Detection and Characterization of Cancer Cells and Pathogenic Bacteria Using Aptamer-Based Nano-Conjugates
Abstract
: Detection and characterization of cells using aptamers and aptamer-conjugated nanoprobes has evolved a great deal over the past few decades. This evolution has been driven by the easy selection of aptamers via in vitro cell-SELEX, permitting sensitive discrimination between target and normal cells, which includes pathogenic prokaryotic and cancerous eukaryotic cells. Additionally, when the aptamer-based strategies are used in conjunction with nanomaterials, there is the potential for cell targeting and therapeutic effects with improved specificity and sensitivity. Here we review recent advances in aptamer-based nano-conjugates and their applications for detecting cancer cells and pathogenic bacteria. The multidisciplinary research utilized in this field will play an increasingly significant role in clinical medicine and drug discovery.1. Introduction
Extensive genetic information on a large panel of diseases has been necessarily accompanied by targeting cells, which is of great significance for early diagnosis and effective therapy. While traditional methods used to identify the differences between normal and abnormal cells or between host and pathogenic cells rely upon the long-standing observation of cell phenotypes or PCR-based molecular diagnosis [1–3], straightforward methods to target aberrant cells have been developed using sensing molecules, such as monoclonal antibodies, smaller antibody fragments, peptides and low-molecular mass ligands [4–7].
As single-stranded nucleic acids (ssDNA or RNA), aptamers offer several advantages over other sensing molecules for diagnostic and therapeutic applications [8]. In addition to being chemically stable, cost-effective and producible on a large scale, aptamers also possess an intermediate size (between antibodies and small peptides) and have comparable or higher binding affinities for their targets. These targets range from small molecules to whole cells [9–11] via an in vitro selection process known as systematic evolution of ligands by exponential enrichment (SELEX) [12]. In particular, the use of aptamers that utilize cell-SELEX have contributed to significant advances in disease diagnosis and drug development on the cellular and tissue levels [13,14] and are superior to antibody-based diagnostic and therapeutic applications. Although there is no prior knowledge of specific targets, a counter-selection strategy using control cells (negative-SELEX) with target cell-SELEX gives rise to cell-specific aptamers with high stability and binding affinity. These allow for the reliable identification of targeted cells at the molecular level in combination with various analysis tools, such as fluorescence-activated cell sorting or the enzyme-linked immunosorbent assay [15,16]. Unlike antibodies based on purified receptors, aptamers are more attainable and responsive in living cells because they are selected from intact cells; it is not necessary to consider the conformational changes of the recognition domains in the cell membrane. Among many other types of cells, bacteria and tumor cells have been of primary interest [17,18], due to their involvement in many human diseases.
To exploit the full potential of aptamer-based cell targeting, aptamers can be combined with nanomaterials, such as gold nanoparticles (AuNPs), silica NPs (SiNP), graphenes, magnetic NPs (MNP) and quantum dots (QDs). This aptamer-nanomaterial hybridization process is easily accomplished due to the simple chemical modification and well-defined structures of aptamers [19,20]. These hybrid materials are expected to improve target diagnosis and therapy with higher sensitivity and selectivity compared to aptamer-only-based targeting strategies. Most importantly, due to the large surface area, multivalent structure, and relevant physiochemical properties of nanomaterials, aptamer-nanomaterial hybrids should provide signal amplification and an increased target binding affinity in a multivalent manner.
Although many reviews discuss aptamer-functionalized nanomaterials [21–24], this review includes recent advances in isolation, detection, and characterization of cells using the aptamer-nanomaterial hybrid systems via cell-SELEX. Furthermore, to avoid redundancy with other recent reports, which provide broad insight into aptamers and their recent applications in cancer diagnosis and therapy [25], we focus on applications based on living bacteria and cancer cells with recent advances in targeting strategies.
2. Cell-SELEX
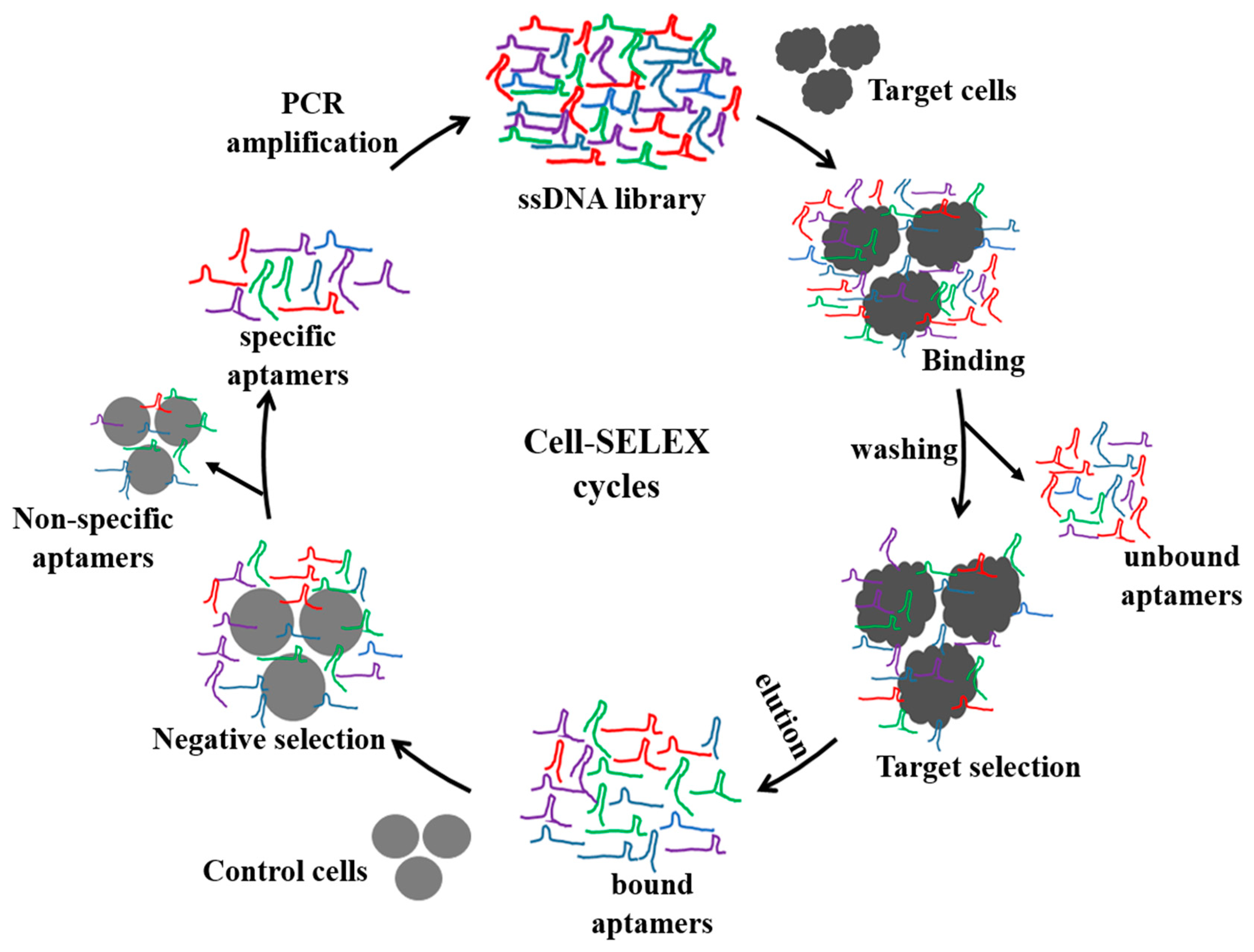
Among living cells, aptamers have been developed to detect whole single cells, where they bind to cell surface protein targets. As summarized in Tables 1 and 2, recent reports of cell-SELEX have mainly focused on specific aptamers against various cancer cells [15,16,26–36] and whole bacteria [18,37–50]. As depicted in Figure 1, to generate a specific aptamer, a random ssDNA/RNA aptamer library is initially incubated with target cells for a specific period of time (from 30 min to 1 h) to allow for target-specific recognition. This is followed by centrifugation to remove the unbound aptamer. The cells are then washed several times, and the aptamers bound to the surfaces of the cells are eluted by heat-induced denaturation. The eluted aptamers are amplified and further subjected to negative selection using control cells; negative selection plays an important role in cell-SELEX to remove aptamers binding to common cell surface proteins. In order to increase the efficiency of negative selection, the concentrations of the control cells are often in five- to ten-fold excesses compared to the target cells. This selection can be performed for several rounds until the produced aptamers have a high binding affinity (nanomolar range Kd values) toward the target cells. Consequently, the aptamers that are evolved from cell-SELEX are implemented for detection of cancer or infectious cells, discovery of new biomarkers, and disease therapy [10,13,14,51–53]. Furthermore, cell-binding aptamers are suitable for the development of in vivo tumor targeting [54,55], receptor-dependent neutralization [56–58], and drug delivery systems [59,60].
3. Aptamer-Conjugated Nanomaterials in Whole-Cell Detection
The hybridization of aptamers with nanomaterials is of particular interest due to the unique properties and utilities of nanomaterials, including their small size, increased surface-to-volume ratio, and a wide range of sensing modules, that include metal and semiconductor core materials. To date, nanomaterials can be easily fashioned with aptamers via direct covalent linkage or by non-covalent interactions. This modification aim to address several shortcomings caused by less than ideal detection sensitivity, assay time, and target specificity [22,61–64]. While several strategies have been reported for conjugating aptamers with nanomaterials [19], nanomaterials are generally used as either supporting reservoirs for immobilizing ligands or as labeling agents for signal amplification. Importantly, aptamer-conjugated-nanoparticles (Apt-NP) are capable of being detected by optical, electrochemical, fluorescence or mass-sensitive analytical techniques, depending on their physical and/or chemical properties. Compared with antibody-based cell assays, aptamers screened from cell-SELEX provide much more versatile strategies for recognizing cells because the smaller size of the aptamers is beneficial for designing nano-hybrid sensors in a highly compact regime, which contributes to the signal amplification effect and increases target binding affinity. Since the aptamers are also directly selected from living cells, Apt-NPs would be more effective for targeting cells, compared to antibodies or their nano-hybrid formats, which often do not reflect the conformational changes of receptor proteins in living cells. In addition, nanomaterials could protect aptamers from being digested by nucleases, indicating that Apt-NPs can act as stealthy carriers for drug or gene delivery. Here we provide an overview of the nanomaterials that are widely used in aptamer-based sensors for detecting cancer cells and pathogenic bacteria via limit of detection (LOD).
3.1. Aptamer-Conjugated AuNPs
AuNPs are widely used in bioanalysis due to their simple synthesis and unique spectral properties. AuNPs have typically been produced from the chemical reduction of AuCl4 ions by agents, such as citric acid, and their size can be easily controlled [65–67]. Due to their biocompatibility and optical properties triggered by the surface plasmon phenomenon, AuNPs conjugated with aptamers from cell-SELEX have been extensively utilized in the fields of cellular diagnostics and imaging. The distance-dependent fluorescence quenching behavior of AuNPs enables distinct biomolecular interactions in close proximity, which is limited to cellular assays, due to the difficulty in target-labeling and the large amount of noise caused by interferants. In contrast, AuNP-based plasmonic assays provide a simpler method to amplify the signal; therefore, plasmon assays prevail in cellular analysis and are utilized depending on the change in localized surface plasmon resonance (LSPR) or enhancement caused by AuNP-triggered self-assembly or AuNP-conjugated probes, respectively [68–72].
