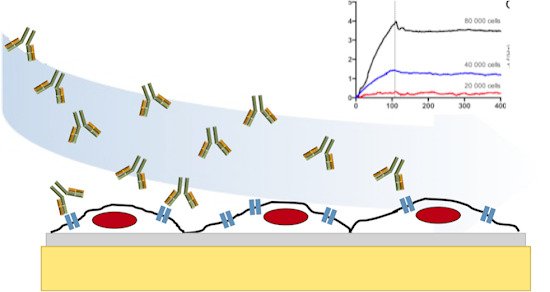
Study of the Interaction of Trastuzumab and SKOV3 Epithelial Cancer Cells Using a Quartz Crystal Microbalance Sensor
Abstract
:1. Introduction
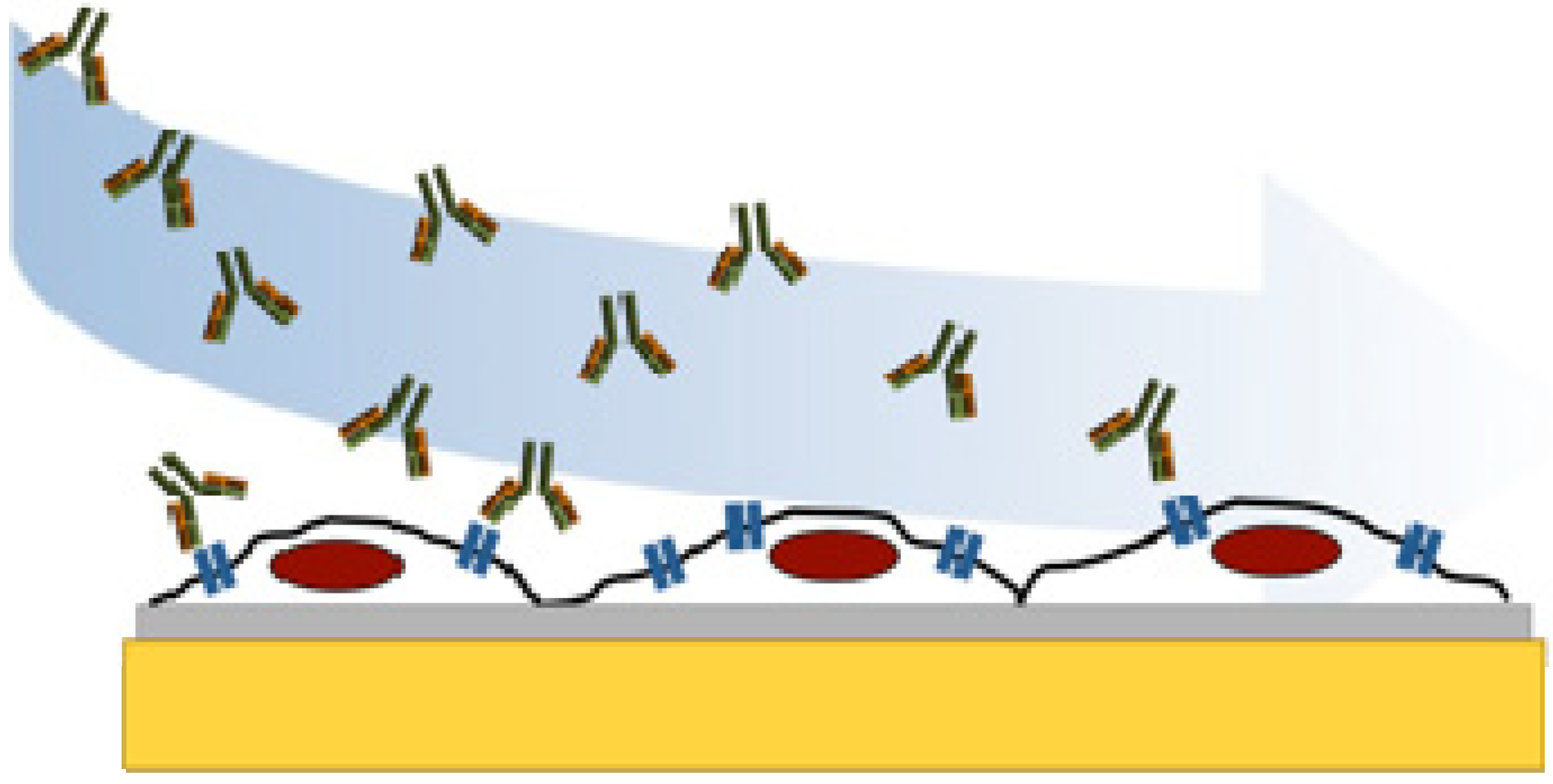
2. Experimental Section
2.1. Chemicals
2.2. Preparation of Cell Chips
2.3. Quartz Crystal Microbalance Studies
3. Results and Discussion
3.1. Fixation of SKOV3 Cells to COP-1 Chips
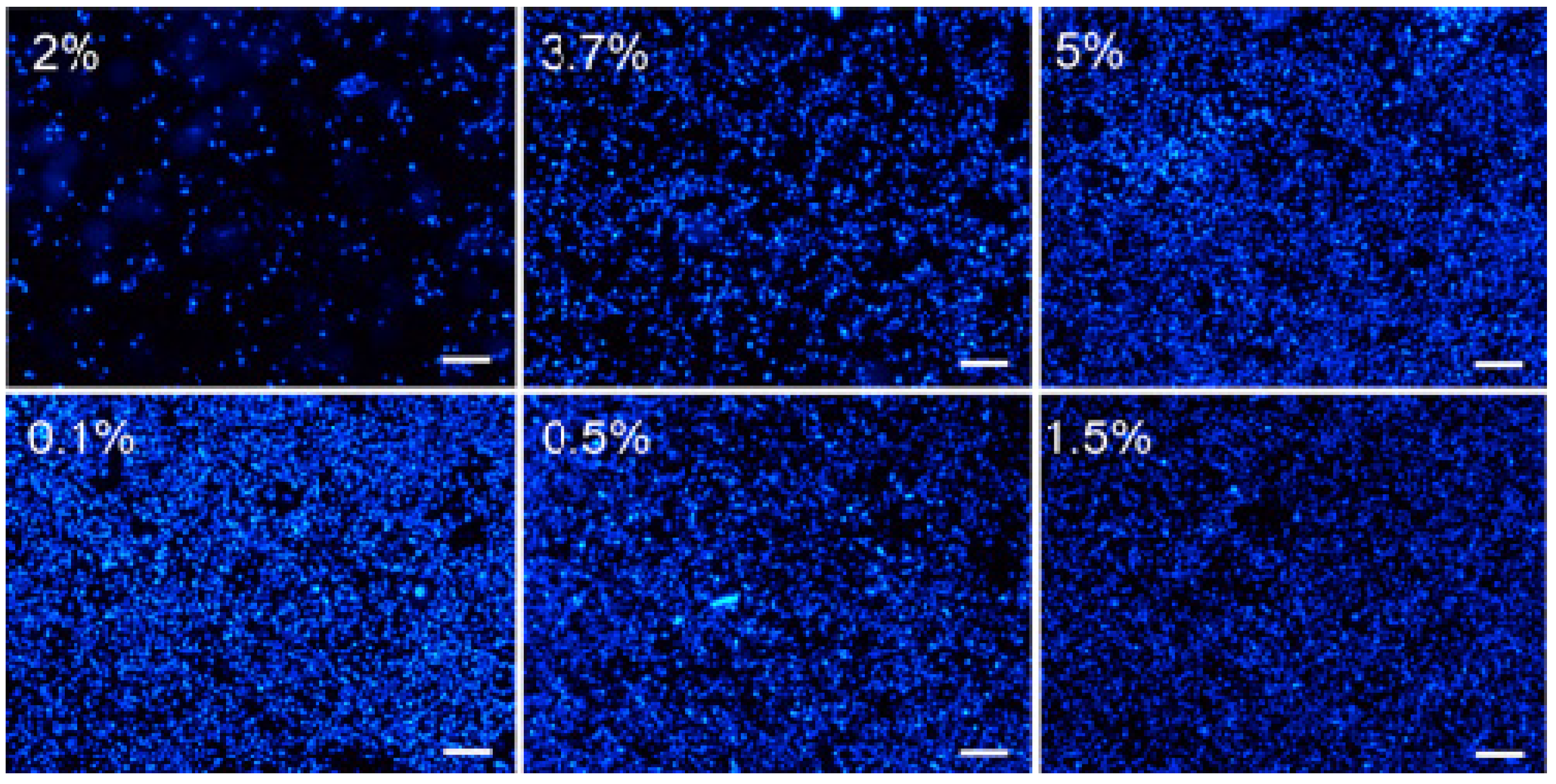
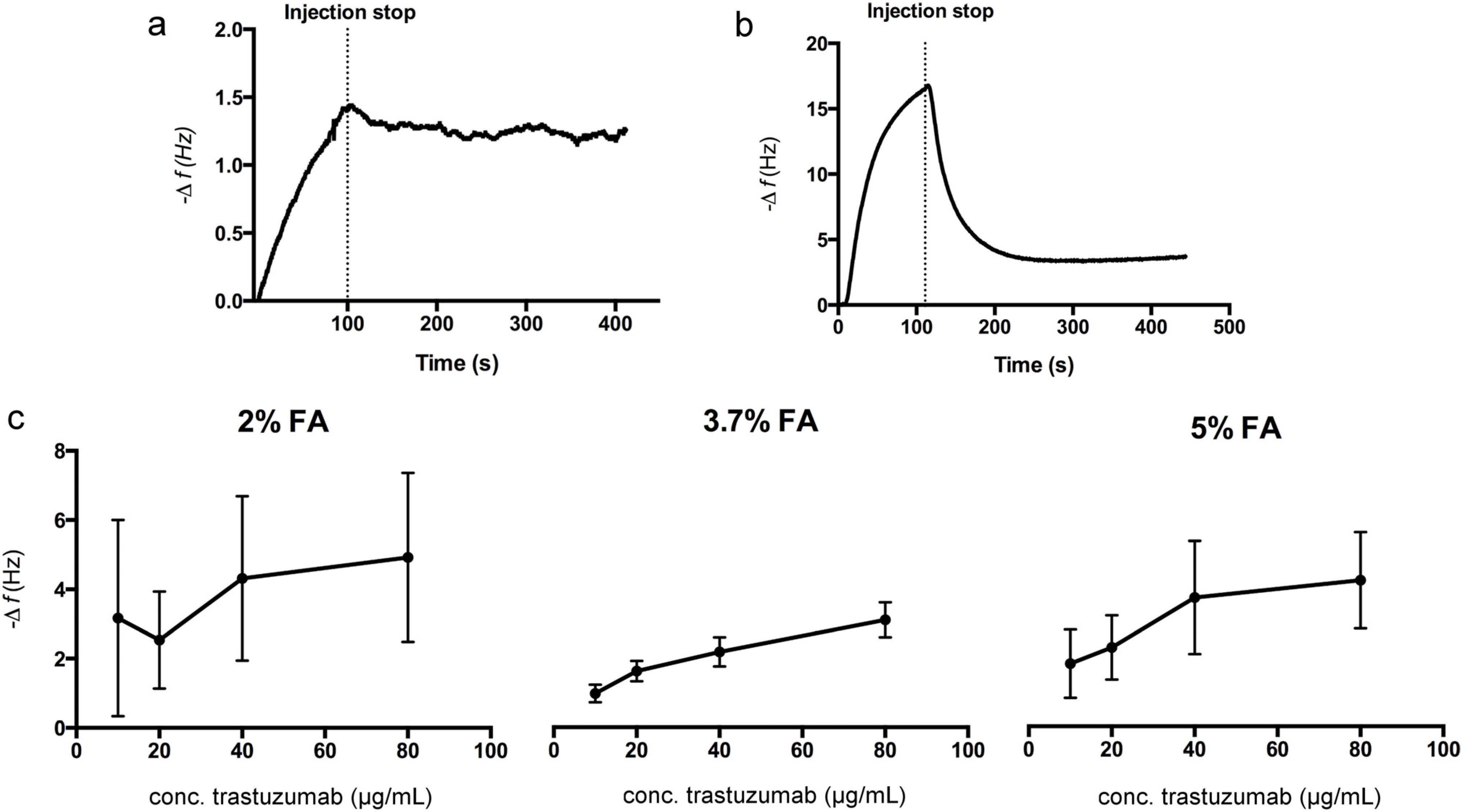
3.2. Seeding Density

3.3. Kinetic Evaluation of Trastuzumab Binding to SKOV3 Cell Chips
4. Conclusions/Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Michelini, E.; Cevenini, L.; Mezzanotte, L.; Coppa, A.; Roda, A. Cell-based assays: Fuelling drug discovery. Anal. Bioanal. Chem. 2010, 398, 227–238. [Google Scholar] [CrossRef] [PubMed]
- An, W.F.; Tolliday, N. Cell-based assays for high-throughput screening. Mol. Biotechnol. 2010, 45, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Tothill, I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar] [CrossRef]
- Hawkins, E.; Cooper, M.; Campbell, I. Acoustic detection technology in the analysis of biomolecular interactions. Innov. Pharm. Technol. 2006, 21, 30–34. [Google Scholar]
- Elebring, T.; Gill, A.; Plowright, A.T. What is the most important approach in current drug discovery: Doing the right things or doing things right? Drug Discov. Today 2012, 17, 1166–1169. [Google Scholar] [CrossRef] [PubMed]
- Núñez, S.; Venhorst, J.; Kruse, C.G. Target–drug interactions: First principles and their application to drug discovery. Drug Discov. Today 2012, 17, 10–22. [Google Scholar] [CrossRef]
- Li, X.; Pei, Y.; Zhang, R.; Shuai, Q.; Wang, F.; Aastrup, T.; Pei, Z. A suspension-cell biosensor for real-time determination of binding kinetics of protein-carbohydrate interactions on cancer cell surfaces. Chem. Commun. 2013, 49, 9908–9910. [Google Scholar] [CrossRef]
