Determination of Ammonium Ion Using a Reagentless Amperometric Biosensor Based on Immobilized Alanine Dehydrogenase
Abstract
: The use of the enzyme alanine dehydrogenase (AlaDH) for the determination of ammonium ion (NH4+) usually requires the addition of pyruvate substrate and reduced nicotinamide adenine dinucleotide (NADH) simultaneously to effect the reaction. This addition of reagents is inconvenient when an enzyme biosensor based on AlaDH is used. To resolve the problem, a novel reagentless amperometric biosensor using a stacked methacrylic membrane system coated onto a screen-printed carbon paste electrode (SPE) for NH4+ ion determination is described. A mixture of pyruvate and NADH was immobilized in low molecular weight poly(2-hydroxyethyl methacrylate) (pHEMA) membrane, which was then deposited over a photocured pHEMA membrane (photoHEMA) containing alanine dehydrogenase (AlaDH) enzyme. Due to the enzymatic reaction of AlaDH and the pyruvate substrate, NH4+ was consumed in the process and thus the signal from the electrocatalytic oxidation of NADH at an applied potential of +0.55 V was proportional to the NH4+ ion concentration under optimal conditions. The stacked methacrylate membranes responded rapidly and linearly to changes in NH4+ ion concentrations between 10–100 mM, with a detection limit of 0.18 mM NH4+ ion. The reproducibility of the amperometrical NH4+ biosensor yielded low relative standard deviations between 1.4–4.9%. The stacked membrane biosensor has been successfully applied to the determination of NH4+ ion in spiked river water samples without pretreatment. A good correlation was found between the analytical results for NH4+ obtained from the biosensor and the Nessler spectrophotometric method.1. Introduction
The accurate measurement of NH4+ ion in an aquatic environment is of significant interest in environmental biological studies and the environmental evaluation of water since it is known to be toxic for aquatic organisms. In this sense, several methods have been proposed, most of them involving ion chromatography [1–3], potentiometry [4,5] or flow injection systems [6–11].
Most of the ion chromatography procedures proposed for its determination involve elaborate pre-column derivatization in some kind of matrix. Mori et al. [12] described a sensitive and rapid ion chromatography method to determine NH4+ ion in river waters. However, baseline separations of NH4+ from alkali and alkaline earth metal ions in water samples were non achievable. For potentiometric detection of NH4+ ion, nonactin has been widely used as sensing material. Even though nonactin-based ion-selective electrodes show good sensitivity toward NH4+ ion, they suffer interference from other ions such as K+ [13,14]. Flow injection systems combined with spectrophotometric methods, e.g., the Berthelot reaction involving a colour change in the presence of NH4+ ion, have very slow reaction kinetics [15], whereas fluorimetric flow injection analysis requires pretreatment of the samples with long diffusion times to avoid background interferences [2,16].
Today, there is a well-recognised trend towards the simplification and miniaturisation of analytical processes [2]. An amperometry approach employing a miniaturised SPE with immobilized enzyme as tranducer considerably improves the operation cost, providing for a simple, reliable, rapid and reproducible analytical procedure. A few biosensors for the amperometric determination of NH4+ ion employing glutamate dehydrogenase (GLDH) have been reported where the enzyme was immobilized onto the working electrode in several ways [17–19]. However, to effect the enzymic GLDH reaction, a substrate and co-factor normally needed to be introduced and this leads to an extra step during the assay of NH4+ ion. In order to obviate the needs for external reagent treatment during measurement, which may also cause contamination of the reference electrode, we describe in this work an approach employing a stacked membranes system for the immobilization of enzyme, co-factor and also substrate that eventually leads to a reagentless biosensor for NH4+ ion determination.
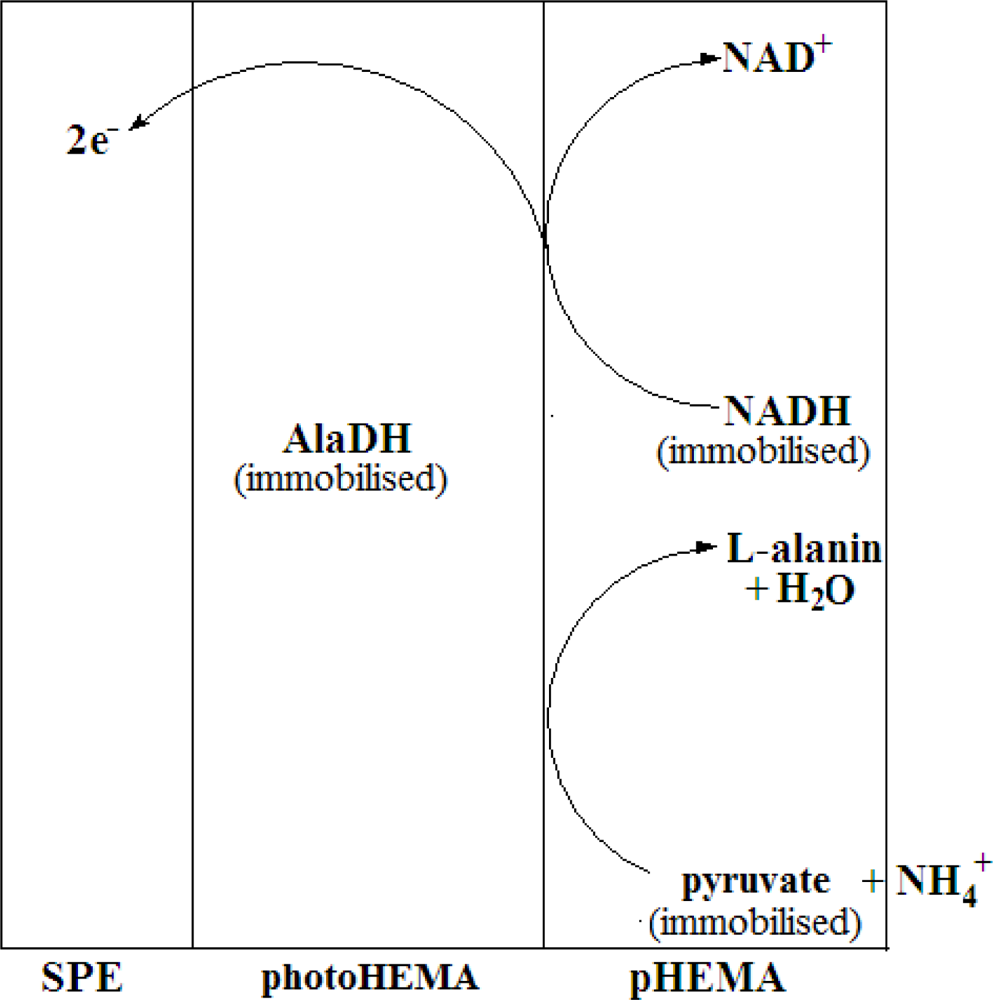
In this work, we have used alanine dehydrogenase (AlaDH) to construct a biosensor for the determination of NH4+ ion. To our knowledge, the use of AlaDH in an NH4+ ion biosensor has not been reported. The concept of the biosensor based on AlaDH is the reversible amination of pyruvate to L-alanine by AlaDH in the presence of NADH co-factor and NH4+ ion (Equation (1)) [20–22]. The current generated from the electrochemical process was measured based on the oxidation of NADH (Equation (2)) whilst the enzyme redox reaction consumed NH4+ ion in the process. Thus, the redox current is proportional to the NH4+ ion concentration changes under optimal conditions at an applied potential of +0.55 V:
To construct the stacked membrane biosensor, AlaDH enzyme was first entrapped in the photoHEMA membrane, whereby the membrane with the entrapped enzyme was formed via UV photopolymerisation of 2-hydroxylethyl methacrylate monomer. Past studies have shown that the use of photoHEMA is compatible with many enzymes without leaching problems. To further immobilize the pyruvate and NADH, a low molecular weight poly(2-hydroxylethyl methacrylate) (pHEMA) membrane containing both of these substances was cast on top of the enzyme membrane after photocuring. The use of this second membrane allowed both pyruvate and NADH to be immobilized but their free diffusion in the membrane would also be ensured to maintain a rapid reaction rate. The biosensor was thus designed to provide a reagentless system specific for NH4+ ion. Figure 1 represents the mechanism of the enzymic reaction involved in the developed NH4+ ion biosensor based on a stacked membrane system.