Taking advantage of these optical properties, Medley et al. developed a simple colorimetric assay for cancer cells using aptamer-conjugated-AuNPs (Apt-AuNP) [73]. A thiol-modified aptamer which was specifically obtained using cell-SELEX for CCRF-CEM acute leukemia cells [15], was conjugated with AuNPs and targeted to assemble on the surfaces of cells through the target recognition ability of the aptamer. The binding and assembly of Apt-AuNPs on the cell surfaces bring AuNPs into close proximity with one another, causing a shift in the extinction spectra, which can then be used to quantify the number of cells [73]. In a similar study, Liu et al. detected Ramos cells using AuNPs and a pair of previously identified aptamers from cell-SELEX [29,74]. Briefly, a sample containing Ramos cells and Apt-AuNPs was applied to a secondary aptamer immobilized on a strip surface. Ramos cells interacted with Apt-AuNPs and migrated over the strip to the region containing the surface-immobilized secondary aptamers. The accumulation of Apt-AuNPs was then visualized as a red color and quantified. With this technique, as few as 4000 Ramos cells could be detected with the naked eye; this sensitivity increased to 800 cells when using a portable strip reader [74]. Wu et al. also demonstrated the salt-induced color change of AuNPs for the detection of E. coli and Salmonella typhimurium [75]. They incubated Apt-AuNPs with the target bacterial cells for 10 min. These were then aggregated upon the addition of NaCl. The subsequent color change from red to purple was simply detected either visually or with UV-vis spectroscopy [76]. This colorimetric method was able to detect 105 CFU·mL−1 of E. coli or S. typhimurium [75] without requiring any expensive instrumentation or labeling process.
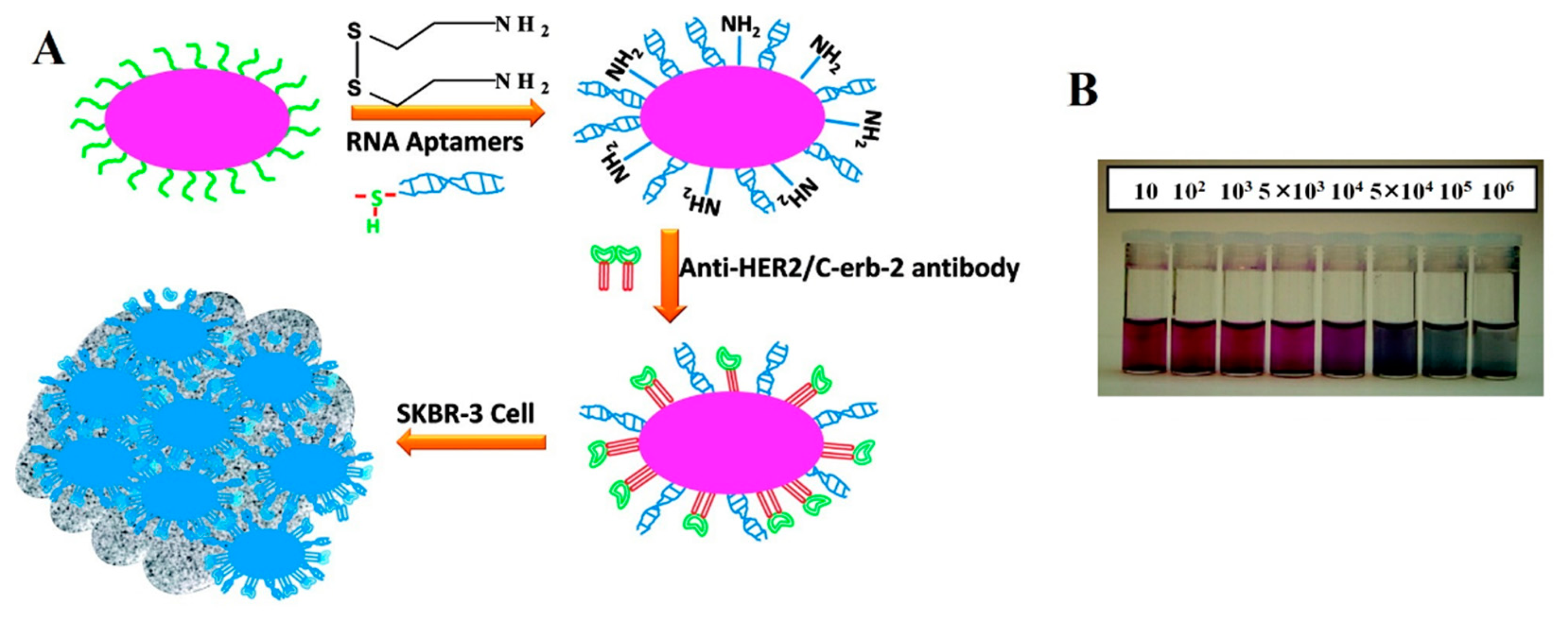
Recently, Lu et al. reported oval-shaped AuNPs conjugated with an aptamer identified through cell-SELEX [30] and a monoclonal anti-HER2/c-erb-2 antibody for the sensitive and selective detection of breast cancer SK-BR-3 cells, respectively [77]. As shown in Figure 2, a colorimetric change was observed from pink to bluish, as AuNPs accumulated on the surfaces of SK-BR-3 cells. The LOD of the colorimetric assay was 104 cells·mL−1. The sensitivity was further improved by employing a two-photon scattering technique (TPS), which was powerful enough to detect the small changes in the sizes of NPs, leading to a LOD improved by as much as two orders of magnitude (100 cells·mL−1) compared to the simple colorimetric assay [77]. In comparison, the TPS intensity was two-fold higher when the cells were incubated with the AuNPs conjugated with both aptamer and antibody compared to the AuNPs conjugated with either aptamer or antibody. The conjugation of anti-HER2/c-erb-2 antibody together with the aptamer clearly detected the various breast cancer cells, which depended on the overexpression levels of HER2 [77]. In another approach, Chang et al. measured the resonance light scattering of Apt-AuNPs for the detection Staphylococcus aureus cells [44]. They employed ssDNA-modified AuNPs, where the ssDNA acted as an adaptor sequence for aptamer hybridization. The Apt-AuNPs were then bound with S. aureus cells and subjected to resonance light-scattering analysis using a laser light source, objective lens, photodiode and digital voltmeter after being eluted with NaOH. The result showed that the sensitive resonance light-scattering analysis was able to detect as few as 312 cells [44].
Improved detection sensitivity at lower target concentrations was accomplished using a signal amplification method in which silver ions are reduced on the AuNP surface by a reductant (e.g., hydrazine). By employing two different target ligands consisting of the antibody and the aptamer, HER2-overexpressing SK-BR-3 breast cancer cells were initially captured by monoclonal anti-HER2 antibodies immobilized onto a nanocomposite, comprised of self-assembled AuNPs [78]. The bound target cells were further conjugated with hydrazine-AuNP-aptamer (Hyd-AuNP-Apt) which was also specific to HER2. The signal transduction was performed via silver nitrate addition, which was selectively reduced to silver metal by hydrazine and specifically deposited onto the Hyd-AuNP-Apt. Therefore, the deposited silver metal was analyzed using square wave stripping voltammetry to determine the amount of HER2-overexpressing cells. Under optimized conditions, the LOD of SK-BR-3 breast cancer cells is 26 cells·mL−1 [78].
A similar strategy with a few modifications, has also been reported to detect S. typhimurium [79]. A biotinylated aptamer immobilized on an avidin-coated microplate was used to capture the target bacteria, the bound bacteria were further detected by secondary Apt-AuNPs, which was followed by the addition of a silver enhancer solution. The combination of Apt-AuNPs and the silver staining method had a detection limit as low as 7 CFU·mL−1 [79].
Yi et al. developed a different electrochemical method for detecting Ramos cancer cells using Apt-AuNPs and enzyme-triggered silver enhancement [80]. They immobilized a thiol-modified aptamer onto the Au electrode to capture the Ramos cells. As a detection probe, a biotinylated secondary aptamer was subsequently employed to amplify target cells with the addition of streptavidin-alkaline phosphatase (ALP); the presence of ALP on the surface promoted enzymatic silver ion reduction and deposition onto the Au-electrode, allowing for simple electrochemical detection. Through two aptamer-based sandwich assays and enzymatic reaction, the LOD of Ramos cells was determined to be as low as 10 cells [80].
3.2. Aptamer-Conjugated Magnetic Nanoparticles
Magnetic nanoparticles (MNPs) are composed of inorganic nanocrystals with metals, metal alloys, and metal oxides as their magnetic cores [81]. Among MNPs, superparamagnetic iron oxide NPs (SPIONs), including Fe3O4 and g-Fe2O3 have been a major research focus [82] due to their reversibly switched response to an external magnetic force. Importantly, the surfaces of MNPs can be modified with other metal atomic layers, such as Au, Ag, and Al2O3, which serve as effective conjugating sites for various ligands and labeling groups (e.g., aptamers, proteins and fluorescent dyes) [83,84]. When combined with cell-specific aptamers, multifunctional MNPs have additional advantages in terms of their ability to detect living cells compared to other nanoparticles conjugated with aptamers. Magnetic control without centrifugation enables aptamer-conjugated-MNPs (Apt-MNP) to be extensively applied to the non-invasive separation, extraction, and enrichment of target cells. This is also relevant to increasing the detection sensitivity by enriching low-abundant targets from complicated samples. In addition, MNPs can act as both a nanostructured active site with large surface area and as an additional signal generator when they are combined with other detecting molecules, such as fluorophores, enzymes, or other metals.
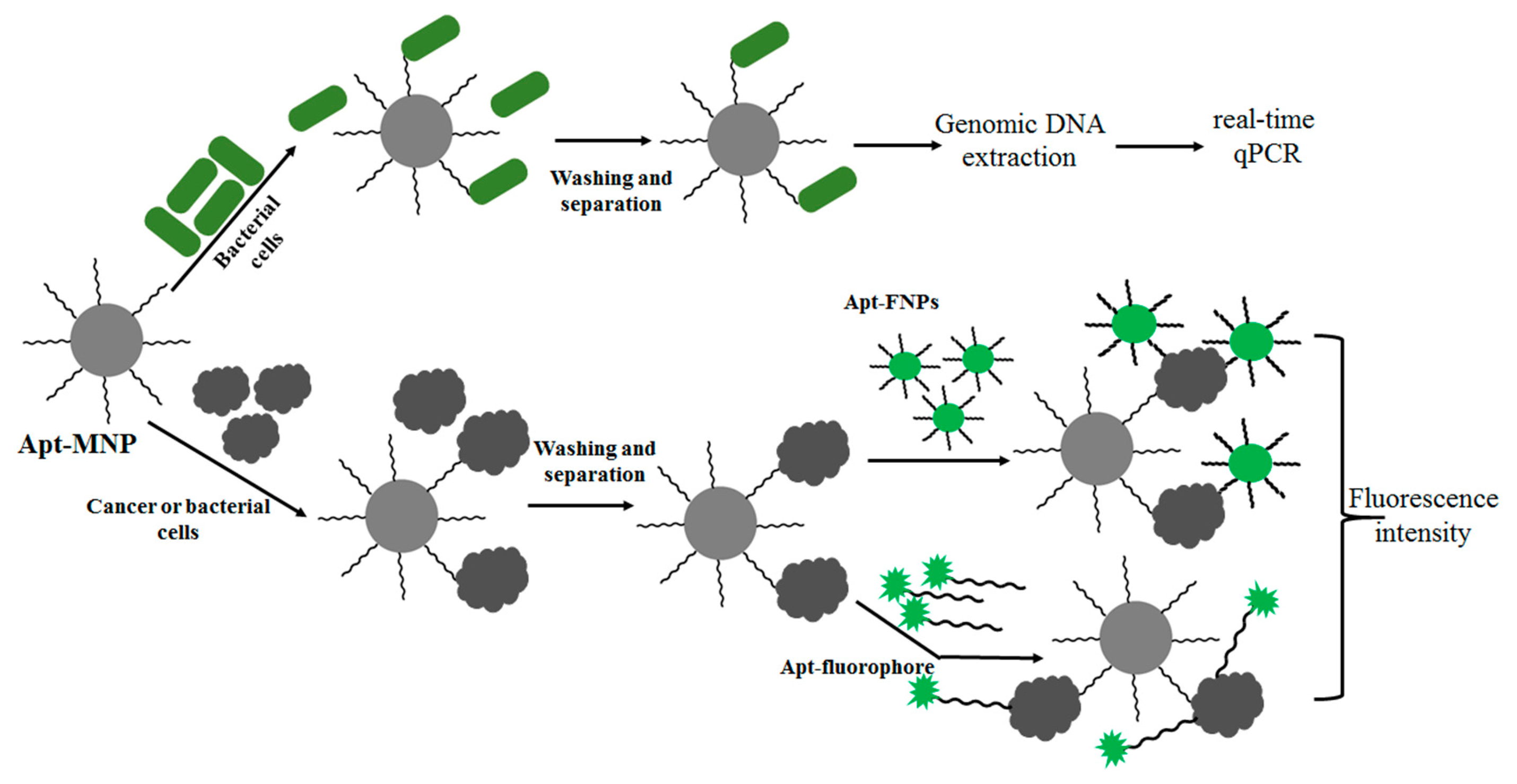
A recent report demonstrated this principle: Apt-MNPs were exploited to rapidly and easily capture and concentrate bacterial cells (Figure 3). The captured bacterial cells were identified with high sensitivity using subsequent detection methods, including fluorophore-conjugated aptamers [46] and real-time qPCR [43,85]. As a result, this strategy was shown to be useful for detecting various bacteria such as S. typhimurium, Listeria and E. coli [43,46,85]. In order to obtain a rapid and efficient method for targeting cancer cells, Tan and colleagues used a similar method for the detection of CCRF-CEM acute leukemia cells using modified Apt-MNPs (silica-coated iron oxide MNPs modified with avidin) and Apt-FNPs (Rubpy-doped silica NPs) [86]. They also demonstrated that it was possible to perform multiple cell type extraction from a complex mixture using Apt-MNPs and Apt-FNPs [87]. As a proof of concept, three different Apt-MNPs and three different dye doped Apt-FNPs (Cy5, Rubpy and TMR), specific to three different cells (Ramos, Toledo and CEM cells), were used for separation and detection, respectively.