- Mejard, R.; Griesser, H.J.; Thierry, B. Optical biosensing for label-free cellular studies. Trac-Trend. Anal. Chem. 2014, 53, 178–186. [Google Scholar] [CrossRef]
- Marcotte, L.; Tabrizian, M. Sensing surfaces: Challenges in studying the cell adhesion process and the cell adhesion forces on biomaterials. IRBM 2008, 29, 77–88. [Google Scholar] [CrossRef]
- Speight, R.E.; Cooper, M.A. A survey of the 2010 quartz crystal microbalance literature. J. Mol. Recognit. 2012, 25, 451–473. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yang, Z.; Lei, C.; Lei, J.; Zhou, Y. An integrated giant magnetoimpedance biosensor for detection of biomarker. Biosens. Bioelectron. 2014, 58, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mohapatra, S.; Fal-Miyar, V.; Cerdeira, A.; García, J.A.; Srikanth, H.; Gass, J.; Kurlyandskaya, G.V. Magnetoimpedance biosensor for fe3o4 nanoparticle intracellular uptake evaluation. App. Phys. Lett. 2007, 91, 143902. [Google Scholar] [CrossRef]
- Su, X.D.; Chew, F.T.; Li, S.F.Y. Design and application of piezoelectric quartz crystal-based immunoassay. Anal. Sci. 2000, 16, 107–114. [Google Scholar] [CrossRef]
- Marx, K.A. Quartz crystal microbalance: A useful tool for studying thin polymer films and complex biomolecular systems at the solution-surface interface. Biomacromolecules 2003, 4, 1099–1120. [Google Scholar] [CrossRef] [PubMed]
- Anderson, H.; Jönsson, M.; Vestling, L.; Lindberg, U.; Aastrup, T. Quartz crystal microbalance sensor design: I. Experimental study of sensor response and performance. Sens. Actuat. B-Chem. 2007, 123, 27–34. [Google Scholar] [CrossRef]