2. Experimental
2.1. Reagents
Chemicals were obtained from commercial sources and were used without further purification. β-Nicotinamide adenine dinucleotide, reduced form (NADH, 98%) was purchased from Sigma. Stock solution of L-alanine dehydrogenase enzyme (AlaDH, E.C. 1.4.1.1, from Bacillus subtillis, Sigma) was prepared by mixing an appropriate amount of AlaDH enzyme solution with 10 mM phosphate buffer pH 7 in an Eppendorf tube and stored at 4 °C [23]. Sodium pyruvate (C3H3NaO3, 99%, Sigma) stock solution was prepared by dissolving an appropriate amount of pyruvate salt in deionised water. pH 7, 10 mM phosphate buffer was prepared by adding 10 mM dipotassium hydrogen phosphate (K2HPO4, 98%, Fluka) to 10 mM potassium dihydrogen phosphate (KH2PO4, 99.5%, Fluka) and adjusting to the required pH value [23]. A standard stock ammonia solution was prepared by dissolving the required amount of concentrated ammonia solution (NH4OH, 25%, Merck) in deionised water. The ammonia solution used was standardised by the Nessler method using ammonium chloride (NH4Cl, 99.5%) salt. A homogeneous stock solution of 2-hydroxyethyl methacrylate monomer (HEMA, C(CH3)COOCH2CH2OH, 97%, Aldrich) was prepared by mixing appropriate amounts of HEMA monomer and initiator (2,2-dimethoxy-2-phenylacetophenone, DMPP, C16H16O3, 98%, Fluka) in a vial wrapped with aluminium foil. The mixture was then stirred gently for a few minutes and stored at 4 °C. Poly(2-hydroxyethyl methacrylate) (pHEMA) was obtained commercially from Aldrich.
2.2. Instrumentation
Electrochemical behaviours of the reagentless biosensor were characterised using a chronoamperometry technique with an Autolab PG12 (AUT 71681) Potentiostat/Galvanostat. A conventional three-electrode electrochemical cell was used with a glassy carbon electrode as an auxiliary electrode. A Ag/AgCl electrode saturated with KCl was used as a reference and a modified carbon paste electrode was used as a working electrode. The SPEs used were designed by Universiti Kebangsaan Malaysia and manufactured by Scrint Print Co. All potentials were measured and reported versus a Ag/AgCl electrode (saturated by KCl). During the constant potential experiments, a magnetic stir bar was used and the background current was allowed to decay to a constant value before NH4+ ion was added to the buffer solution. Measurements of pH were made with a pH-meter (MeterLab PHM 210). AlaDH enzyme-containing photoHEMA membrane was prepared by UV-initiated photopolymerisation with an UV-exposure unit (RS Components 196-5251).
2.3. Construction of Biosensor
Reagentless NH4+ biosensor was constructed by depositing 3 μL of 2.98 mg AlaDH/g of HEMA monomer mixture onto the SPE and exposing it to long-wave ultraviolet radiation for 500 s with extensive nitrogen gas purging. Next, appropriate amounts of NADH and pyruvate were dissolved into an appropriate amount of pHEMA solution prepared by dissolving 50 mg of the polymer in 20% 1,4-dioxane in water. This was then deposited on the photocured membrane containing AlaDH enzyme and left to dry at 4 °C for 24 h to form the second membrane layer.
2.4. Optimisation of Biosensor Responses
All electrochemical experiments were performed at room temperature in an undivided three-electrode cell containing supporting electrolyte solution (4 mL of 10 mM phosphate buffer pH 7) under constant stirring conditions (100 rpm). NH4+ ion was injected at the electrode surface in the measurement cell after stabilisation of the baseline current. The measurements taken were expressed as the current difference in the absence and presence of NH4+ ion.
Biosensor response was studied using the cyclic voltammetry technique between −1.00–1.00 V versus Ag/AgCl at a scan rate of 0.02 V/s. The dependence of the amperometric signal on the applied potential was examined in the range of 0.45–0.65 V versus Ag/AgCl. The pH effect was studied by varying the pH of the electrolyte solution in the range of pH 5.8–8.0 using 10 mM potassium phosphate buffer. For the optimisation of enzyme loading in the photoHEMA membrane, enzyme membranes were prepared from 5 μL mixtures of AlaDH and monomer HEMA (1:1) in different enzyme loadings (0.44–4.10 mgAlaDH/g photoHEMA).
Enzyme electrodes with different membrane thicknesses were prepared by changing the mixture volumes of monomer HEMA and AlaDH (1:1) in the range of 2–6 μL while the enzyme loading was kept constant at 2.98 mg/g photoHEMA. The thicknesses of the membranes were measured using a digital Vernier caliper after exposure of the sensing membranes to UV radiation.
Biosensor response to temperature changes in the 10–50 °C range was determined by varing the temperature of the electrolyte solution in a thermostatic bath. For this study, about 21 biosensors were prepared a day before and stored overnight at 4 °C before testing was performed.
For the optimisation of the respective NADH and pyruvate loadings in the pHEMA layer, various loadings of pyruvate (3.74–163.11 mg/g pHEMA) and NADH (22.98–1,149.11 mg/g pHEMA) were used. After optimisation of the experimental conditions, the effect on the reagentless biosensor’s performance was investigated against different NH4+ ion concentrations in the range of 10–600 mM.
The reproducibility of the biosensor was assessed using different biosensors and each biosensor was tested once by using 30 mM NH4+ ion, whereas the repeatability of the biosensor was examined using the same biosensor tested in a concentration of 30 mM NH4+ ion and five consecutive measurements were taken for every biosensor.
The shelf life of the biosensor was examined by determining 30 mM NH4+ ion over a period of 30 days. All of the electrodes were tested once on the first day and kept at 4 °C while not in use. After that, each of these biosensors was tested twice only according to a specified date.