The conjugated aptamers were individually obtained through whole cell-SELX and are known for their specific binding towards target cell [15,29]. The cells bound to the respective Apt-MNPs were validated by flow cytometer analysis with the dye-doped Apt-FNPs after magnetic separation. Surprisingly, the combination of two different Apt-NPs resulted in an LOD with very low detection sensitivity (250 cells). Another experiment by the same group showed that among different MNP sizes, 60-nm particles resulted in the greatest measurable intensity with an LOD of 152 cells. Interestingly, the detection sensitivity against the same target cells was further improved by conjugating the MNPs with multiple aptamers, such as two-, three- and four-Apt-MNPs, leading to an LOD as low as 45, 97 and 6.6 cells, respectively [88]. Tan's group also utilized the multiple aptamer-MNPs to detect CCRF-CEM cells by measuring the change in spin-spin relaxation time (ΔT2) [89]. This assay method was very effective and could detect as few as 10 cells. They suggested that this method would be suitable for differentiating various cell types through arrayed-type pattern recognition based on the specific signature between the target cells and the Apt-MNPs.
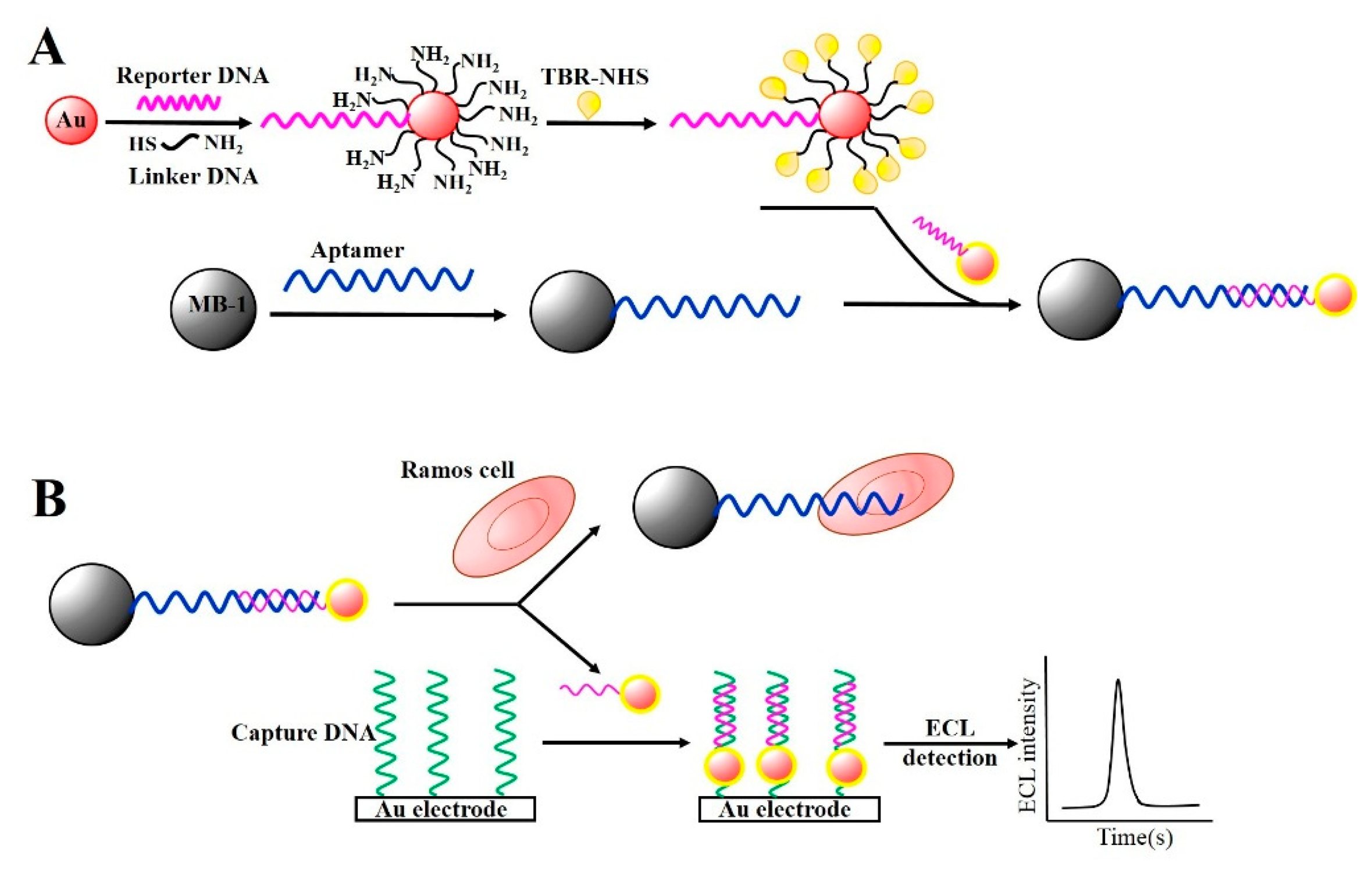
Other attempts have been made utilizing aptamer-conjugated magnetic beads (Apt-MBs) in combination with AuNPs for signal amplification because AuNPs are responsible for generating electrochemical or electrochemiluminescence (ECL) signals. Ding et al. reported the construction of a magnetic biocomplex consisting of Apt-MBs and reporter DNA-AuNPs, where the AuNPs were modified with a signal DNA labeled with tris(2,2′-bipyridyl)ruthenium(II) (TBR) and a linker DNA that can partially hybridize with the aptamers. This hybrid complex was referred to as AuNP-Apt-MB (Figure 4A) [90].
When the AuNP-Apt-MB was applied to detect Ramos cells, the DNA-linked AuNP was used as a single reporter. In the presence of target cells in the biocomplex, the complexes of AuNP-Apt-MBs were separated because the target cells were bound to the aptamer on the MBs. This released the DNA-AuNPs due to the loss of binding sites on the MBs. Consequently, the reporter DNA in the Apt-AuNPs was hybridized with the capture DNA on the Au-electrode, contributing to the ECL generation via the TBR loaded on the AuNPs (Figure 4B). They showed that the ECL intensity of the TBR was directly proportional to the amount of Ramos cells in the biocomplex, and the LOD was determined to be 50 cells·mL−1 under optimal conditions. They also examined whether magnetic nanocomposites, as opposed to micro-sized magnetic beads, showed better efficiency for capturing target cells. As a result, magnetic nanocomposites provided easier separation and increased ECL signals [91], lowering the LOD to five cells·mL−1. Zang et al. demonstrated a similar strategy to extract HL-60 cancer cells using Apt-MBs and AuNP signal amplification [92]. However, instead of TBR, they used a CdS nanocluster film to provide the ECL signal. This strategy was well-suited for the detection of different cancer cells at concentrations as low as 20 cells·mL−1.
Zhang et al. demonstrated the electrochemical detection of CCRF-CEM acute leukemia cells using the Fe3O4 MNPs and AuNP-catalyzed silver deposition enhancement [93]. Due to their large surface-to-volume ratio, the Apt-MNPs were used as carriers and reservoirs for loading a large amount of AuNPs. A competitive binding assay in the presence of target cells enabled the Apt-MNP-AuNP complexes to be separated by the magnetic support. The released AuNPs were subsequently treated via silver deposition. The AuNP-catalyzed silver deposition enhancement showed high sensitivity with an LOD of 10 cells·mL−1 [93].
3.3. Aptamer-Conjugated Silica Nanoparticles
Silica nanoparticles (SiNPs) have emerged as promising candidates for characterizing cells due to their excellent biocompatibility, easy separation, broad size range (5−1000 nm) with large surface area, and superb carrier ability with versatile labeling techniques [94]. When SiNPs were conjugated with aptamers (Apt-SiNPs), target analytes were easily separated from the biocomplex through simple centrifugation [95]. The large surface area of SiNPs also allowed for the encapsulation of a large number of fluorophores in order to generate intense fluorescence [96], enabling them to function as a signal enhancer for the detection of cancer cells. For example, Wang et al. developed fluorescence resonance energy transfer (FRET)-based SiNPs doped with three different dyes that exhibit multiple colors at one wavelength [97]. These SiNPs exhibited excellent fluorescence intensity with weak photobleaching properties when targeting CEM cancer cells after being conjugated with aptamers [97].
Tan and his colleagues demonstrated FRET-based SiNPs labeled with several fluorophores and aptamers, and exhibited the feasibility of multiplexed detection of cancer cells [98,99]. Three different SiNPs contained a single dye (FAM), two dyes (FAM and R6G), and three dyes (FAM, R6G and ROX) that were conjugated with aptamers (FAM-T1-SiNP, FAM-R6G-sgc8-SiNP, FAM-R6G-ROX-TD05-SiNP for Toledo, CEM and Ramos cells, respectively) via neutravidin-biotin interaction [98]. These SiNPs improved the specificity of the as-selected aptamers against the corresponding cell types with a nanomolar range in the binding affinity [99]. As shown in Figure 5, three different fluorophore/aptamer-conjugated SiNPs specifically bound to their corresponding target cells in a three-cell mixture, as evidenced by the distinct colors of the attached SiNPs. Recently, Cai et al. synthesized Rubpy-doped SiNPs for MCF-7 cell imaging using a MUC-1 specific aptamer, showing that these Apt-SiNPs have better photostability and selectivity than the dye-labeled MUC-1 aptamer [100]. Although such SiNPs with aptamers have excellent photostability and possess a great potential for separating and imaging both individual and multiple mixtures of cancer cells, they mostly rely on fluorescence-based methods that may suffer from inevitable autofluorescence in a real mixture or in vivo. To attain their full potential, further studies are necessary. Near-infrared dye or bioluminescence-based approaches are needed to expand the utility of SiNPs. In addition, the binding affinity of aptamer-conjugated SiNPs against living cells should be compared with that of other aptamer-NPs or antibody-conjugated SiNPs.