- Funari, R.; Della Ventura, B.; Carrieri, R.; Morra, L.; Lahoz, E.; Gesuele, F.; Altucci, C.; Velotta, R. Detection of parathion and patulin by quartz-crystal microbalance functionalized by the photonics immobilization technique. Biosens. Bioelectron. 2014, 67, 224–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, R.; Jeong, B.; Rahman, M.A. Stimulated mass enhancement strategy-based highly sensitive detection of a protein in serum using quartz crystal microbalance technique. Analyst 2015, 140, 995–998. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.K.; Guilbault, G.G. Commercial quartz crystal microbalances–theory and applications. Biosens. Bioelectron. 1999, 14, 663–670. [Google Scholar] [CrossRef]
- Bunde, R.L.; Jarvi, E.J.; Rosentreter, J.J. Piezoelectric quartz crystal biosensors. Talanta 1998, 46, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Shons, A.; Dorman, F.; Najarian, J. An immunospecific microbalance. J. Biomed. Mater. Res. 1972, 6, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Whitcombe, M.J.; Chianella, I.; Larcombe, L.; Piletsky, S.A.; Noble, J.; Porter, R.; Horgan, A. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2011, 40, 1547–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmlund, L.; Suriyanarayanan, S.; Wiklander, J.G.; Aastrup, T.; Nicholls, I.A. Biotin selective polymer nano-films. J. Nanobiotechnology. 2014, 12. Available online: http://www.biomedcentral.com/content/pdf/1477-3155-12-8.pdf (accessed on 2 March 2015). [CrossRef]
- Madani, S.Y.; Tan, A.; Dwek, M.; Seifalian, A.M. Functionalization of single-walled carbon nanotubes and their binding to cancer cells. Int. J. Nanomed. 2012, 7, 905–914. [Google Scholar]