Interferences with the NH4+ ion biosensor were investigated by using interferents such as Na+, K+, methylamine (CH3NH2) and ethylamine (C2H5NH2). The molar ratio of interferents to NH4+ ion was varied in the range of 0.01–10. The degree of interference of these interferents was evaluated separately with amperometric measurements recorded in the presence and absence of the interferent in the determination of 30 mM NH4+ ion. Statistical t-tests were employed to compare the responses obtained in the absence and presence of interferent.
Analysis of water samples was performed using five real water samples collected at different points along a river. Water samples presenting suspended particles were filtered through a 0.45 μm cellulose acetate membrane filter (Whatman) and the pH was adjusted to pH 7 using 10 mM K2HPO4 and KH2PO4. Recovery tests were conducted by adding standard concentrations of NH4+ ion in the linear range to the water samples. The results were then validated using established procedure of Nesslers’ method.
3. Results and Discussion
3.1. The Electrochemistry of Immobilized AlaDH
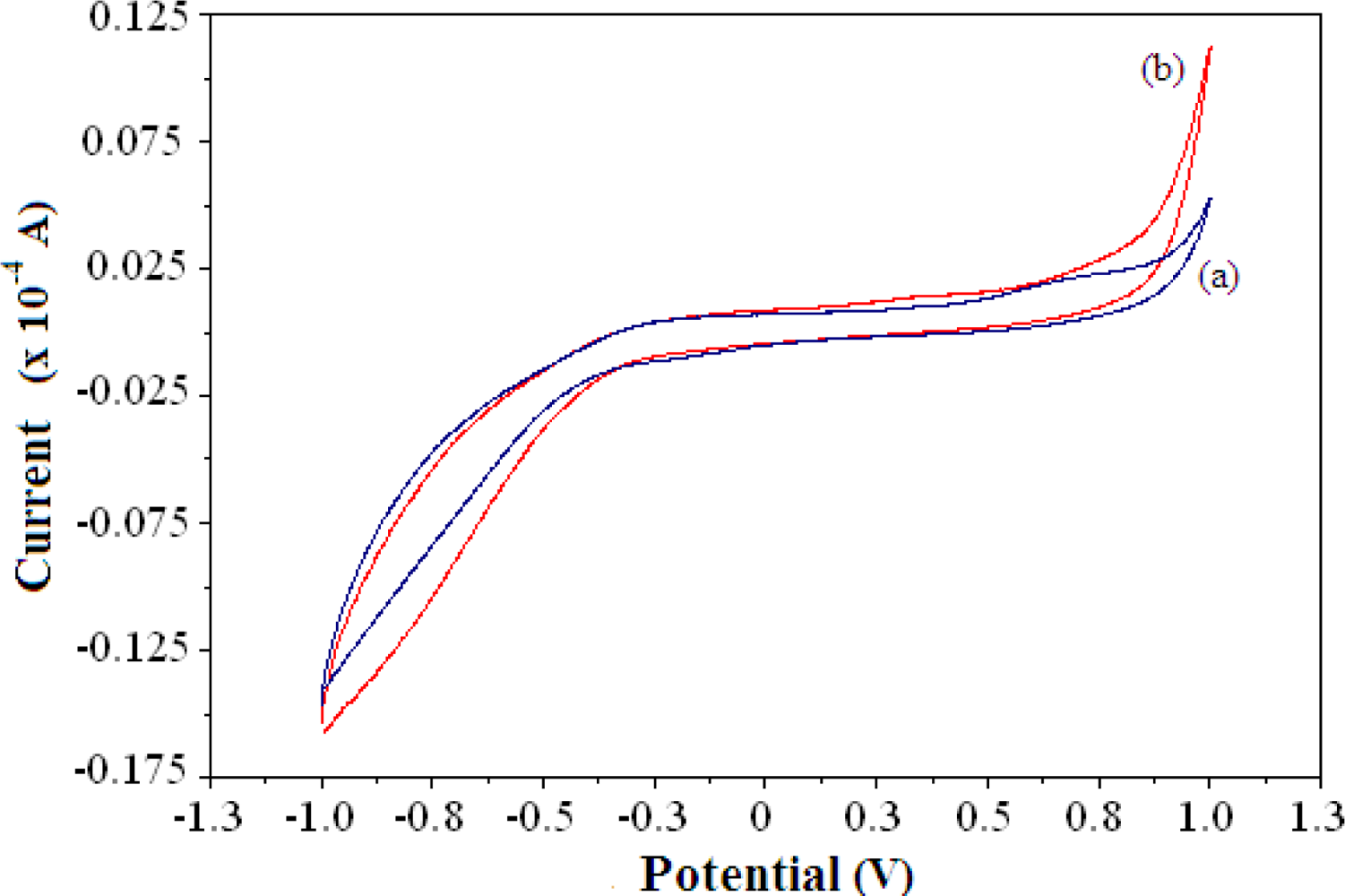
Due to the enzymatic reaction of AlaDH and the pyruvate substrate, the signal from the electrocatalytic oxidation of NADH was proportional to the NH4+ ion concentration. Figure 2 shows a typical cyclic voltammogram of the reagentless NH4+ ion biosensor demonstrating the electrocatalytic activity of biosensor in the absence and presence of 100 mM NH4+ ion. The oxidation current of the biosensor increases after NH4+ ion addition in the positive potential region. Although no clearly defined cyclic voltammetric peak appears, the biosensor shows an increase in current in response to NH4+ ion in the potential range of 0.4–0.9, as indicated by the cyclic voltammogram.
