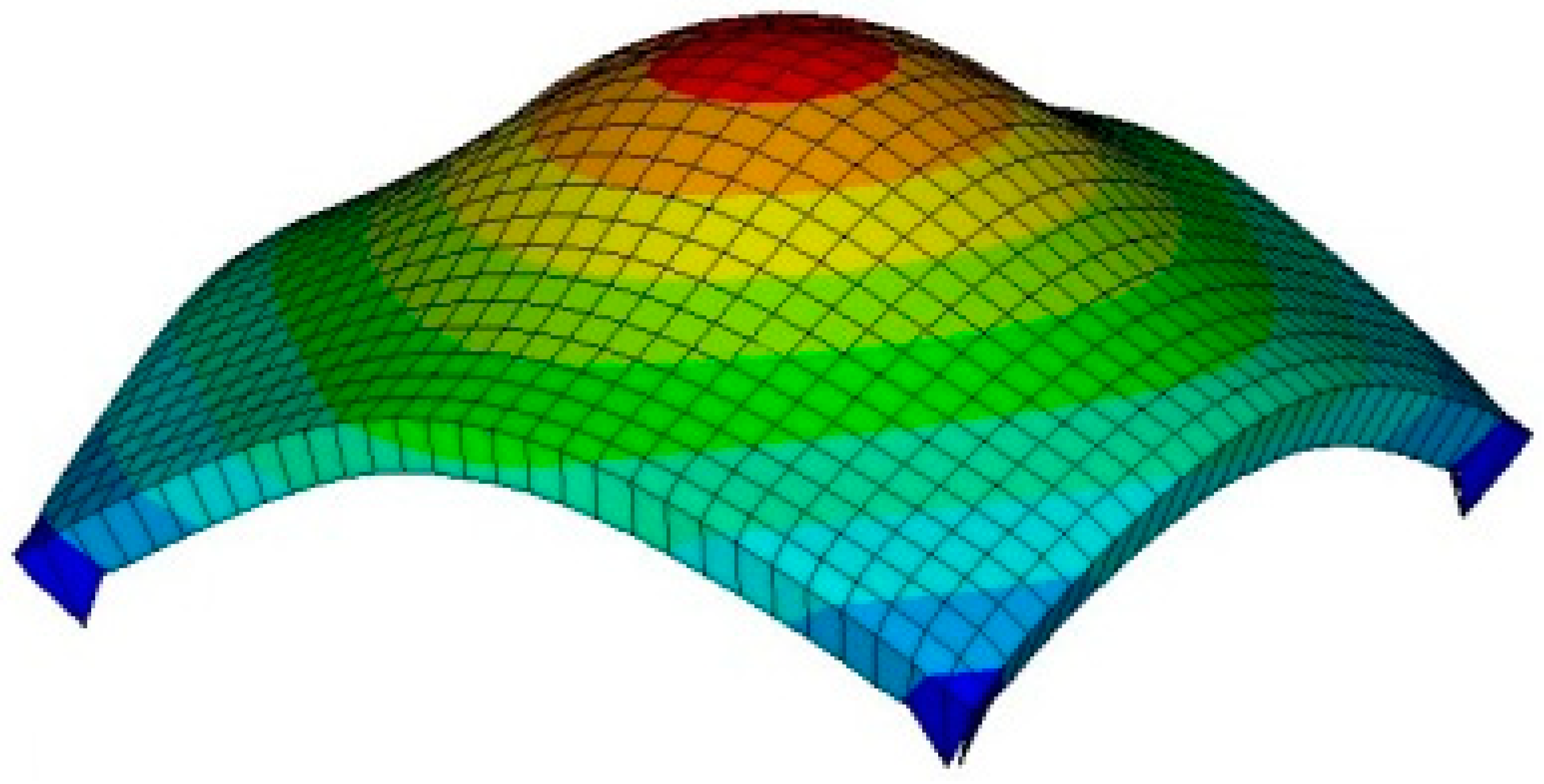

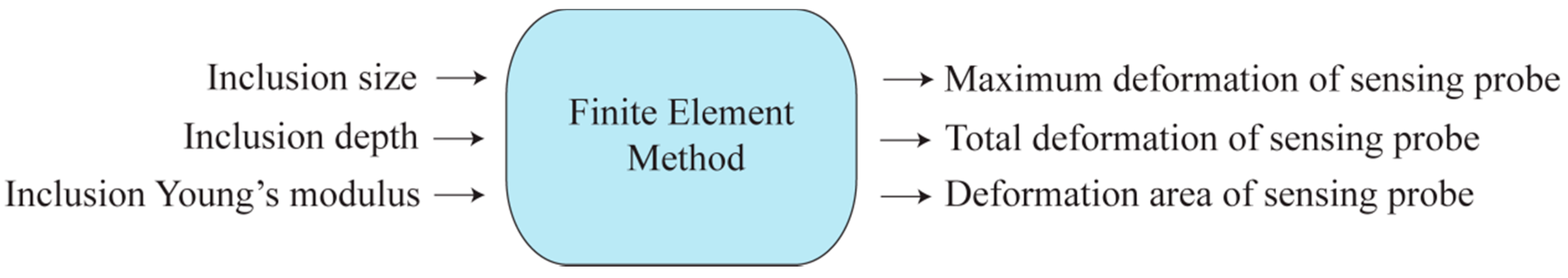
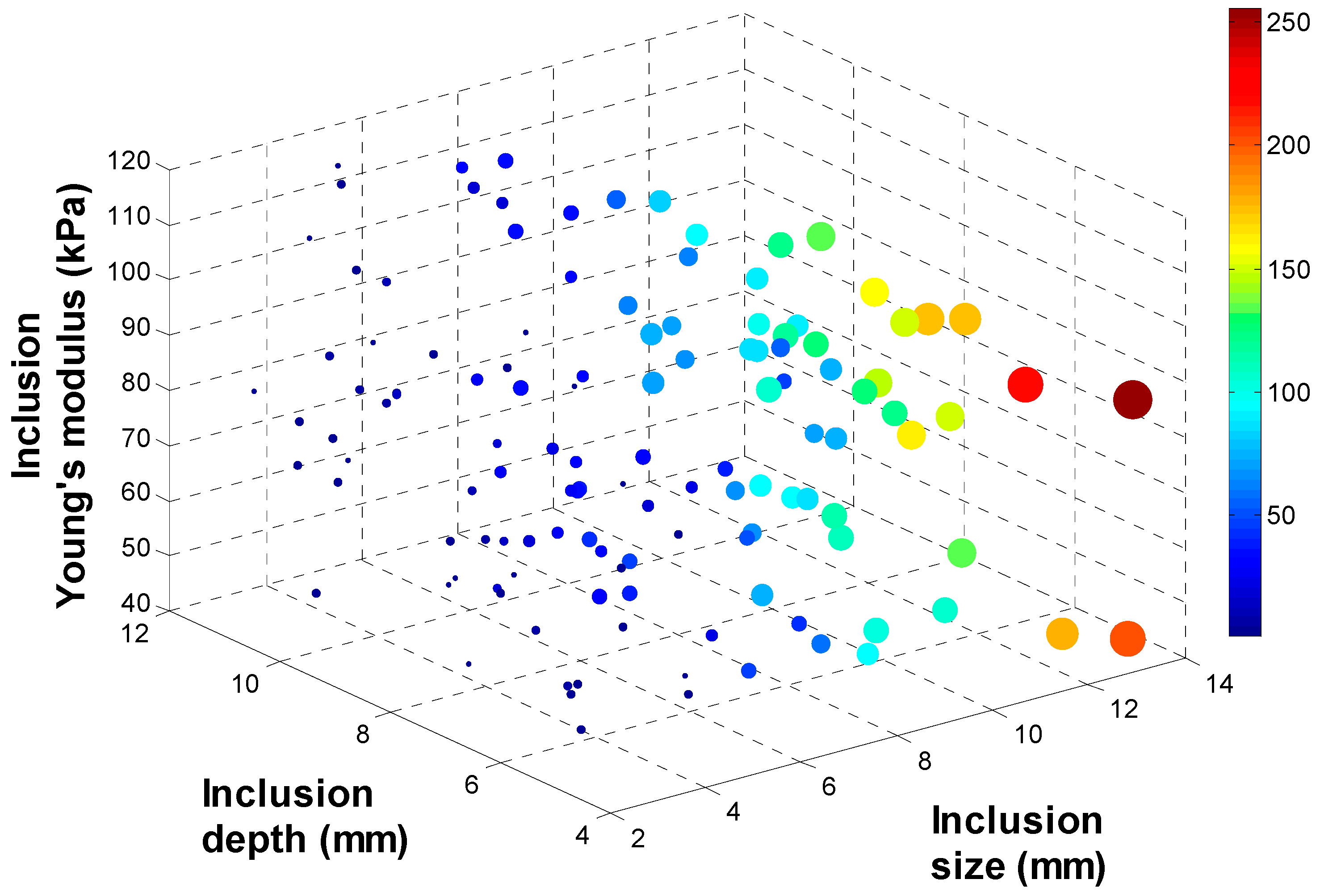

3.3. Tactile Data from TSIS
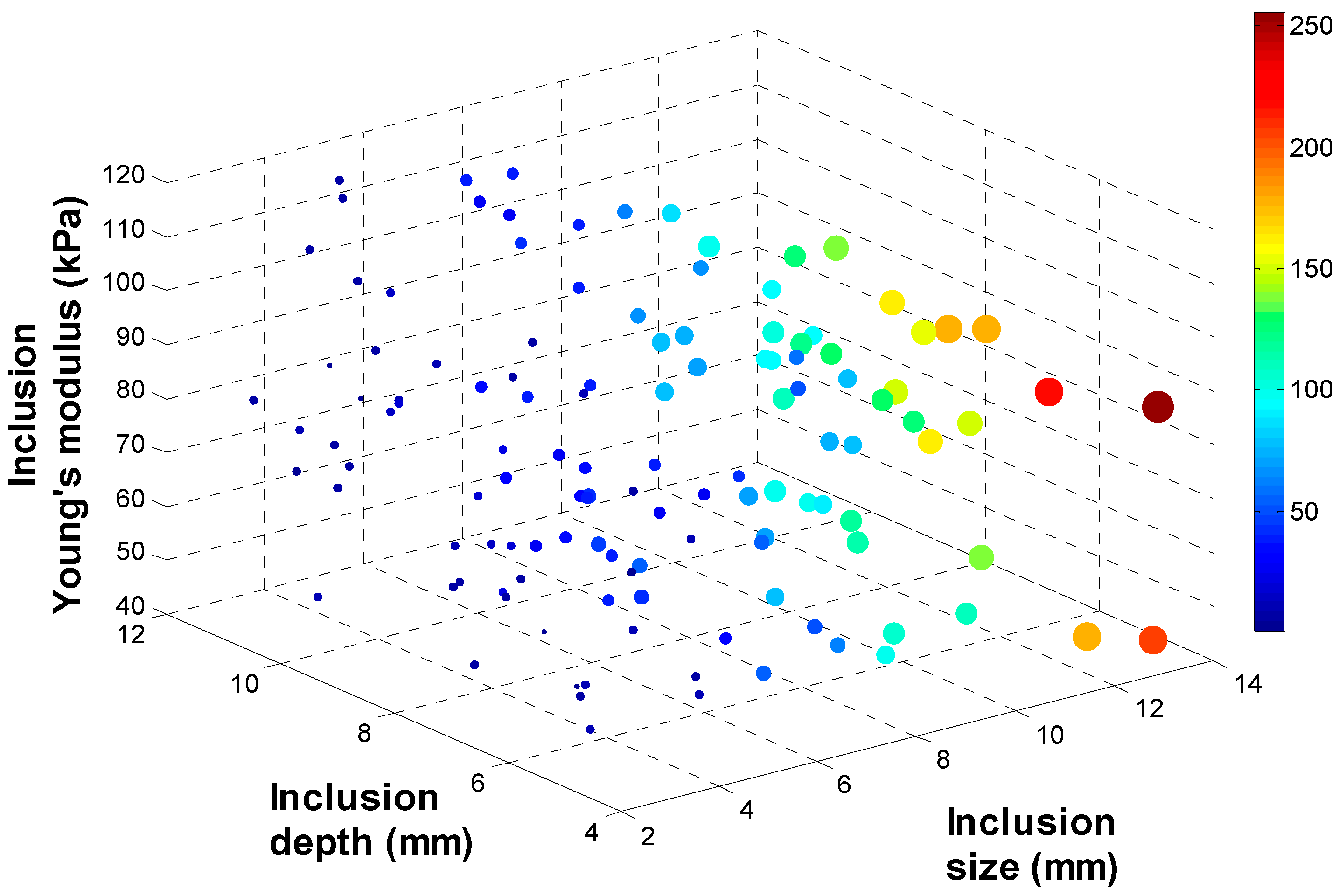
3.4. Calibration: Mapping TSIS Tactile Data to FEM Tactile Data



3.5. Inversion Algorithm


4. Experimental Results
4.1. Validation Method
4.2. Test Results
| Iterations | Hold-Out Validation | Leave-One-Out Cross Validation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 50 | 100 | 10 | 50 | 100 | ||||||||
| Output # | Train | Test | Train | Test | Train | Test | Train | Test | Train | Test | Train | Test | |
| SCGA | 1 (size) | 0.79 | 1.58 | 0.3 | 1.03 | 0.27 | 0.99 | 0.93 | 1.72 | 0.32 | 0.79 | 0.29 | 0.58 |
| 2 (depth) | 2.7 | 4.08 | 2.59 | 3.99 | 2.56 | 4.09 | 2.78 | 4.1 | 2.66 | 3.87 | 2.62 | 3.82 | |
| 3 (modulus) | 3.16 | 6.6 | 3.1 | 6.58 | 3.09 | 4.6 | 3.36 | 6.59 | 3.28 | 6.64 | 3.24 | 6.9 | |
| LMA | 1 (size) | 0.28 | 1.06 | 0.26 | 3.19 | 0.26 | 6.84 | 0.29 | 0.52 | 0.26 | 1.03 | 0.25 | 3.08 |