3.4. Aptamer-Conjugated Carbon Nanomaterials
Carbon nanomaterials, including graphenes and single-wall carbon nanotubes (SWCNTs), have attracted a lot of interest in the detection of cancer cells and bacteria [101–103]. One of the interesting properties of nanomaterials is their ability to act as a transducer or quencher with a tunable band gap and high elasticity [104–106]. Additionally, due to their hydrophobic surfaces, ssDNA molecules preferentially adsorb onto graphenes or SWCNTs by means of π-stacking interactions between the nucleotide bases and the sidewalls of the carbon nanomaterials [107].
In terms of the energy-absorbing ability, Wei et al. reported that graphene oxide (GO) could serve as an acceptor in the electrochemiluminescence (ECL) resonance energy transfer dubbed ERET and was designated to be a target recognition probe in combination with mucin 1 protein (MUC1) aptamer labeled with bis(2,2′-bipyridine)-(5-aminophenanthroline)ruthenium (II) (Ru1) (Apt-Ru1) [108]. Since Ru1 is a Ru(bpy)32+ derivative, it possesses ECL behavior; efficient quenching of ECL occurred when the Apt-Ru1 bound with GO. However, in the presence of either MUC1 or MCF-7 cells, the ECL increased significantly due to the folded formation of the target-bound aptamer, which led to the release of the Ru1-aptamer from the surface of GO. Based on this strategy, they achieved a detection limit of 40 nM for purified MUC1 and 30 cells·mL−1 for MCF-7. Likewise, bacterial cells were measurable on the GO surface using a FAM (carboxyfluorescein)-aptamer which was specific for S. typhimurium. The increased fluorescence in the presence of the target bacteria was quantified as a function of bacterial cells, and the aptamer-based sensor reached an LOD as low as 100 CFU·mL−1 [101]. Cao et al. incorporated this FAM aptamer-immobilized GO (FAM-Apt-GO) into a 33-channel microfluidic chip for the sensitive detection of CCRF-CEM cancer cells [102]. The FRET between FAM-Apt and GO exhibited quenched fluorescence, whereas increased fluorescence intensity was observed when the target cells were present. The multichannel microfluidic chip based system was able to detect as few as 25 cells·mL−1 in a simultaneous and multiplexing manner.
Feng et al. reported a reusable graphene sensor functionalized with aptamer using electrochemical detection [109]. Briefly, the NH2-modified aptamer, specific to nucleolin of tumor cell, was conjugated with tetracarboxylic acid-functionalized graphene via a carbodiimide-mediated chemistry. The resulting graphene surface was used as a nanoscale anchorage substrate to effectively capture HeLa cells on the electrode. Based on the results of electrochemical impedance spectroscopy (EIS), which targeted the cells by monitoring the change in electron-transfer resistance (Ret) on the electrode, this method was able to detect low concentrations of HeLa cells with an LOD of 794 cells·mL−1. Furthermore, the strong association between the aptamer and the target cell was disrupted by the hybridization of the aptamer with its complementary DNA, which made this biosensor reusable after mild washing.
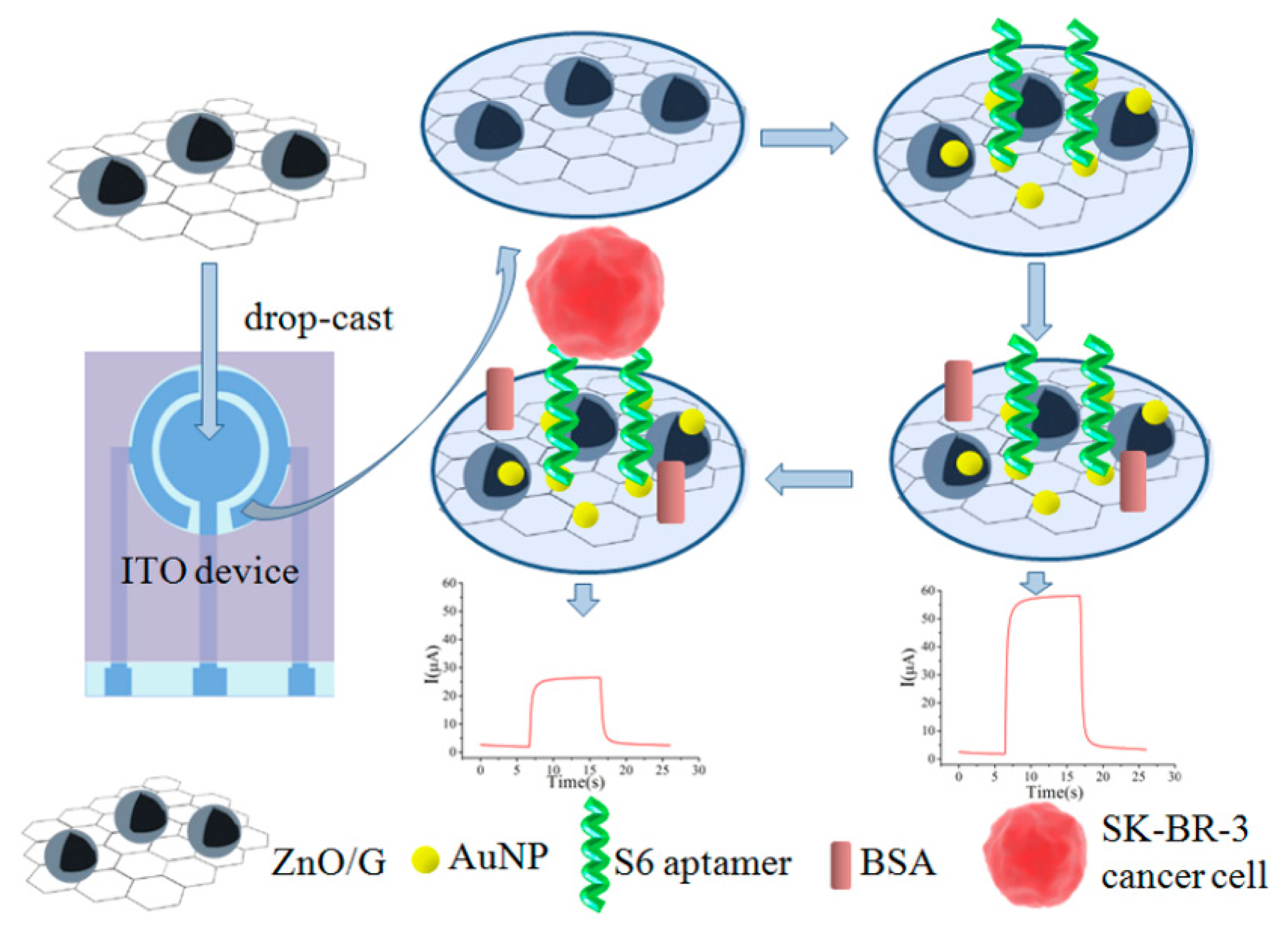
Recently, Liu et al. demonstrated a ZnO/graphene (ZnO/G)-based Apt-AuNP composite for targeting SK-BR-3 cells on a portable indium tin oxide micro-device using photoelectrochemical detection (PEC) (Figure 6) [110]. In this assay, the AuNPs were electrodeposited onto the ZnO/G composite, followed by immobilization of the aptamer.
The PEC measurement of SK-BR-3 cancer cells captured by S6 aptamers was performed by the addition of ascorbic acid. As a result, the LOD was found to be 58 cells·mL−1. Wang and colleagues applied a similar approach to detect Salmonella. In this scenario, the GO and AuNPs were coated onto a glassy carbon electrode for EIS analysis [111]. In the presence of Salmonella, the aptamer immobilized on the AuNPs specifically captured the target, leading to retardation of electron transfer between the electrode and the electrolyte, producing a higher resistance. Using EIS, the detection sensitivity reached 3 CFU·mL−1.
For the synthesis of improved nano-composites, Yan et al. combined a porous GO/Au/aptamer composite with a thionine-functionalized porous PtFe alloy to target MCF-7 breast cancer cells [112], because the porous GO composites have several advantages compared to the conventional graphene, such as a large surface area, fast electron transportation, and good biocompatibility [113,114]. For the typical sandwich type assay, the GO/Au composite was deposited on a glassy carbon electrode, followed by conjugating the aptamer to recognize MUC1. Upon adding the nanoporous PtFe alloy conjugated with multiple anti-MUC1 aptamers, the final composite allowed for the sensitive detection of target MCF-7 cells with MUC1 overexpression. Using this amplification strategy, the detection limit was as low as 38 cells·mL−1.
Potentiometric analysis using SWCNTs as efficient ion-to-electron transduces was demonstrated for the ultra-sensitive detection [115]. Zelada-Guillen et al. reported an aptamer-conjugated SWCNT (Apt-SWCNT) for the potentiometric detection of pathogenic bacteria, where the Apt-SWCNT played dual roles in sensing and transducing (Figure 7) [116].
For the preparation of this device, a layer of SWCNTs was sprayed onto a glassy carbon rod that was electrically connected to a potentiometer and the NH2-modified aptamer against S. typhimurium was covalently conjugated with the SWCNTs to capture the target bacteria. While the aptamers were being self-assembled through π-π stacking interactions between the bases and the SWCNT walls, the binding of the target bacteria to the aptamers triggered conformational and charge changes in the aptamer, leading to a subsequent change in the potential. Such Apt-SWCNT-based methods are rapid and sensitive enough to detect 1 CFU·mL−1 of S. typhimurium. Additionally, the aptamer can be easily regenerated and reused after dissociating and reconditioning steps. Similar potentiometric methods based on Apt-SWCNT have been reported for the sensitive detection of various pathogenic bacteria including, E. coli, S. aureus, and Salmonella paratyphi A [117,118].
3.5. Aptamer-Conjugated Quantum Dots
Quantum dots (QDs) are semiconducting nanocrystals that exhibit exceptional optical and electrical behavior in cellular imaging and analysis [119–121]. Compared to conventional fluorophores, QDs have narrow and size-tunable emission spectra, high quantum yield, and great photostability [122,123]. Most importantly, since QDs exhibit multiplex emissions when excited with a single wavelength, QDs conjugated with aptamers (Apt-QDs) have been broadly used for the detection of various cancer cells and bacteria with high specificity, as well as being implemented for signal enhancement with high assay stability.
It was reported by Lian et al. that poly(ethylene glycol)-phospholipid micelles in conjunction with CdSe/ZnS QDs modified with a thiol-ended aptamer, which specifically binds with nucleolin in cancer cell surface [124], were useful to specifically recognize breast cancer cells [125]. Lian et al. showed that the Apt-QDs successfully targeted MCF-7 and MDA-MB-231 cells with a strong red fluorescence compared to normal cells (NIH-3T3). Furthermore, the Apt-QDs exhibited no apparent cytotoxicity at the tested concentrations, indicating good biocompatibility and suitability for targeting cells. Similarly, Duan et al. used two QDs with different colors (green and orange) at a single excitation for the simultaneous detection of Vibrio parahaemolyticus and S. typhimurium [126]. After the QDs were conjugated with an aptamer specific for either V. parahaemolyticus or S. typhimurium, the Apt-QDs allowed for the individual or collective detection of two bacterial cells with a flow cytometer (Figure 8A–C). It was also reported that the cocktails of Apt-QDs, which contained three different types of aptamer-conjugated QDs specific to E. coli cells, were used to enhance the binding and detection sensitivity [127]. Compared to any individual aptamer or other combinations of aptamers, the cocktail mixture in the Apt-QDs exhibited an 18-fold higher LOD (371 CFU·mL−1). It should be noteworthy that aptamers evolved from cell-SELEX promote the binding affinity to target cells by recognizing multiple sites on the cell surface.