The increase of the current was attributed to the electrocatalytic oxidation of NADH during enzymatic conversion of pyruvate to L-alanine in the presence of NH4+ ion. The electrode reaction at the electrode surface is governed by a single one-electron step in an overall two-electron process, where NADH is irreversibly oxidized through loss of an electron to produce a cation radical NADH•+ and is expressed by Equation (3):
NAD• is immediately oxidized to NAD+ at the electrode surface at the positive potential involved as described by Equation (5):
The net electrochemical oxidation of NADH could be described by Equation (2) [24]:
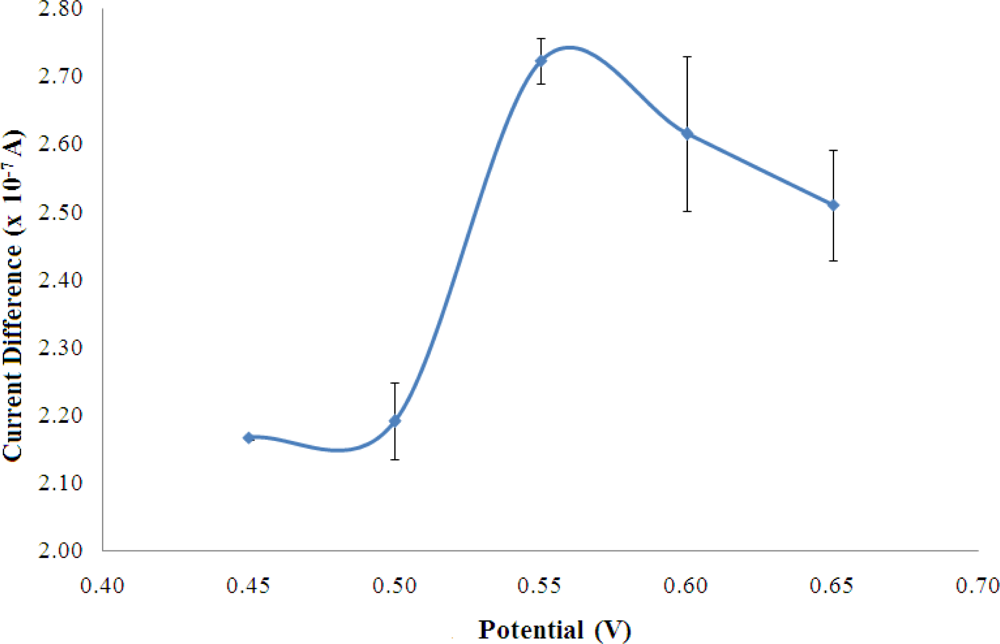
Figure 3 shows the biosensor response of a fixed amount of NH4+ ion when the potential was varied between 0.45–0.65 V and optimum applied potential was observed at +0.55 V, where the highest current was obtained. The working potential was found like the NADH oxidation potential using a 4,6-diamino-2-mercaptopyrimidine modified gold (Au) electrode [25] and a thiocytosine modified Au electrode [26]. However, the oxidation of NADH at +0.55 V was found lower compared to the electrochemical oxidation of NADH employing boron-doped diamond electrode (+0.58 V) [27]. The potential of +0.55 V was used in this work for the amperometric detection of NH4+ ion.
3.2. Optimization Studies on Biosensor Operation
3.2.1. Optimisation of pH
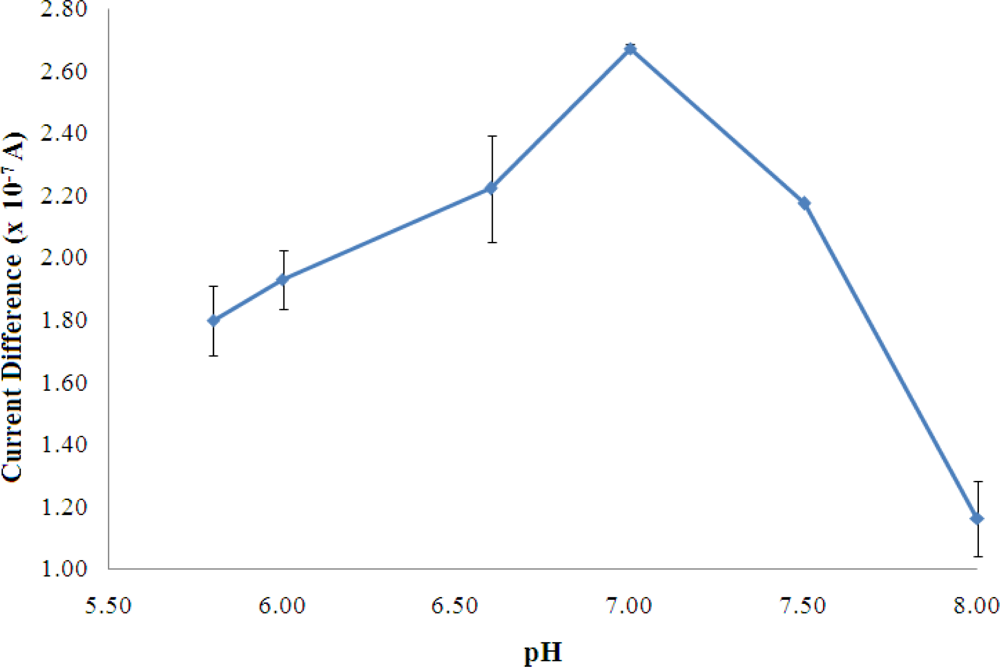
For the optimisation of buffer pH, as observed in Figure 4, the maximum enzyme reaction occurred at pH 7 and it is the optimum pH for reversible behavior of AlaDH as reported in the literature [28].
The rate of an enzyme reaction, in most cases, passes through a maximum as a function of pH. From Figure 3, at pH above 7, the enzyme may undergo an irreversible denaturation where the activity may not be restored even after readjustment of the pH to the optimal value [29].
3.2.2. Optimisation of Enzyme Loading for Biosensor
The effect of enzyme loading on the biosensor response is shown in Figure 5. The response increased with increasing AlaDH enzyme loading in the membrane until saturation was achieved at an enzyme loading of 2.98 mg AlaDH/g fotoHEMA. However, the response was lower after this optimal loading.
Typically, the rate of the enzymatic reaction is directly proportional to the enzyme loading. There are several conditions under which this proportionality may not occur. When large amounts of enzyme are used the added cofactor NADH may bind to the enzyme and not be available for reaction [29]. High enzyme loading also results in the blocking of the enzyme active sites, especially for enzyme that is located far from the surface of the membrane, thus making some of the enzyme not participate in enzymatic reactions. Furthermore, excessive loading of enzyme will also create a diffusion barrier to the movement of substrates and reaction products, which will also reduce the response obtained. Thus, the optimised enzyme loading of 2.98 mg/g photoHEMA was used throughout the experiments. Kwan et al. [30] have reported using the same enzyme entrapped by a poly(carbamoyl) sulfonate hydrogel on a Teflon membrane for amperometric determination of alanine. The backward enzyme reaction involved specific dehydrogenation of alanine that consumed NAD+. The reaction products, pyruvate and NADH were used to initiate the two other reactions that follow, which involved salicylate hydroxylase and pyruvate oxidase [30].