| 2 (depth) | 2.54 | 5.21 | 2.36 | 39.1 | 2.25 | 105 | 2.59 | 3.86 | 2.42 | 6.25 | 2.29 | 33.8 | |
| 3 (modulus) | 3.05 | 6.53 | 2.91 | 12.1 | 2.77 | 46.4 | 3.16 | 6.9 | 2.83 | 13.3 | 2.68 | 43.1 | |
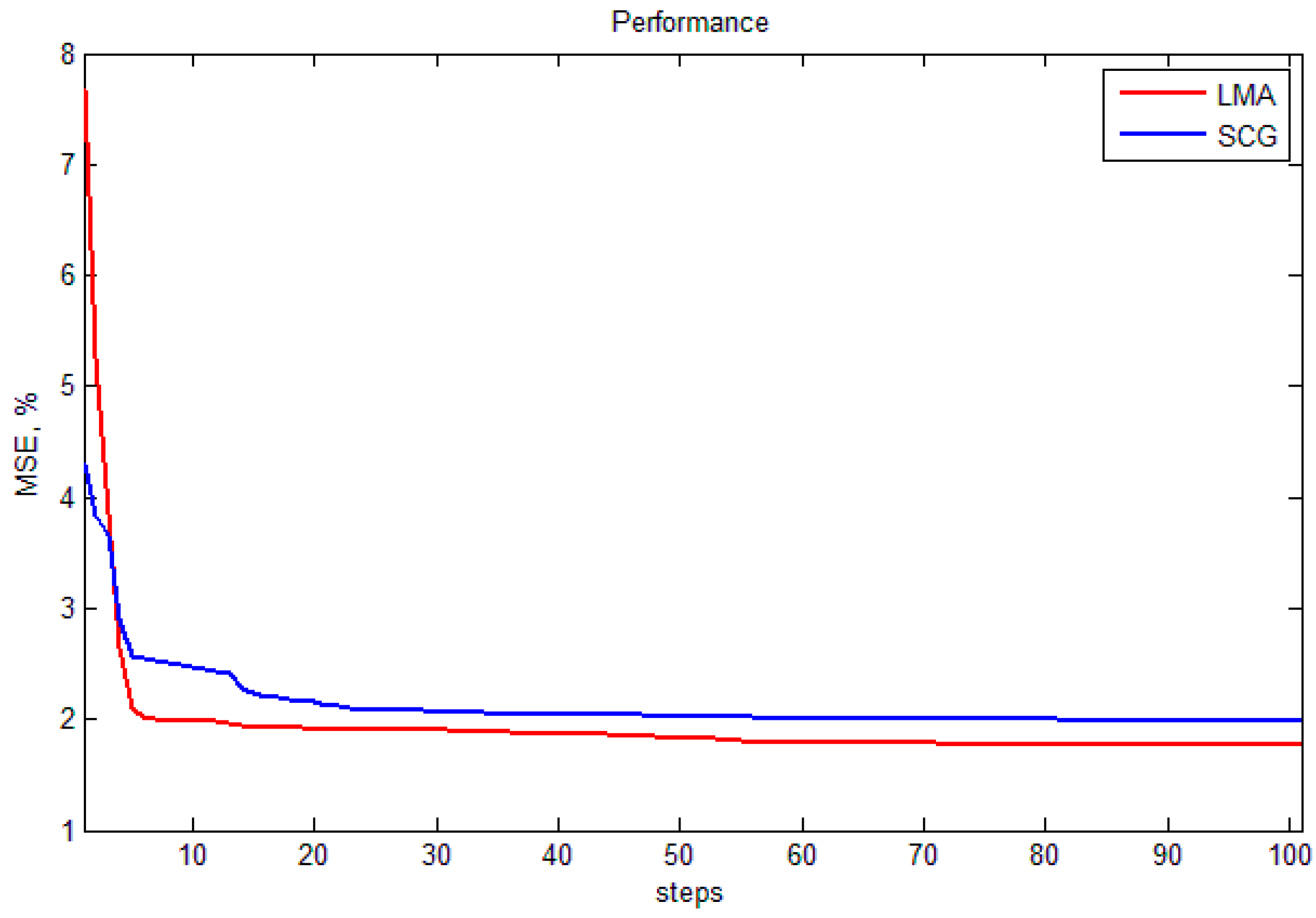
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures 2010; American Cancer Society: Atlanta, GA, USA, 2010. [Google Scholar]
- Ehman, E.C.; Rossman, P.J.; Kruse, S.A.; Sahakian, A.V.; Glaser, K.J. Vibration safety limits for magnetic resonance elastography. Phys. Med. Biol. 2008, 53, 925. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Parker, K.J.; Lerner, R.; Levinson, S. Imaging of the elastic properties of tissue—A review. Ultrasound Med. Biol. 1996, 22, 959–977. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Cokkinides, V.; Eyre, H.J. American cancer society guidelines for the early detection of cancer. CA: A Cancer J. Clin. 2006, 56, 11–25. [Google Scholar] [CrossRef]
- Kopans, D.B. Clinical breast examination for detecting breast cancer. J. Am. Med. Assoc. 2000, 283, 1270–1280. [Google Scholar]