To further increase the number of QDs on the target, Hua et al. fabricated QD-decorated SiNPs (SiNP-QD) [128]. Along with two different aptamers targeting MUC1 or nucleolin in the tumor cell surface, they prepared two different sensing probes consisting of MUC1 aptamer-conjugated MBs and nucleolin aptamer-conjugated SiNP-QDs. Using two probes, the MCF-7 breast cancer cells were sensitively detected at a concentration as low as 85 cells·mL−1.
In addition to fluorescent detection based on QDs, Li et al. applied Apt-QDs to electrochemical stripping voltammetry for the detection of MCF-7 breast cancer cells [129]. After anti-MUC1 aptamers were hybridized with a complementary DNA anchored on the Au-electrode surface, the carboxyl QDs were conjugated with NH2-modified aptamers via EDC/NHS reaction, which was followed by the addition of MCF-7 breast cancer cells. Since the MUC1 on the MCF-7 cell surface could compete with cDNA, the Apt-QDs were released from the Au-electrode surface in the presence of the target cells. The number of QDs on the electrode was inversely proportional to the concentration of target cells, which was determined by the electrochemical stripping method. The competitive electrochemical cytosensor was able to detect concentrations as low as 100 MCF-7 cells·mL−1.
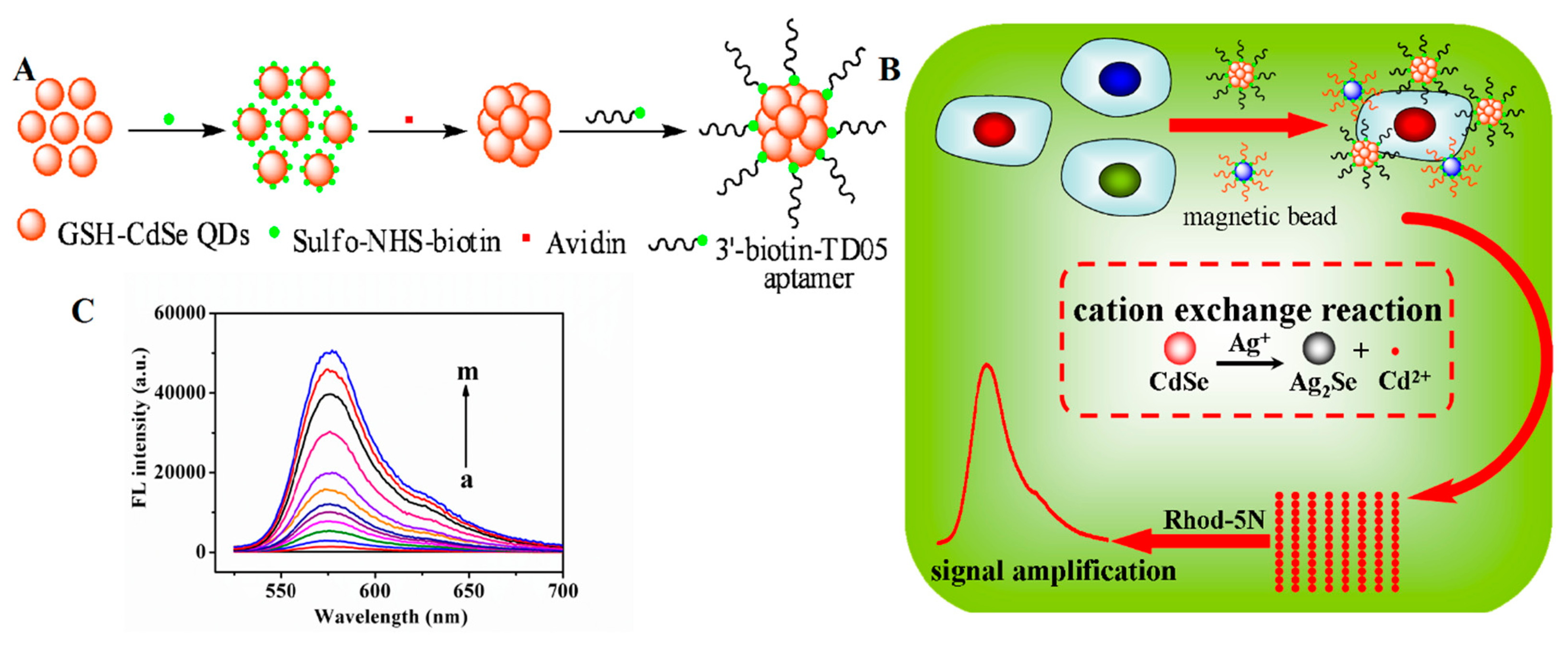
A similar study was described by Sheng et al., who used QD nanoclusters and MBs to develop a cation exchange reaction-based fluorescence method for the sensitive detection of Ramos cells (Figure 9) [130]. A biotinylated aptamer was conjugated with avidin-modified CdSe QDs and used for targeting cells. At the same time, a secondary aptamer that binds with a different site on the target cells was conjugated with MBs. The target cells were captured in a sandwich by the two probes (i.e., Apt-QDs and Apt-MBs), allowing for magnetic separation. Upon the addition of Ag+ ions and the Rhod-5N dye (a nonfluorescent metal-sensitive dye), a large number of Cd2+ ions were released from the QD nanoclusters, resulting in fluorescence of the Rhod-5N dye. Under optimal conditions, the cation exchange-based method was able to detect concentrations as low as 50 cells·mL−1.
3.6. Aptamer-Incorporated DNA Nanostructures
Due to the ability of DNA to generate a myriad of folding structures, the aptamer-DNA conjugate is capable of carrying drug-like molecules to target cells by utilizing the target-specific aptamer and the drug-binding DNA structure [131,132]. It has been reported that two- and three-dimensional nanostructures based on the DNA origami provided distinct binding sites with various drug components [133,134]. The folding ability of DNA to produce many shapes is mainly relied on the Watson-Crick base-pairing between short nucleic acid sequences. Such a modified DNA nanostructure is easy controllable, biocompatible, and cell-permeable. In addition, the conjugation of aptamers to the DNA structures can improve the target specificity.
Chang et al. reported a distinct aptamer-DNA structure with icosahedral shape containing six-point-star motif, which was used as a nanocarrier with doxorubicin for cancer chemotherapy [135]. The DNA icosahedra was constructed using six individual DNA single strands, where five of the strands bended to a specific angle by hybridizing adjacent strands and the sixth strand was used for aptamer conjugation. For this, they employed a MUC1 DNA aptamer which specifically binds to MUC1 expressed in breast cancer cells (MCF-7) [136]. The doxorubicin was then intercalated into the aptamer-DNA icosahedral structure to construct Doxo-Apt-DNA-icosa. Under optimized conditions, the Doxo-Apt-DNA-icosa was not only internalized into MCF-7 cells but also showed a significantly higher cytotoxicity than free doxorubicin. This Apt-DNA-based nanostructure provides a controlled release of cancer drugs with site-specific targeting. Similarly, Zhu et al. demonstrated an aptamer-guided DNA nanotrain (Apt-NTr) for targeted transport of cancer drugs [137]. The aptamer in the Apt-NTr was screened from cell-SELELX using CEM cells [15] and was linked to two repeated hairpin DNA sequences at one end. This structure can be easily polymerized through Watson-Crick base-pairing. Furthermore, the Apt-NTr loaded with doxorubicin acts as an anticancer drug selectively to tumor CEM cells, whereas no cytotoxicity was observed for control cells [137].
Recently, Hu et al. reported an aptamer-conjugated FRET nanoflower (Apt-NF) which exhibited multi-fluorescence emissions by a single wavelength excitation [138]. Unlike the traditional Watson-Crick base-pairing, the multi-fluorescence NFs were achieved by incorporating three different dyes (FAM, Cy3 and ROX) into rolling circle replicon that can rapidly synthesize DNA. The template used for rolling circle replication consists of complementary aptamer toward CEM cells [15] and drug-loading sequence. The aptamer in NFs preserved its binding affinity and exhibited extremely bright fluorescence upon binding with CEM cells, but not with control Ramos cells. In addition to the imaging ability, the apt-NFs were also suitable for targeted drug delivery [138].
4. Conclusions
To generate high binding affinity without the knowledge of cell receptors, cell-SELEX is very useful for providing specific aptamers against various cancer cells and bacteria. This review shows that by utilizing cell-SELEX, recent advances in the development of aptamer-nano hybrid sensors have led to remarkable improvement in targeting cell. Since aptamers can be easily generated and modified with various nanomaterials, the traditional limits, related to low sensitivity, poor stability, and high cost can be overcome. Aptamer-nanomaterial hybrids show tremendous potential as robust diagnostic and therapeutic reagents for detecting and characterizing different types of cells. Given the complexity of cancer, the aptamers identified through cell-SELEX would be more advantageous than the conventional probes to simultaneously detect and differentiate normal and abnormal cells. Future efforts should focus on designing multimeric aptamers which will contribute to boosting the early detection of detrimental cells related to human diseases with high binding affinity and target specificity.
Acknowledgments
This work was supported by Mid-career Researcher Program (No. NRF-2013R1A2A2A03015161), Nano Material Technology Development Program (No. 2012M3A7B4035286), and Bio-Signal Analysis Technology Innovation Program (No. NRF-2006-2005074) through the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MSIP). This work was also supported by Basic Science Research Programs (No. 20120008222 and No. 2012R1A6A1029029) through the NRF funded by the Ministry of Education.
Author Contributions
V.G. drafted the main review article; Y.-P.K. contributed to revising the article for scientific content and supervised the figure collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Paredes-Aguilera, R.; Romero-Guzman, L.; Lopez-Santiago, N.; Burbano-Ceron, L.; Camacho-Del Monte, O.; Nieto-Martinez, S. Flow cytometric analysis of cell-surface and intracellular antigens in the diagnosis of acute leukemia. Am. J. Hematol. 2001, 68, 69–74. [Google Scholar]
- Belov, L.; de la Vega, O.; dos Remedios, C.G.; Mulligan, S.P.; Christopherson, R.I. Immunophenotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res. 2001, 61, 4483–4489. [Google Scholar]
- Eltahir, E.M.; Mallinson, D.S.; Birnie, G.D.; Hagan, C.; George, W.D.; Purushotham, A.D. Putative markers for the detection of breast carcinoma cells in blood. Br. J. Cancer. 1998, 77, 1203–1207. [Google Scholar]
- Moscovici, M.; Bhimji, A.; Kelley, S.O. Rapid and specific electrochemical detection of prostate cancer cells using an aperture sensor array. Lab Chip 2013, 13, 940–946. [Google Scholar]
- Xu, X.D.; Cheng, H.; Chen, W.H.; Cheng, S.X.; Zhuo, R.X.; Zhang, X.Z. In situ recognition of cell-surface glycans and targeted imaging of cancer cells. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Bajaj, A.; Miranda, O.R.; Kim, I.B.; Phillips, R.L.; Jerry, D.J.; Bunz, U.H.F.; Rotello, V.M. Detection and differentiation of normal, cancerous, and metastatic cells using nanoparticle-polymer sensor arrays. Proc. Natl. Acad. Sci. USA 2009, 106, 10912–10916. [Google Scholar]
- Feng, L.Y.; Wu, L.; Wang, J.S.; Ren, J.S.; Miyoshi, D.; Sugimoto, N.; Qu, X.G. Detection of a Prognostic Indicator in Early-Stage Cancer Using Functionalized Graphene-Based Peptide Sensors. Adv. Mater. 2012, 24, 125–131. [Google Scholar]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar]
- Wilson, D.S.; Szostak, J.W. In vitro selection of functional nucleic acids. Ann. Rev. Biochem. 1999, 68, 611–647. [Google Scholar]
- Cerchia, L.; de Franciscis, V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010, 28, 517–525. [Google Scholar]
- Lee, S.; Kang, J.; Ren, S.; Laurell, T.; Kim, S.; Jeong, O.C. A cross-contamination-free SELEX platform for a multi-target selection strategy. Biochip. J. 2013, 7, 38–45. [Google Scholar]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar]
- Fang, X.H.; Tan, W.H. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc. Chem. Res. 2010, 43, 48–57. [Google Scholar]
- Tan, W.H.; Donovan, M.J.; Jiang, J.H. Aptamers from Cell-Based Selection for Bioanalytical Applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar]
- Shangguan, D.; Li, Y.; Tang, Z.W.; Cao, Z.H.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.Y.J.; Tan, W.H. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006, 103, 11838–11843. [Google Scholar]
- Sefah, K.; Meng, L.; Lopez-Colon, D.; Jimenez, E.; Liu, C.; Tan, W.H. DNA Aptamers as Molecular Probes for Colorectal Cancer Study. PLoS One. 2010. [Google Scholar] [CrossRef]
- Sefah, K.; Shangguan, D.; Xiong, X.L.; O'Donoghue, M.B.; Tan, W.H. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar]
- Chen, F.; Zhou, J.; Luo, F.L.; Mohammed, A.B.; Zhang, X.L. Biochem. Biophys. Res. Commun. 2007, 357, 743–748.