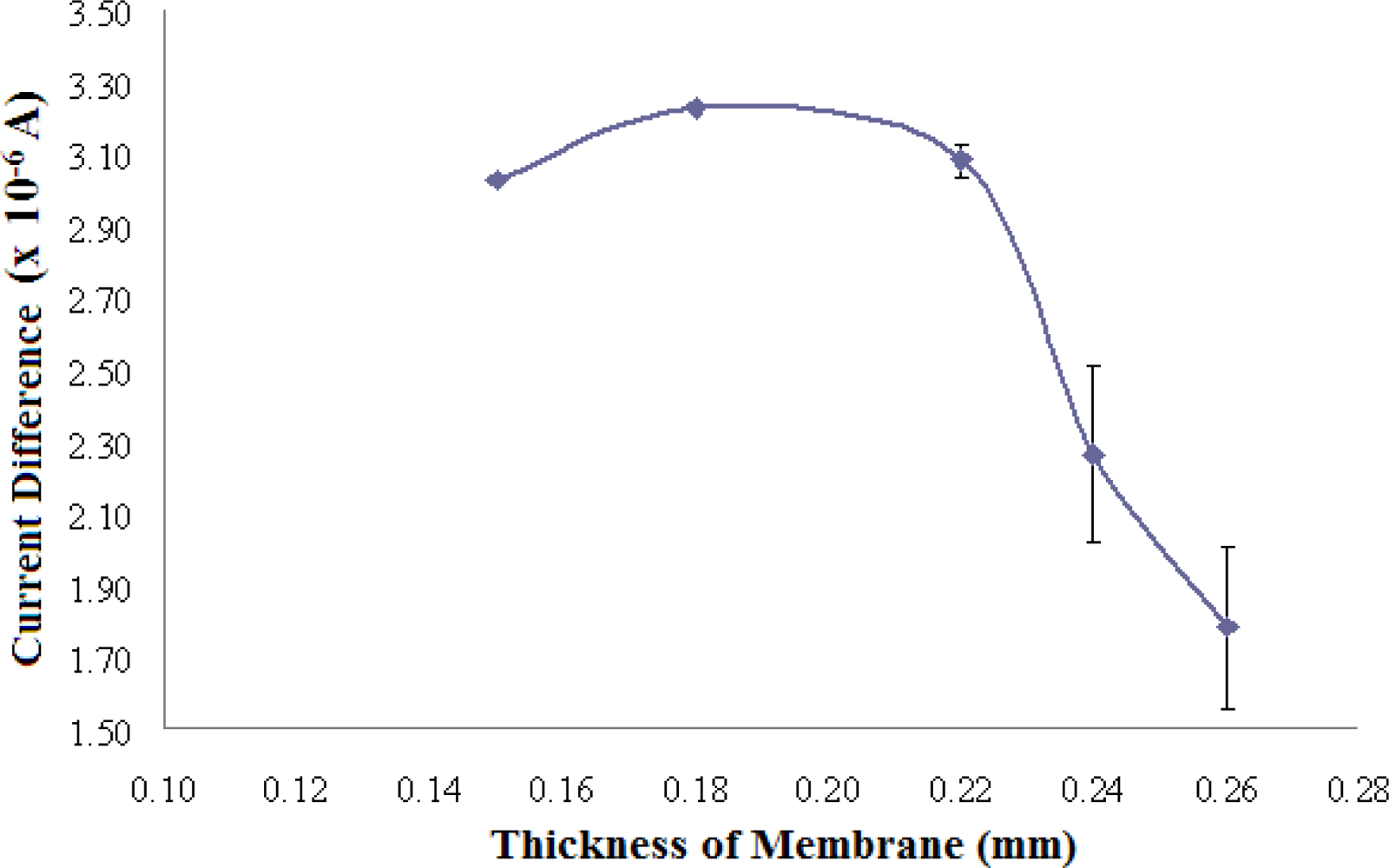
3.2.3. Optimisation of Membrane Thickness for Biosensor Fabrication
The effect of the thickness of photoHEMA membranes containing 2.98 mg AlaDH/g photo HEMA on the biosensor response is presented in Figure 6. The optimum thickness of AlaDH enzyme-containing photoHEMA membrane was found to be 0.18 mm because of the consistent high signal attained for the same NH4+ ion concentration (RSD = 5.6%). A membrane thickness of 0.15 mm gave a slightly lower response due to the low reproducibility of preparing a thin membrane (RSD = 14.2%). Moreover, a thin membrane may lead to the leaching out of the immobilized enzyme. Thicker membranes, between 0.22–0.26 mm, gave inconsistently low responses as these membranes resulted in the active sites of the enzyme becoming less accessible, in addition to hindering the diffusion of rectants and products.
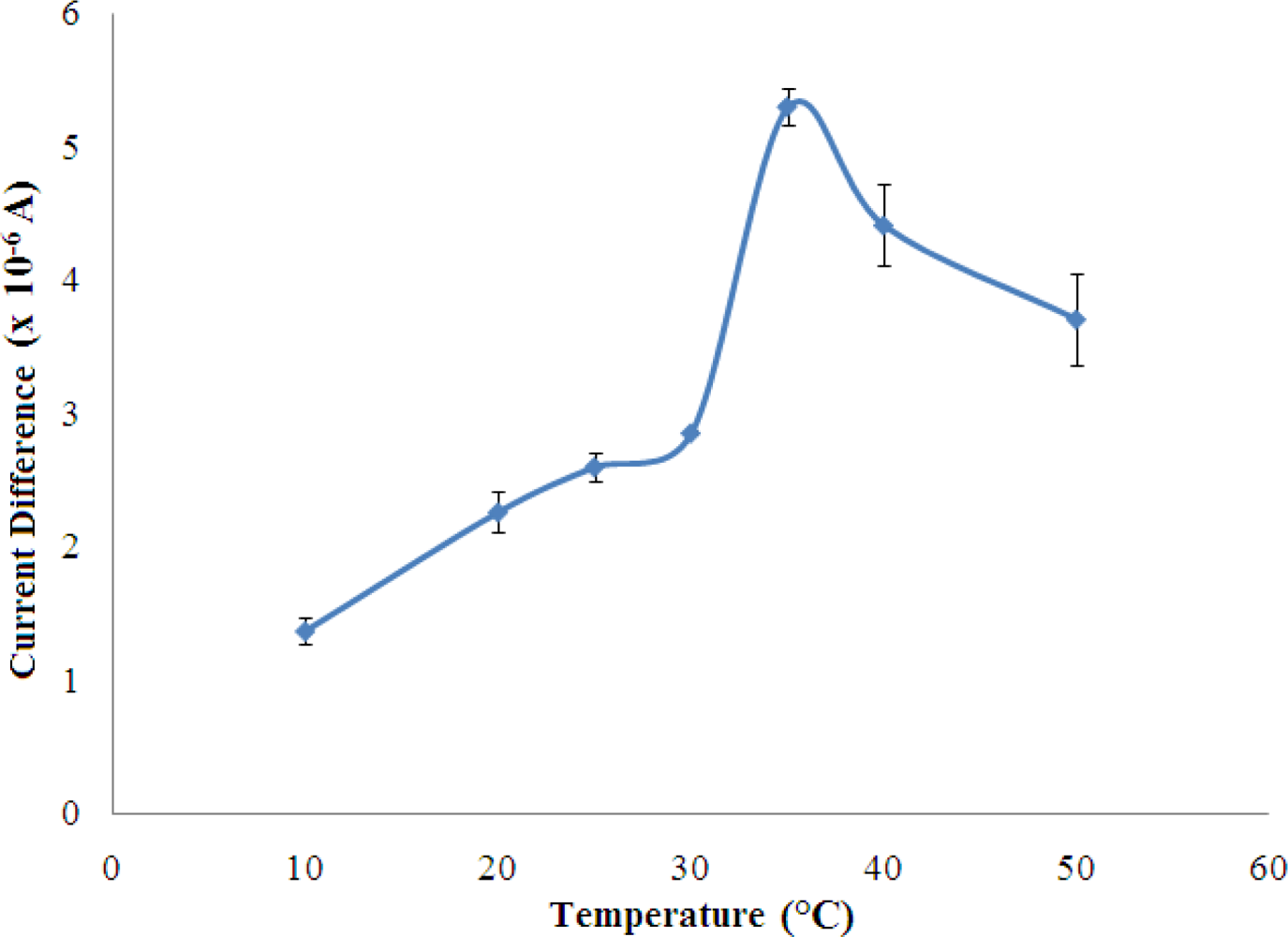
3.2.4. Optimisation of Temperature of Biosensor Operation
The biosensor response increased rapidly with temperature and reached a maximum at the temperature of 35 °C (Figure 7). This is explained by the increase in the rate of an enzyme reaction with increasing temperature [29].