- Pei, Z.C.; Saint-Guirons, J.; Kack, C.; Ingemarsson, B.; Aastrup, T. Real-time analysis of the carbohydrates on cell surfaces using a qcm biosensor: A lectin-based approach. Biosens. Bioelectron. 2012, 35, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Peiris, D.; Markiv, A.; Curley, G.P.; Dwek, M.V. A novel approach to determining the affinity of protein–carbohydrate interactions employing adherent cancer cells grown on a biosensor surface. Biosens. Bioelectron. 2012, 35, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Braunhut, S.J.; McIntosh, D.; Vorotnikova, E.; Zhou, T.; Marx, K.A. Detection of apoptosis and drug resistance of human breast cancer cells to taxane treatments using quartz crystal microbalance biosensor technology. Assay Drug Dev. Technol. 2005, 3, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Marx, K.A.; Zhou, T.; Montrone, A.; McIntosh, D.; Braunhut, S.J. A comparative study of the cytoskeleton binding drugs nocodazole and taxol with a mammalian cell quartz crystal microbalance biosensor: Different dynamic responses and energy dissipation effects. Anal. Biochem. 2007, 361, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Chen, J.; Zhang, Y.; Chen, K.; Pan, C.; Yao, S. Enhanced adhesion/spreading and proliferation of mammalian cells on electropolymerized porphyrin film for biosensing applications. Biosens. Bioelectron. 2008, 23, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.P.; Jia, X.E.; Tan, L.; Xie, Q.J.; Lei, L.H.; Yao, S.Z. Magnetically enhanced cytotoxicity of paramagnetic selenium-ferroferric oxide nanocomposites on human osteoblast-like mg-63 cells. Biosens. Bioelectron. 2010, 25, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.W.; Muramatsu, H.; Lee, B.J.; Kwon, Y.S. Monitoring of anticancer effect of cisplatin and 5-fluorouracil on hepg2 cells by quartz crystal microbalance and micro ccd camera. Biosens. Bioelectron. 2010, 26, 1576–1581. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Guo, M.; Nie, Z.; Huang, Y.; Pan, C.; Zeng, K.; Zhang, Y.; Yao, S. Selective collection and detection of leukemia cells on a magnet-quartz crystal microbalance system using aptamer-conjugated magnetic beads. Biosens. Bioelectron. 2010, 25, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhao, L.B.; Guo, S.S.; Shi, B.X.; Lam, T.L.; Leung, Y.C.; Chen, Y.; Zhao, X.Z.; Chan, H.L.W.; Wang, Y. A microfluidic system with surface modified piezoelectric sensor for trapping and detection of cancer cells. Biosens. Bioelectron. 2010, 26, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Lin, P.; Chisti, M.M.; Rehman, A.; Zeng, X. Real time analysis of binding between rituximab (anti-cd20 antibody) and b lymphoma cells. Anal. Chem. 2013, 85, 8543–8551. [Google Scholar] [CrossRef] [PubMed]
- Davoli, A.; Hocevar, B.A.; Brown, T.L. Progression and treatment of her2-positive breast cancer. Cancer Chemoth. Pharm. 2010, 65, 611–623. [Google Scholar] [CrossRef]
- Wegener, J.; Janshoff, A.; Galla, H.J. Cell adhesion monitoring using a quartz crystal microbalance: Comparative analysis of different mammalian cell lines. Eur. Biophys. J. Biophy. 1998, 28, 26–37. [Google Scholar] [CrossRef]
- Fohlerova, Z.; Turanek, J.; Skladal, P. The cell adhesion and cytotoxicity effects of the derivate of vitamin e compared for two cell lines using a piezoelectric biosensor. Sens. Actuat. B-Chem. 2012, 174, 153–157. [Google Scholar] [CrossRef]
- Tanaka, K.A.; Suzuki, K.G.; Shirai, Y.M.; Shibutani, S.T.; Miyahara, M.S.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T.K.; et al. Membrane molecules mobile even after chemical fixation. Nat. Methods 2010, 7, 865–866. [Google Scholar] [CrossRef]