- Jatoi, I. Breast Cancer Screening; Chapman and Hall: New York, NY, USA, 1997. [Google Scholar]
- Wellman, P.S. Tactile Imaging; Division of Engineering and Applied Sciences, Harvard University: Cambridge, MA, USA, 1999. [Google Scholar]
- Wellman, P.S.; Dalton, E.P.; Krag, D.; Kern, K.A.; Howe, R.D. Tactile imaging of breast masses: First clinical report. Arch. Surg. 2001, 136, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.L. Taber’s Cyclopedic Medical Dictionary; F.A. Davis Co.: Philadelphia, PA, USA, 1997. [Google Scholar]
- Fung, Y.C. Biomechanics: Mechanical Properties of Living Tissues; Springer Verlag: New York, NY, USA, 1993. [Google Scholar]
- Parker, K.J.; Huang, S.R.; Musulin, R.A.; Lerner, R.M. Tissue response to mechanical vibrations for sonoelasticity imaging. Ultrasound Med. Biol. 1990, 16, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Vinckier, A.; Semenza, G. Measuring elasticity of biological materials by atomic force microscopy. Feder. Eur. Biochem. Soc. Lett. 1998, 430, 12–16. [Google Scholar] [CrossRef]
- Insana, M.F.; Pellot-Barakat, C.; Sridhar, M.; Lindfors, K.K. Viscoelastic imaging of breast tumor microenvironment with ultrasound. J. Mammary Gland Biol. Neoplasia 2004, 9, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Stravros, A.; Thickman, D.; Dennis, M.; Parker, S.; Sisney, G. Solid breast nodules: Use of sonography to distinguish between benign and malignant lesions. Radiology 1995, 196, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.J.W.; Merritt, C.; Piccoli, C.; Schmidt, R.; Rouse, G.; Fornage, B.; Rubin, E.; Georgian-Smith, D.; Winsberg, F.; Goldberg, B.; et al. Ultrasound as a complement to mammography and breast examination to characterize breast masses. Ultrasound Med. Biol. 2002, 28, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ophir, J.; Cespedes, I.; Pennekanti, H.; Yazdi, Y.; Li, X. Elastography, a quantitative method for imaging the elasticity of biological tissues. Ultrason. Imag. 1991, 13, 111–134. [Google Scholar] [CrossRef]