However, the biosensor response decreased when the temperature increased above 35 °C. In this state, the denaturation of the enzyme occurs where the weak bonding such as hydrogen and ionic bonds are broken at high temperatures [31]. In the present work, 25 °C was chosen as working temperature for all subsequent experiments since the sensitivity of the biosensors were sufficiently good for further studies.
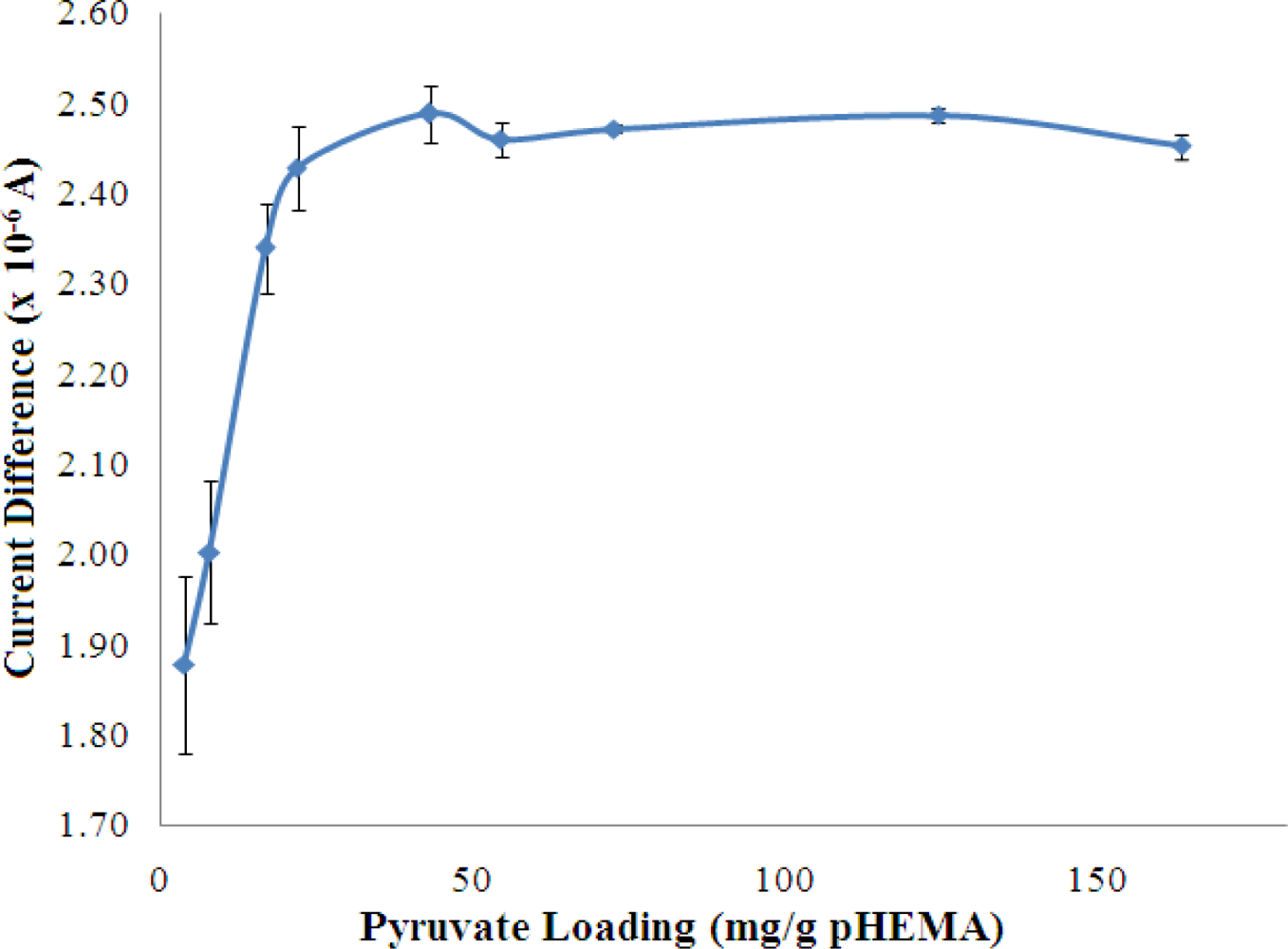
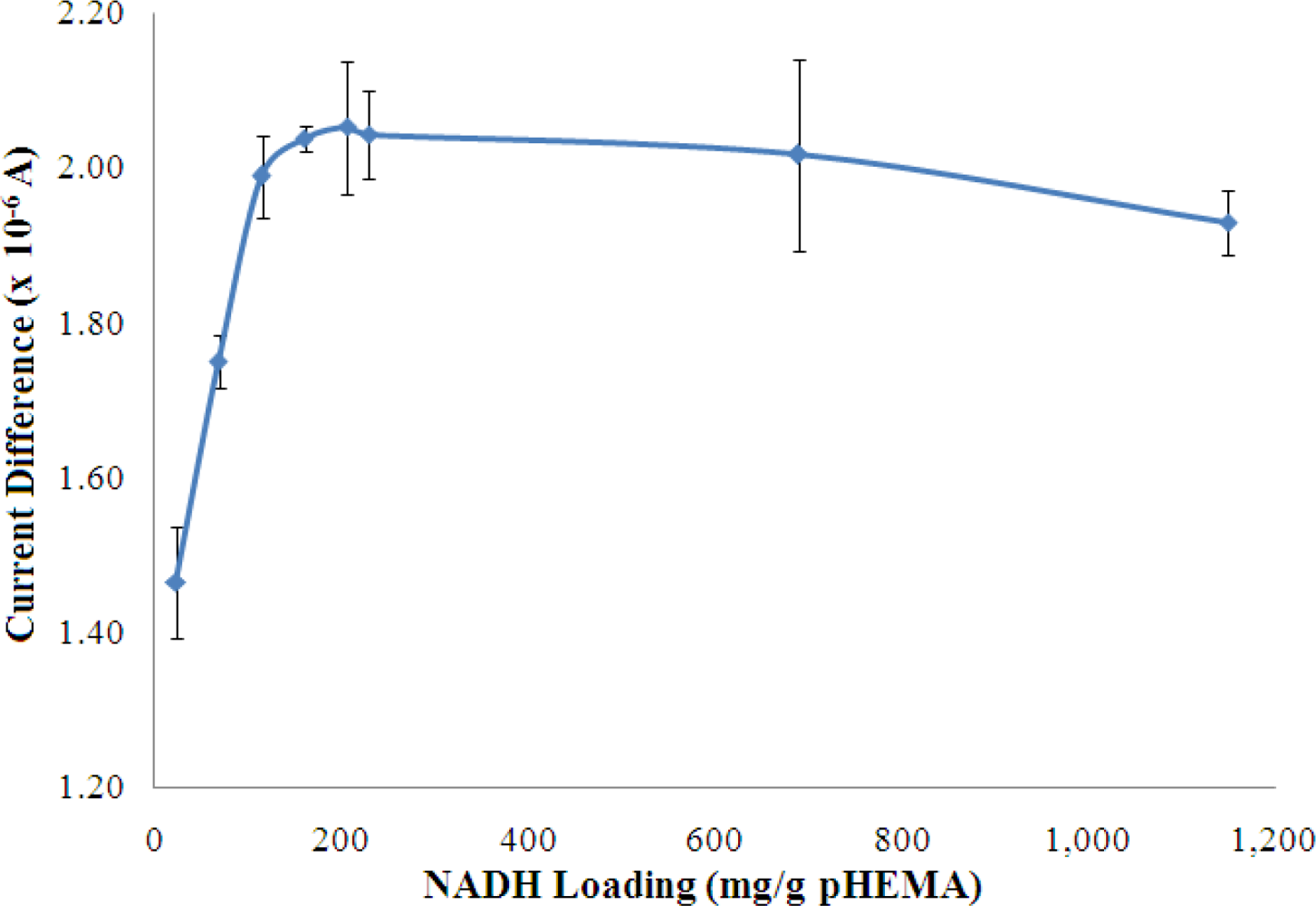
3.2.5. Optimisation of Loading for Pyruvate and NADH
The concentrations of both pyruvate and NADH, which were immobilized in the pHEMA membrane were optimized to yield the best biosensor response to NH4+ ion. The results are shown in Figures 8 and 9, which indicate that the current response increases with increasing pyruvate and NADH loadings and becomes saturated at loadings of 42.99 mg pyruvate/g pHEMA and 160.87 mg NADH/g pHEMA, respectively.