- Lee, J.H. Conjugation approaches for construction of aptamer-modified nanoparticles for application in imaging. Curr. Top. Med. Chem. 2013, 13, 504–512. [Google Scholar]
- Yu, M.K.; Park, J.; Jon, S. Targeting strategies for multifunctional nanoparticles in cancer imaging and therapy. Theranostics 2012, 2, 3–44. [Google Scholar]
- Meyer, C.; Hahn, U.; Rentmeister, A. Cell-specific aptamers as emerging therapeutics. J. Nucl. Acids 2011, 2011, 904750. [Google Scholar]
- Yang, L.; Zhang, X.B.; Ye, M.; Jiang, J.H.; Yang, R.H.; Fu, T.; Chen, Y.; Wang, K.M.; Liu, C.; Tan, W.H. Aptamer-conjugated nanomaterials and their applications. Adv. Drug Deliv. Rev. 2011, 63, 1361–1370. [Google Scholar]
- Xiao, Z.Y.; Farokhzad, O.C. Aptamer-Functionalized Nanoparticles for Medical Applications: Challenges and Opportunities. ACS Nano 2012, 6, 3670–3676. [Google Scholar]
- Zhang, H.; Stockley, P.G.; Zhou, D. Development of smart nanoparticle-aptamer sensing technology. Faraday Discuss. 2011, 149, 319–332. [Google Scholar]
- Liu, Q.L.; Jin, C.; Wang, Y.Y.; Fang, X.H.; Zhang, X.B.; Chen, Z.; Tan, W.H. Aptamer-conjugated nanomaterials for specific cancer cell recognition and targeted cancer therapy. NPG Asia Mater. 2014, 6. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Ma, Y.; Pei, X.; Liu, Q.; Lu, B.; Jin, L.; Wang, J.; Liu, J. A cell-based single-stranded DNA aptamer specifically targetsgastric cancer. Int. J. Biochem. Cell Biol. 2014, 46, 1–8. [Google Scholar]
- Ray, P.; Rialon-Guevara, K.L.; Veras, E.; Sullenger, B.A.; White, R.R. Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker. J. Clin. Invest. 2012, 122, 1734–1741. [Google Scholar]
- Jimenez, E.; Sefah, K.; Lopez-Colon, D.; Van Simaeys, D.; Chen, H.W.; Tockman, M.S.; Tan, W.H. Generation of Lung Adenocarcinoma DNA Aptamers for Cancer Studies. PLoS One 2012, 7, e4622. [Google Scholar]
- Tang, Z.W.; Shangguan, D.; Wang, K.M.; Shi, H.; Sefah, K.; Mallikratchy, P.; Chen, H.W.; Li, Y.; Tan, W.H. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907. [Google Scholar]
- Kang, H.S.; Huh, Y.M.; Kim, S.; Lee, D. Isolation of RNA Aptamers Targeting HER-2-Overexpressing Breast Cancer Cells Using Cell-SELEX. Bull. Korean Chem. Soc. 2009, 30, 1827–1831. [Google Scholar]
- Sefah, K.; Tang, Z.W.; Shangguan, D.H.; Chen, H.; Lopez-Colon, D.; Li, Y.; Parekh, P.; Martin, J.; Meng, L.; Phillips, J.A.; et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia 2009, 23, 235–244. [Google Scholar]
- Chen, H.W.; Medley, C.D.; Sefah, K.; Shangguan, D.; Tang, Z.W.; Meng, L.; Smith, J.E.; Tan, W.H. Molecular recognition of small-cell lung cancer cells using aptamers. Chemmedchem 2008, 3, 991–1001. [Google Scholar]
- Kang, D.Z.; Wang, J.J.; Zhang, W.Y.; Song, Y.L.; Li, X.L.; Zou, Y.; Zhu, M.T.; Zhu, Z.; Chen, F.Y.; Yang, C.J. Selection of DNA Aptamers against Glioblastoma Cells with High Affinity and Specificity. PLoS One 2012, 7, e42731. [Google Scholar]
- Zhao, Z.L.; Xu, L.; Shi, X.L.; Tan, W.H.; Fang, X.H.; Shangguan, D.H. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst 2009, 134, 1808–1814. [Google Scholar]
- Wang, Y.; Luo, Y.; Bing, T.; Chen, Z.; Lu, M.; Zhang, N.; Shangguan, D.; Gao, X. DNA Aptamer Evolved by Cell-SELEX for Recognition of Prostate Cancer. PLoS One 2014, 9, e100243. [Google Scholar]
- Van Simaeys, D.; Lopez-Colon, D.; Sefah, K.; Sutphen, R.; Jimenez, E.; Tan, W.H. Study of the Molecular Recognition of Aptamers Selected through Ovarian Cancer Cell-SELEX. PLoS One 2010, 5, e13770. [Google Scholar]
- Cao, X.X.; Li, S.H.; Chen, L.C.; Ding, H.M.; Xu, H.; Huang, Y.P.; Li, J.; Liu, N.L.; Cao, W.H.; Zhu, Y.J.; et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucl. Acids Res. 2009, 37, 4621–4628. [Google Scholar]
- Hyeon, J.Y.; Chon, J.W.; Choi, I.S.; Park, C.; Kim, D.E.; Seo, K.H. Development of RNA aptamers for detection of Salmonella enteritidis. J. Microbiol. Meth. 2012, 89, 79–82. [Google Scholar]
- Moon, J.; Kim, G.; Lee, S.; Park, S. Identification of Salmonella typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J. Microbiol. Meth. 2013, 95, 162–166. [Google Scholar]
- Duan, N.; Ding, X.Y.; Wu, S.J.; Xia, Y.; Ma, X.Y.; Wang, Z.P.; Chen, J. In vitro selection of a DNA aptamer targeted against Shigella dysenteriae. J. Microbiol. Meth. 2013, 94, 170–174. [Google Scholar]
- Lee, Y.J.; Han, S.R.; Maeng, J.S.; Cho, Y.J.; Lee, S.W. In vitro selection of Escherichia coli O157:H7-specific RNA aptamer. Biochem. Biophys. Res. Commun. 2012, 417, 414–420. [Google Scholar]
- Kim, Y.S.; Song, M.Y.; Jurng, J.; Kim, B.C. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell-systematic evolution of ligands by exponential enrichment approach. Anal. Biochem. 2013, 436, 22–28. [Google Scholar]
- Suh, S.H.; Jaykus, L.A. Nucleic acid aptamers for capture and detection of Listeria spp. J. Biotechnol. 2013, 167, 454–461. [Google Scholar]
- Chang, Y.C.; Yang, C.Y.; Sun, R.L.; Cheng, Y.F.; Kao, W.C.; Yang, P.C. Rapid single cell detection of Staphylococcus aureus by aptamer- conjugated gold nanoparticles. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Liu, G.Q.; Yu, X.F.; Xue, F.; Chen, W.; Ye, Y.K.; Yang, X.J.; Lian, Y.Q.; Yan, Y.; Zong, K. Screening and preliminary application of a DNA aptamer for rapid detection of Salmonella O8. Microchim. Acta 2012, 178, 237–244. [Google Scholar]
- Duan, N.; Wu, S.J.; Chen, X.J.; Huang, Y.K.; Xia, Y.; Ma, X.Y.; Wang, Z.P. Selection and Characterization of Aptamers against Salmonella typhimurium Using Whole-Bacterium Systemic Evolution of Ligands by Exponential Enrichment (SELEX). J. Agric. Food Chem. 2013, 61, 3229–3234. [Google Scholar]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 2010, 87, 2323–2334. [Google Scholar]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.A. Selection of DNA aptamers for capture and detection of Salmonella typhimurium using a whole-cell SELEX approach in conjunction with cell sorting. Appl. Microbiol. Biotechnol. 2013, 97, 3677–3686. [Google Scholar]
- Duan, N.; Ding, X.Y.; He, L.X.; Wu, S.J.; Wei, Y.X.; Wang, Z.P. Selection, identification and application of a DNA aptamer against Listeria monocytogenes. Food Control. 2013, 33, 239–243. [Google Scholar]
- Wang, K.Y.; Zeng, Y.L.; Yang, X.Y.; Li, W.B.; Lan, X.P. Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. 2011, 30, 273–278. [Google Scholar]
- Pu, Y.; Zhu, Z.; Liu, H.X.; Zhang, J.N.; Liu, J.; Tan, W.H. Using aptamers to visualize and capture cancer cells. Anal. Bioanal. Chem. 2010, 397, 3225–3233. [Google Scholar]
- Shangguan, D.; Cao, Z.H.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W.H. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008, 7, 2133–2139. [Google Scholar]
- Van Simaeys, D.; Turek, D.; Champanhac, C.; Vaizer, J.; Sefah, K.; Zhen, J.; Sutphen, R.; Tan, W.H. Identification of Cell Membrane Protein Stress-Induced Phosphoprotein 1 as a Potential Ovarian Cancer Biomarker Using Aptamers Selected by Cell Systematic Evolution of Ligands by Exponential Enrichment. Anal. Chem. 2014, 86, 4521–4527. [Google Scholar]
- Shi, H.; Tang, Z.W.; Kim, Y.; Nie, H.L.; Huang, Y.F.; He, X.X.; Deng, K.; Wang, K.M.; Tan, W.H. In vivo Fluorescence Imaging of Tumors using Molecular Aptamers Generated by Cell-SELEX. Chem.-Asian J. 2010, 5, 2209–2213. [Google Scholar]
- Shi, H.; Cui, W.S.; He, X.X.; Guo, Q.P.; Wang, K.M.; Ye, X.S.; Tang, J.L. Whole Cell-SELEX Aptamers for Highly Specific Fluorescence Molecular Imaging of Carcinomas in Vivo. PLoS One 2013, 8, e70476. [Google Scholar]
- Cerchia, L.; Duconge, F.; Pestourie, C.; Boulay, J.; Aissouni, Y.; Gombert, K.; Tavitian, B.; de Franciscis, V.; Libri, D. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005, 3, e123. [Google Scholar]
- Esposito, C.L.; Passaro, D.; Longobardo, I.; Condorelli, G.; Marotta, P.; Affuso, A.; de Franciscis, V.; Cerchia, L. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One 2011, 6, e24071. [Google Scholar]
- Camorani, S.; Esposito, C.L.; Rienzo, A.; Catuogno, S.; Iaboni, M.; Condorelli, G.; de Franciscis, V.; Cerchia, L. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRbeta aptamer. Mol. Ther. 2014, 22, 828–841. [Google Scholar]
- Wu, Y.R.; Sefah, K.; Liu, H.P.; Wang, R.W.; Tan, W.H. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 5–10. [Google Scholar]
- Gao, H.L.; Qian, J.; Yang, Z.; Pang, Z.Q.; Xi, Z.J.; Cao, S.J.; Wang, Y.C.; Pan, S.Q.; Zhang, S.; Wang, W.; et al. Whole-cell SELEX aptamer-functionalised poly(ethyleneglycol)-poly(epsilon-caprolactone) nanoparticles for enhanced targeted glioblastoma therapy. Biomaterials 2012, 33, 6264–6272. [Google Scholar]
- Kong, R.M.; Zhang, X.B.; Chen, Z.; Tan, W.H. Aptamer-Assembled Nanomaterials for Biosensing and Biomedical Applications. Small 2011, 7, 2428–2436. [Google Scholar]
- Song, S.P.; Wang, L.H.; Li, J.; Zhao, J.L.; Fan, C.H. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar]