- O’Donnell, M.; Skovoroda, A.R.; Shapo, B.M.; Emelianov, S.Y. Internal displacement and strain imaging using ultrasonic speckle tracking. IEEE Trans. Biol. Ultrason. Ferroelectr. Freq. Control. 1994, 41, 314–325. [Google Scholar] [CrossRef]
- Krouskop, T.; Wheeler, T.; Kallel, F.; Garra, B. Elastic moduli of breast and prostate tissues under compression. Ultrason. Imaging 1998, 20, 260–274. [Google Scholar] [CrossRef] [PubMed]
- Lerner, R.M.; Parker, K.J.; Holen, J.; Gremiak, R.; Waag, R.C. “Sonoelasticity”: Medical elasticity images derived from ultrasound signals in mechanically vibrated targets. Ultrasound Med. Biol. 1988, 16, 231–239. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Sato, J.; Sato, T. Ultrasonic imaging of internal vibration of soft tissue under forced vibration. IEEE Trans. Ultrason. Ferroelec. Freq. Contr. 1990, 17, 45–53. [Google Scholar] [CrossRef]
- Zienkiewicz, O.C.; Taylor, R.L. The Finite Element Method, 5th ed.; Butterworth-Heinemann: Portsmouth, NH, USA, 2000; Volume 1. [Google Scholar]