3.3. Analytical Performance of NH4+ Ion Biosensor
3.3.1. The Response Range of Biosensor
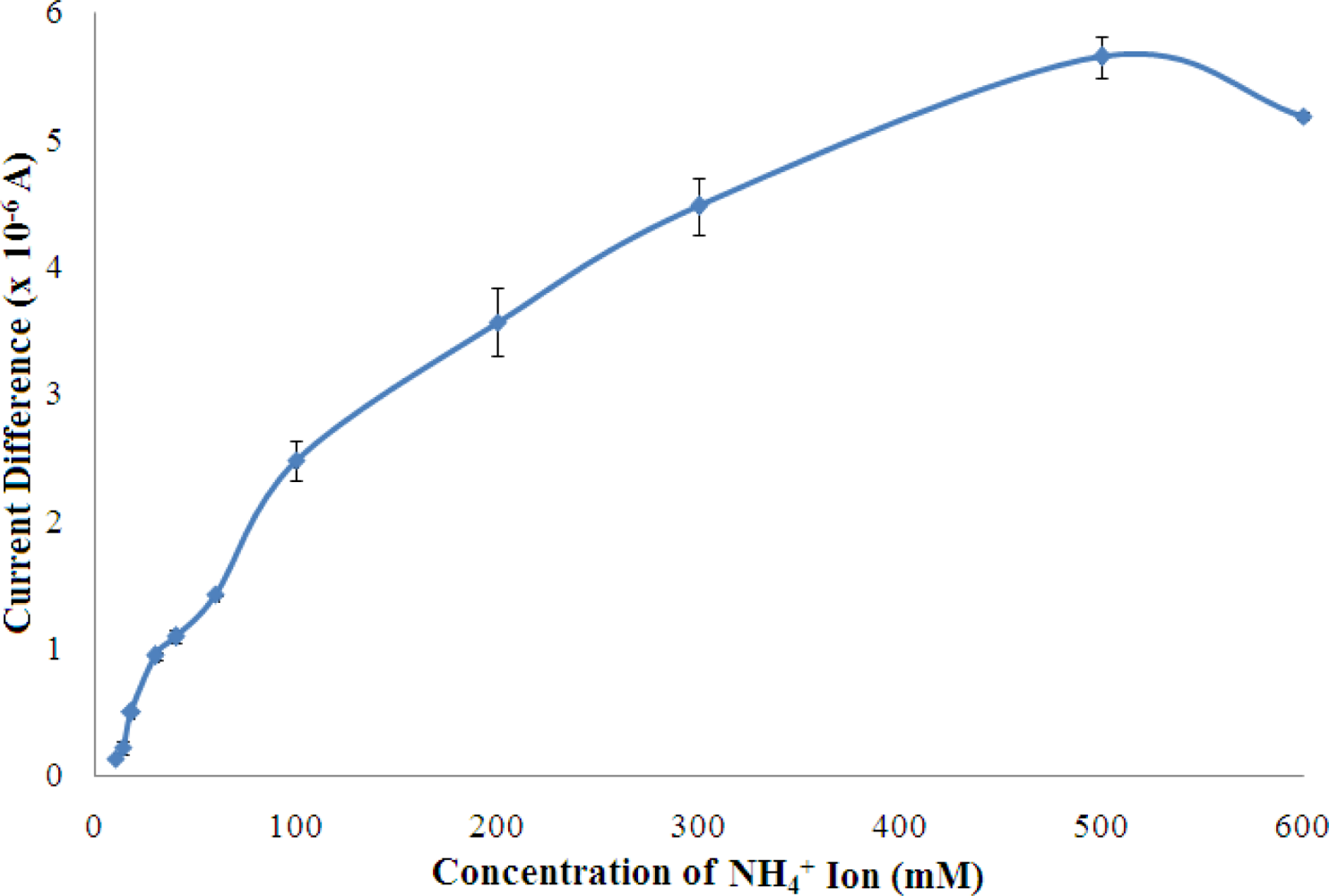
The response of the biosensor was examined from 10–600 mM of NH4+ ion and the results are shown in Figure 10.
The biosensor sensor response was found increased with increasing NH4+ ion concentration from 10–500 mM, and reached saturation beyond 500 mM NH4+ ion. This phenomenon is due to the fact that available pyruvate, cofactor NADH, and enzyme active sites have been gradually consumed with increasing NH4+ ion concentration until a saturation condition is achieved. Furthermore, at high levels of NH4+ ion, the ionic strength of the reaction medium increases and thereby reduces the enzymatic reaction which is also influenced by the charge at the enzyme active sites. A linear response range was observed at 10–100 mM (R2 = 0.978, n = 7) with a limit of detection of 0.18 mM. The apparent Km value estimated from the Lineweaver-Burk plot was 34.01 mM (Figure 11).