- Han, K.; Liang, Z.Q.; Zhou, N.D. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar]
- Mok, W.; Li, Y.F. Recent Progress in Nucleic Acid Aptamer-Based Biosensors and Bioassays. Sensors 2008, 8, 7050–7084. [Google Scholar]
- Daniel, M.C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar]
- Sau, T.K.; Pal, A.; Jana, N.R.; Wang, Z.L.; Pal, T. Size controlled synthesis of gold nanoparticles using photochemically prepared seed particles. J. Nanopart Res. 2001, 3, 257–261. [Google Scholar]
- Frenkel, A.I.; Nemzer, S.; Pister, I.; Soussan, L.; Harris, T.; Sun, Y.; Rafailovich, M.H. Size-controlled synthesis and characterization of thiol-stabilized gold nanoparticles. J. Chem. Phys. 2005, 123, 184701. [Google Scholar]
- Ghosh, S.K.; Pal, T. Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: From theory to applications. Chem. Rev. 2007, 107, 4797–4862. [Google Scholar]
- Yang, Y.; Hu, Y.; Du, H.; Wang, H. Intracellular gold nanoparticle aggregation and their potential applications in photodynamic therapy. Chem. Commun. 2014, 50, 7287–7290. [Google Scholar]
- Xiao, Z.; Ji, C.; Shi, J.; Pridgen, E.M.; Frieder, J.; Wu, J.; Farokhzad, O.C. DNA self-assembly of targeted near-infrared-responsive gold nanoparticles for cancer thermo-chemotherapy. Angew. Chem. 2012, 51, 11853–11857. [Google Scholar]
- Park, S.J.; Lazarides, A.A.; Mirkin, C.A.; Letsinger, R.L. Directed Assembly of Periodic Materials from Protein and Oligonucleotide-Modified Nanoparticle Building Blocks. Angew. Chem. 2001, 40, 2909–2912. [Google Scholar]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 1997, 277, 1078–1081. [Google Scholar]
- Medley, C.D.; Smith, J.E.; Tang, Z.; Wu, Y.; Bamrungsap, S.; Tan, W.H. Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal. Chem. 2008, 80, 1067–1072. [Google Scholar]
- Liu, G.D.; Mao, X.; Phillips, J.A.; Xu, H.; Tan, W.H.; Zeng, L.W. Aptamer-Nanoparticle Strip Biosensor for Sensitive Detection of Cancer Cells. Anal. Chem. 2009, 81, 10013–10018. [Google Scholar]
- Wu, W.H.; Li, M.; Wang, Y.; Ouyang, H.X.; Wang, L.; Li, C.X.; Cao, Y.C.; Meng, Q.H.; Lu, J.X. Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Res. Lett. 2012, 7, 658–664. [Google Scholar]
- Shipway, A.N.; Lahav, M.; Gabai, R.; Willner, I. Investigations into the electrostatically induced aggregation of Au nanoparticles. Langmuir 2000, 16, 8789–8795. [Google Scholar]
- Lu, W.T.; Arumugam, R.; Senapati, D.; Singh, A.K.; Arbneshi, T.; Khan, S.A.; Yu, H.T.; Ray, P.C. Multifunctional Oval-Shaped Gold-Nanoparticle-Based Selective Detection of Breast Cancer Cells Using Simple Colorimetric and Highly Sensitive Two-Photon Scattering Assay. ACS Nano 2010. [Google Scholar]
- Zhu, Y.; Chandra, P.; Shim, Y.B. Ultrasensitive and selective electrochemical diagnosis of breast cancer based on a hydrazine-Au nanoparticle-aptamer bioconjugate. Anal. Chem. 2013, 85, 1058–1064. [Google Scholar]
- Yuan, J.L.; Tao, Z.; Yu, Y.; Ma, X.Y.; Xia, Y.; Wang, L.; Wang, Z.P. A visual detection method for Salmonella typhimurium based on aptamer recognition and nanogold labeling. Food Control 2014, 37, 188–192. [Google Scholar]
- Yi, Z.; Li, X.Y.; Gao, Q.; Tang, L.J.; Chu, X. Aptamer-aided target capturing with biocatalytic metal deposition: An electrochemical platform for sensitive detection of cancer cells. Analyst. 2013, 138, 2032–2037. [Google Scholar]
- Lu, A.H.; Salabas, E.L.; Schuth, F. Magnetic nanoparticles: Synthesis, protection, functionalization, and application. Angew. Chem. 2007, 46, 1222–1244. [Google Scholar]
- Laurent, S.; Forge, D.; Port, M.; Roch, A.; Robic, C.; Vander Elst, L.; Muller, R.N. Magnetic iron oxide nanoparticles: Synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications. Chem. Rev. 2008, 108, 2064–2110. [Google Scholar]
- Berry, C.C.; Curtis, A.S.G. Functionalisation of magnetic nanoparticles for applications in biomedicine. J. Phys. D Appl. Phys. 2003, 36, R198–R206. [Google Scholar]
- Berry, C.C. Progress in functionalization of magnetic nanoparticles for applications in biomedicine. J. Phys. D Appl. Phys. 2009, 42, 224003. [Google Scholar]
- Ozalp, V.C.; Bayramoglu, G.; Kavruk, M.; Keskin, B.B.; Oktem, H.A.; Arica, M.Y. Pathogen detection by core-shell type aptamer-magnetic preconcentration coupled to real-time PCR. Anal. Biochem. 2014, 447, 119–125. [Google Scholar]
- Herr, J.K.; Smith, J.E.; Medley, C.D.; Shangguan, D.H.; Tan, W.H. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal. Chem. 2006, 78, 2918–2924. [Google Scholar]
- Smith, J.E.; Medley, C.D.; Tang, Z.W.; Shangguan, D.; Lofton, C.; Tan, W.H. Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells. Anal. Chem. 2007, 79, 3075–3082. [Google Scholar]
- Medley, C.D.; Bamrungsap, S.; Tan, W.H.; Smith, J.E. Aptamer-Conjugated Nanoparticles for Cancer Cell Detection. Anal. Chem. 2011, 83, 727–734. [Google Scholar]
- Bamrungsap, S.; Chen, T.; Shukoor, M.I.; Chen, Z.; Sefah, K.; Chen, Y.; Tan, W. Pattern recognition of cancer cells using aptamer-conjugated magnetic nanoparticles. ACS Nano 2012, 6, 3974–3981. [Google Scholar]
- Ding, C.F.; Zheng, Q.; Wang, N.N.; Yue, Q.F. An electrochemiluminescence strategy based on aptamers and nanoparticles for the detection of cancer cells. Anal. Chim. Acta 2012, 756, 73–78. [Google Scholar]
- Ding, C.F.; Wei, S.; Liu, H.T. Electrochemiluminescent Determination of Cancer Cells Based on Aptamers, Nanoparticles, and Magnetic Beads. Chem.-Eur. J. 2012, 18, 7263–7268. [Google Scholar]
- Zhang, H.R.; Xia, X.H.; Xu, J.J.; Chen, H.Y. Sensitive cancer cell detection based on Au nanoparticles enhanced electrochemiluminescence of CdS nanocrystal film supplemented by magnetic separation. Electrochem. Commun. 2012, 25, 112–115. [Google Scholar]
- Zhang, K.; Tan, T.T.; Fu, J.J.; Zheng, T.T.; Zhu, J.J. A novel aptamer-based competition strategy for ultrasensitive electrochemical detection of leukemia cells. Analyst 2013, 138, 6323–6330. [Google Scholar]
- Bitar, A.; Ahmad, N.M.; Fessi, H.; Elaissari, A. Silica-based nanoparticles for biomedical applications. Drug Discov. Today 2012, 17, 1147–1154. [Google Scholar]
- Hilliard, L.R.; Zhao, X.J.; Tan, W.H. Immobilization of oligonucleotides onto silica nanoparticles for DNA hybridization studies. Anal. Chim. Acta 2002, 470, 51–56. [Google Scholar]
- Smith, J.E.; Wang, L.; Tan, W.T. Bioconjugated silica-coated nanoparticles for bioseparation and bioanalysis. TrAC. Trends Anal. Chem. 2006, 25, 848–855. [Google Scholar]
- Wang, L.; Tan, W.H. Multicolor FRET silica nanoparticles by single wavelength excitation. Nano Lett. 2006, 6, 84–88. [Google Scholar]
- Estevez, M.C.; O'Donoghue, M.B.; Chen, X.L.; Tan, W.H. Highly Fluorescent Dye-Doped Silica Nanoparticles Increase Flow Cytometry Sensitivity for Cancer Cell Monitoring. Nano Res. 2009, 2, 448–461. [Google Scholar]
- Chen, X.L.; Estevez, M.C.; Zhu, Z.; Huang, Y.F.; Chen, Y.; Wang, L.; Tan, W.H. Using Aptamer-Conjugated Fluorescence Resonance Energy Transfer Nanoparticles for Multiplexed Cancer Cell Monitoring. Anal. Chem. 2009, 81, 7009–7014. [Google Scholar]
- Cai, L.; Chen, Z.Z.; Chen, M.Y.; Tang, H.W.; Pang, D.W. MUC-1 aptamer-conjugated dye-doped silica nanoparticles for MCF-7 cells detection. Biomaterials 2013, 34, 371–381. [Google Scholar]
- Duan, Y.F.; Ning, Y.; Song, Y.; Deng, L. Fluorescent aptasensor for the determination of Salmonella typhimurium based on a graphene oxide platform. Microchim. Acta 2014, 181, 647–653. [Google Scholar]
- Cao, L.L.; Cheng, L.W.; Zhang, Z.Y.; Wang, Y.; Zhang, X.X.; Chen, H.; Liu, B.H.; Zhang, S.; Kong, J.L. Visual and high-throughput detection of cancer cells using a graphene oxide-based FRET aptasensing microfluidic chip. Lab Chip 2012, 12, 4864–4869. [Google Scholar]
- Maurer, E.I.; Comfort, K.K.; Hussain, S.M.; Schlager, J.J.; Mukhopadhyay, S.M. Novel Platform Development Using an Assembly of Carbon Nanotube, Nanogold and Immobilized RNA Capture Element towards Rapid, Selective Sensing of Bacteria. Sensors 2012, 12, 8135–8144. [Google Scholar]
- White, C.T.; Mintmire, J.W. Fundamental properties of single-wall carbon nanotubes. J. Phys. Chem. B 2005, 109, 52–65. [Google Scholar]
- Huang, X.; Yin, Z.Y.; Wu, S.X.; Qi, X.Y.; He, Q.Y.; Zhang, Q.C.; Yan, Q.Y.; Boey, F.; Zhang, H. Graphene-Based Materials: Synthesis, Characterization, Properties, and Applications. Small 2011, 7, 1876–1902. [Google Scholar]
- Zhu, Z.; Yang, R.H.; You, M.X.; Zhang, X.L.; Wu, Y.R.; Tan, W.H. Single-walled carbon nanotube as an effective quencher. Anal. Bioanal. Chem. 2010, 396, 73–83. [Google Scholar]
- Zheng, M.; Jagota, A.; Semke, E.D.; Diner, B.A.; Mclean, R.S.; Lustig, S.R.; Richardson, R.E.; Tassi, N.G. DNA-assisted dispersion and separation of carbon nanotubes. Nat. Mater. 2003, 2, 338–342. [Google Scholar]
- Wei, W.; Li, D.F.; Pan, X.H.; Liu, S.Q. Electrochemiluminescent detection of Mucin 1 protein and MCF-7 cancer cells based on the resonance energy transfer. Analyst 2012, 137, 2101–2106. [Google Scholar]