- Garra, B.; Cespedes, E.; Ophir, J.; Spratt, S.; Zuurbier, R.; Magnant, C.; Pennanen, M. Elastography of breast lesions: Initial clinical results. Radiology 1997, 22, 959–977. [Google Scholar]
- Rogowska, J.; Patel, N.A.; Fujimoto, J.G.; Brezinski, M.E. Optical coherence tomographic elastography technique for measuring deformation and strain of atherosclerotic tissues. Heart 2004, 90, 556–562. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-H.; Kim, Y.N.; Park, H.-J. Bio-Optics Based Sensation Imaging for Breast Tumor Detection Using Tissue Characterization. Sensors 2015, 15, 6306-6323. https://doi.org/10.3390/s150306306
Lee J-H, Kim YN, Park H-J. Bio-Optics Based Sensation Imaging for Breast Tumor Detection Using Tissue Characterization. Sensors. 2015; 15(3):6306-6323. https://doi.org/10.3390/s150306306
Chicago/Turabian StyleLee, Jong-Ha, Yoon Nyun Kim, and Hee-Jun Park. 2015. "Bio-Optics Based Sensation Imaging for Breast Tumor Detection Using Tissue Characterization" Sensors 15, no. 3: 6306-6323. https://doi.org/10.3390/s150306306