- Walter, H.; Krob, E.J. Fixation with even small quantities of glutaraldehyde affects red-blood-cell surface-properties in a cell-dependent and species-dependent manner-studies by cell partitioning. Bioscience Rep. 1989, 9, 727–735. [Google Scholar] [CrossRef]
- Kiernan, J. Formaldehyde, formalin, paraformaldehyde and glutaraldehyde: What they are and what they do. Microsc. Today 2000, 00–1, 8–12. [Google Scholar]
- Rich, R.L.; Myszka, D.G. Survey of the year 2007 commercial optical biosensor literature. J. Mol. Recognit. 2008, 21, 355–400. [Google Scholar] [CrossRef] [PubMed]
- Carter, P.; Presta, L.; Gorman, C.M.; Ridgway, J.B.B.; Henner, D.; Wong, W.L.T.; Rowland, A.M.; Kotts, C.; Carver, M.E.; Shepard, H.M. Humanization of an anti-p185her2 antibody for human cancer-therapy. Proc. Natl. Acad. Sci. USA 1992, 89, 4285–4289. [Google Scholar] [CrossRef] [PubMed]
- Bostrom, J.; Haber, L.; Koenig, P.; Kelley, R.F.; Fuh, G. High affinity antigen recognition of the dual specific variants of herceptin is entropy-driven in spite of structural plasticity. PLoS One 2011, 6, e17887. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elmlund, L.; Käck, C.; Aastrup, T.; Nicholls, I.A. Study of the Interaction of Trastuzumab and SKOV3 Epithelial Cancer Cells Using a Quartz Crystal Microbalance Sensor. Sensors 2015, 15, 5884-5894. https://doi.org/10.3390/s150305884
Elmlund L, Käck C, Aastrup T, Nicholls IA. Study of the Interaction of Trastuzumab and SKOV3 Epithelial Cancer Cells Using a Quartz Crystal Microbalance Sensor. Sensors. 2015; 15(3):5884-5894. https://doi.org/10.3390/s150305884
Chicago/Turabian StyleElmlund, Louise, Camilla Käck, Teodor Aastrup, and Ian A. Nicholls. 2015. "Study of the Interaction of Trastuzumab and SKOV3 Epithelial Cancer Cells Using a Quartz Crystal Microbalance Sensor" Sensors 15, no. 3: 5884-5894. https://doi.org/10.3390/s150305884