- Feng, L.; Chen, Y.; Ren, J.; Qu, X. A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials 2011, 32, 2930–2937. [Google Scholar]
- Liu, F.; Zhang, Y.; Yu, J.H.; Wang, S.W.; Ge, S.G.; Song, X.R. Application of ZnO/graphene and S6 aptamers for sensitive photoelectrochemical detection of SK-BR-3 breast cancer cells based on a disposable indium tin oxide device. Biosens. Bioelectron. 2014, 51, 413–420. [Google Scholar]
- Ma, X.Y.; Jiang, Y.H.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z.P. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Meth. 2014, 98, 94–98. [Google Scholar]
- Yan, M.; Sun, G.Q.; Liu, F.; Lu, J.J.; Yu, J.H.; Song, X.R. An aptasensor for sensitive detection of human breast cancer cells by using porous GO/Au composites and porous PtFe alloy as effective sensing platform and signal amplification labels. Anal. Chim. Acta. 2013, 798, 33–39. [Google Scholar]
- Liu, X.J.; Qi, X.; Zhang, Z.; Ren, L.; Hao, G.L.; Liu, Y.D.; Wang, Y.; Huang, K.; Wei, X.L.; Li, J.; et al. Electrochemically reduced graphene oxide with porous structure as a binder-free electrode for high-rate supercapacitors. RSC Adv. 2014, 4, 13673–13679. [Google Scholar]
- Yong, Y.C.; Dong, X.C.; Chan-Park, M.B.; Song, H.; Chen, P. Macroporous and Monolithic Anode Based on Polyaniline Hybridized Three-Dimensional Graphene for High-Performance Microbial Fuel Cells. ACS Nano 2012, 6, 2394–2400. [Google Scholar]
- Crespo, G.A.; Macho, S.; Rius, F.X. Ion-selective electrodes using carbon nanotubes as ion-to-electron transducers. Anal. Chem. 2008, 80, 1316–1322. [Google Scholar]
- Zelada-Guillen, G.A.; Riu, J.; Duzgun, A.; Rius, F.X. Immediate Detection of Living Bacteria at Ultralow Concentrations Using a Carbon Nanotube Based Potentiometric Aptasensor. Angew. Chem. Int. Edit. 2009, 48, 7334–7337. [Google Scholar]
- Zelada-Guillen, G.A.; Sebastian-Avila, J.L.; Blondeau, P.; Riu, J.; Rius, F.X. Label-free detection of Staphylococcus aureus in skin using real-time potentiometric biosensors based on carbon nanotubes and aptamers. Biosens. Bioelectron. 2012, 31, 226–232. [Google Scholar]
- Zelada-Guillen, G.A.; Blondeau, P.; Rius, F.X.; Riu, J. Carbon nanotube-based aptasensors for the rapid and ultrasensitive detection of bacteria. Methods 2013, 63, 233–238. [Google Scholar]
- Pisanic, T.R., 2nd; Zhang, Y.; Wang, T.H. Quantum dots in diagnostics and detection: Principles and paradigms. Analyst 2014, 139, 2968–2981. [Google Scholar]
- Gao, X.; Yang, L.; Petros, J.A.; Marshall, F.F.; Simons, J.W.; Nie, S. In vivo molecular and cellular imaging with quantum dots. Curr. Opin. Biotechnol. 2005, 16, 63–72. [Google Scholar]
- Smith, A.M.; Duan, H.; Mohs, A.M.; Nie, S. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv. Drug Deliv. Rev. 2008, 60, 1226–1240. [Google Scholar]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435–446. [Google Scholar]
- Chan, W.C.; Maxwell, D.J.; Gao, X.; Bailey, R.E.; Han, M.; Nie, S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr. Opin. Biotechnol. 2002, 13, 40–46. [Google Scholar]
- Ireson, C.R.; Kelland, L.R. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006, 5, 2957–2962. [Google Scholar]
- Lian, S.; Zhang, P.; Gong, P.; Hu, D.; Shi, B.; Zeng, C.; Cai, L. A universal quantum dots-aptamer probe for efficient cancer detection and targeted imaging. J. Nanosci. Nanotechnol. 2012, 12, 7703–7708. [Google Scholar]
- Duan, N.; Wu, S.J.; Yu, Y.; Ma, X.Y.; Xia, Y.; Chen, X.J.; Huang, Y.K.; Wang, Z.P. A dual-color flow cytometry protocol for the simultaneous detection of Vibrio parahaemolyticus and Salmonella typhimurium using aptamer conjugated quantum dots as labels. Anal. Chim. Acta 2013, 804, 151–158. [Google Scholar]
- Kim, Y.S.; Chung, J.; Song, M.Y.; Jurng, J.; Kim, B.C. Aptamer cocktails: Enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens. Bioelectron. 2014, 54, 195–198. [Google Scholar]
- Hua, X.; Zhou, Z.X.; Yuan, L.; Liu, S.Q. Selective collection and detection of MCF-7 breast cancer cells using aptamer-functionalized magnetic beads and quantum dots based nano-bio-probes. Anal. Chim. Acta 2013, 788, 135–140. [Google Scholar]
- Li, J.; Xu, M.; Huang, H.; Zhou, J.; Abdel-Halimb, E.S.; Zhang, J.R.; Zhu, J.J. Aptamer-quantum dots conjugates-based ultrasensitive competitive electrochemical cytosensor for the detection of tumor cell. Talanta 2011, 85, 2113–2120. [Google Scholar]
- Sheng, Z.; Hu, D.; Zhang, P.; Gong, P.; Gao, D.; Liu, S.; Cai, L. Cation exchange in aptamer-conjugated CdSe nanoclusters: A novel fluorescence signal amplification for cancer cell detection. Chem. Commun. 2012, 48, 4202–4204. [Google Scholar]
- Wang, Z.G.; Ding, B. DNA-based self-assembly for functional nanomaterials. Adv. Mater. 2013, 25, 3905–3914. [Google Scholar]
- Charoenphol, P.; Bermudez, H. Design and application of multifunctional DNA nanocarriers for therapeutic delivery. Acta Biomater. 2014, 10, 1683–1691. [Google Scholar]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. [Google Scholar]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Hogberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar]
- Chang, M.; Yang, C.S.; Huang, D.M. Aptamer-Conjugated DNA Icosahedral Nanoparticles As a Carrier of Doxorubicin for Cancer Therapy. ACS Nano 2011, 5, 6156–6163. [Google Scholar]
- Ferreira, C.S.M.; Matthews, C.S.; Missailidis, S. DNA aptamers that bind to MUC1 tumour marker: Design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor. Biol. 2006, 27, 289–301. [Google Scholar]
- Zhu, G.Z.; Zheng, J.; Song, E.Q.; Donovan, M.; Zhang, K.J.; Liu, C.; Tan, W.H. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. USA 2013, 110, 7998–8003. [Google Scholar]
- Hu, R.; Zhang, X.B.; Zhao, Z.L.; Zhu, G.Z.; Chen, T.; Fu, T.; Tan, W.H. DNA Nanoflowers for Multiplexed Cellular Imaging and Traceable Targeted Drug Delivery. Angew. Chem. 2014, 53, 5821–5826. [Google Scholar]



| Aptamer | Kd a (nM) | Target Cell | Control Cell Used for Negative Selection | Reference |
|---|---|---|---|---|
| DNA | 16.49 ± 0.40 | gastric cancer cells, HGC-27 | gastric epithelial cell, GES-1 | [26] |
| 28.2 ± 5.5 | non-small cell lung cancer (NSCLC) | HLAMP | [34] | |
| 0.7 ± 0.2 | colorectal cancer cells, (DLD-1) | HCT 116 | [16] | |
| 60 ± 8 | H23 lung adenocarcinoma cells | HBE 135-E6/E7 normal epithelial lung cells | [28] | |
| 0.76 ± 0.13 | Ramos cells | - | [29] | |
| 0.8 | T-cell acute lymphoblastic leukemia (T-ALL) cells, CCRF-CEM | Ramos cells | [15] | |
| 4.5 ± 1.6 | acute myeloid leukemia (AML) cells (HL60) | NB4 | [31] | |
| ∼38 | small-cell lung cancer (SCLC) cells, NCIH69 | NSCLC cells, NCI-H661 | [32] | |
| 20 ± 10 | human glioblastoma cells, U118-MG | human astroglial cells, SVGp12 | [33] | |
| 73.6 ± 11.01 | PC-3 cells | RWPE-1, SMMC-7721 and Hela cells | [35] | |
| 0.25 ± 0.08 | TOV-21G | Hela cells | [36] | |
| RNA | 94.6 | HER-2-overexpressing human breast cancer cells, SK-BR-3 | HER-2 negative cells, (MDA-MB-231) and siHER-2-transfected SK-BR-3 cells | [30] |
| 1.5 ± 0.02 | pancreatic cancer cell line (Mia-PaCa-2) | normal pancreatic cell line (HPDE) | [27] |
aThe Kd values obtained here correspond to the most potent binding aptamer.
| Aptamer | Target Bacteria | Control | Kd a (nM) | Reference |
|---|---|---|---|---|
| DNA | Mycobacterium tuberculosis | Mycobacterium bovis | nd | [18] |
| Staphylococcus aureus | Streptococcus and S. epidermidis | 35 | [37,44] | |
| Shigella dysenteriae | a mixture of related intact pathogenic bacteria | 23.47 ± 2.48 | [40] | |
| Escherichia coli O157:H7 | E. coli K12 cells | ∼110 | [41] | |
| E. coli | a mixture of other bacterial species | 12.4 | [42] | |
| Campylobacter jejuni | non-C. jejuni strains | 292.8 ± 53.1 | [47] | |
| Listeria monocytogenes | a mixture of related intact pathogenic bacteria | 48.74 ± 3.11 | [49] | |
| Pseudomonas aeruginosa | S. maltophilia and A. baumannii | 17.27 ± 5 | [50] | |
| Salmonella O8 | E. coli and S. choleraesuis | 32.04 | [45] | |
| RNA and DNA | Salmonella Enteritidis | other Salmonella serovars | 1.73 ± 0.54 μM | [38,46,48] |
aThe Kd values obtained here correspond to the most potent binding aptamer.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gedi, V.; Kim, Y.-P. Detection and Characterization of Cancer Cells and Pathogenic Bacteria Using Aptamer-Based Nano-Conjugates. Sensors 2014, 14, 18302-18327. https://doi.org/10.3390/s141018302
Gedi V, Kim Y-P. Detection and Characterization of Cancer Cells and Pathogenic Bacteria Using Aptamer-Based Nano-Conjugates. Sensors. 2014; 14(10):18302-18327. https://doi.org/10.3390/s141018302
Chicago/Turabian StyleGedi, Vinayakumar, and Young-Pil Kim. 2014. "Detection and Characterization of Cancer Cells and Pathogenic Bacteria Using Aptamer-Based Nano-Conjugates" Sensors 14, no. 10: 18302-18327. https://doi.org/10.3390/s141018302