3.3.2. The Biosensor Reproducibility and Repeatability
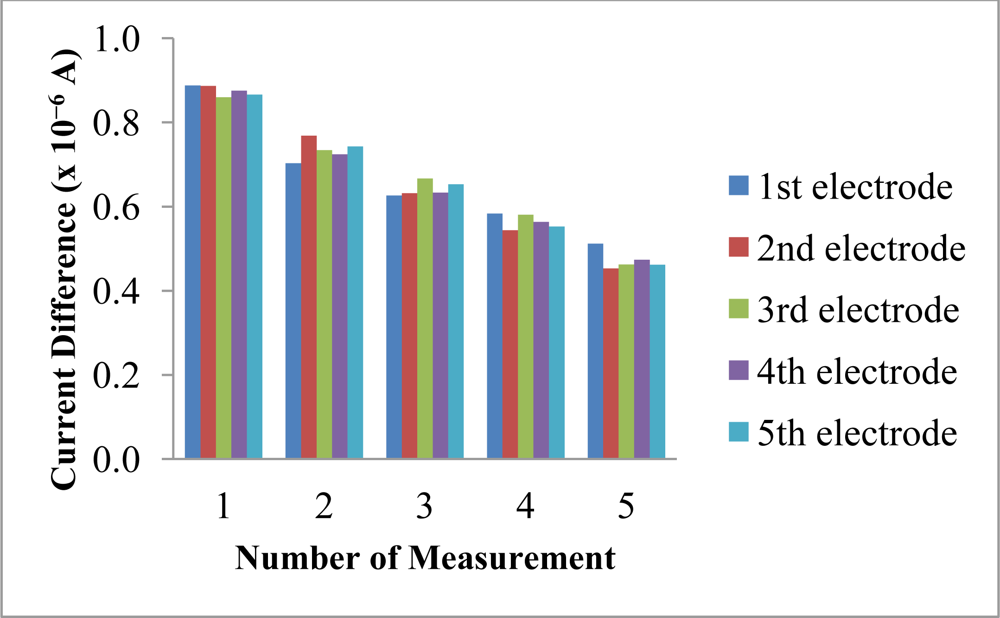
The reproducibility and repeatability of the reagentless biosensor was evaluated at 30 mM NH4+ ion. The current difference measurements obtained using different electrodes gave low RSD values from 1.4–4.9% (n = 5). These values were low enough to consider that the membranes were reproducible as fabricated by the proposed method. However, the slightly higher repeatability RSD values (21.7–26.4%, n = 5) obtained using the same biosensor was attributed to the irreversible oxidation of immobilized NADH at the electrode surface, which resulted in a decreasing current difference being observed for five consecutive measurements over the same electrode (Figure 12). The poor repeatability is an inherent effect from the leaching of the immobilized NADH and pyruvate. Both of these reactants are deliberately allowed to leach out from the membrane in order for them to diffuse into the enzymatic membrane to react with the AlaDH enzyme. Therefore, the poor repeatability is attributed to the loss and consumption of these reactants when the biosensor is reused many times. The biosensor is mainly used for on-site analysis and because it is made from SPE that can be manufactured at low cost, thus one can aim for a single-use and disposable electrode.
3.3.3. Biosensor Stability
In terms of biosensor stability, over a period of 30 days and using 30 mM NH4+ ion, the reagentless biosensor first showed a decrease in sensitivity of approximately 67% during the first four days, after which the sensitivity was maintained for up to 20 days before further deterioration of the response began. After 30-day storage period, the biosensor still retained about 50% of the initial sensitivity. (Table 1). An amperometric biosensor employing AlaDH enzyme for alanine determination reported an operational stability of only two days and the sensor response dropped to 50% of the initial sensitivity after day-13 [30].
3.3.4. Effects of Interferents on Biosensor Response
The possible interferences in the determination of NH4+ ion by the reagentless biosensor were investigated and the results are depicted in Table 2. The reagentless biosensor did not suffer any interference from alkali metal ions at levels occurring in natural waters [32]. But approaching a 1:1 molar ratio, interference was observed because at high concentration of ions, and high ionic strength of the measurement medium that appears to reduce the enzymatic reaction [31]. The stability of AlaDH is higher in the presence of NaCl salt compared to KCl [22], hence Na+ ion did not demonstrate any observable interference when compared with K+ ion, even at a molar ratio of 1:1. CH3NH2 and C2H5NH2 interfered significantly at molar ratios of 0.1 and above. The interference increases the biosensor response as some physicochemical properties of amines are similar to NH4+ ion. However, such interference by amines is not expected in natural water samples due to their low level, usually 100 times lower than NH4+ ion [10].
3.3.5. Performance of Biosensor for Water Sample Analysis
To assess the accuracy of the biosensor, recovery studies were performed using river water samples which had been spiked with known NH4+ ion concentrations within the linear response range of the biosensor. The results are shown in Table 3. The recovery values obtained were close to 100% in the five different NH4+ ion concentrations spiked into river water samples. The analytical performance of the reagentless biosensor for water analysis was compared with the standard method of Nessler method using spectrophotometric method. The t-test applied to examine whether the two methods gave results that both methods were not statistically different for NH4+ ion determination. This indicates that the reagentless biosensor can be used for NH4+ ion determination.
3.3.6. Comparison with Similar Biosensors
Amperometric based enzymic biosensors for NH4+ ion determination using GLDH enzyme are commonly reported in the literature [17–19] (Table 4), however, they all require that the substrates involved in the enzymic reaction with GLDH enzyme, be introduced into the assay solutions. The linear detection ranges of NH4+ ion for biosensors using GLDH were somehow narrower compared to the use of AlaDH enzyme. The GLDH enzyme system has also been used for the optical detection of NH4+ ion in water samples at μM level based on NADH oxidation [33]. A chitosan matrix was used for the immobilization of GLDH enzyme on a supporting glass slide but this optical biosensor system for NH4+ ion determination suffered interference from other possible cations present in the water samples.
4. Conclusions
A new reagentless amperometric biosensor for the determination of NH4+ ions based on the enzyme AlaDH and involving the use of a stacked membrane system for the immobilization of all reagents was successfully developed. The use of a layer of low molecular weight pHEMA for NADH and pyruvate immobilization and a photocured photoHEMA membrane with entrapped AlaDH enzyme offers a simple and rapid biosensor fabrication procedure in addition to convenient NH4+ ion analysis, particularly for performing on-site analysis without the need of any addition of reagents. The reagentless biosensor demonstrated a wide linear response range between 10–100 mM NH4+ ion with a detection limit of 0.18 mM and good selectivity over some major cations. Such a biosensor has a potential to overcome the problem of on-site analysis of NH4+ ion because it can be used for sewage NH4+ ion analysis without further dilution (NH4+ ion concentrations of sewage are typically from 65–170 mM).
Acknowledgments
We gratefully acknowledged Universiti Kebangsaan Malaysia for a research operation grant (UKM-OUP-NBT-29-151/2011) and a Postgraduate Fellowship awarded to Tan Ling Ling.
References
- Kuo, CT; Wang, PY; Wu, CH. Fluorometric determination of ammonium ion by ion chromatography using postcolumn derivatization with o-phthaldialdehyde. J. Chromatogr. A 2005, 1085, 91–97. [Google Scholar]
- Martinez, YM; Hernandez, RH; Falco, PC. Improved detection limit for ammonium/ammonia achieved by Berhelot’s reaction by use of solid-phase extraction coupled to diffuse reflectance spectroscopy. Anal. Chim. Acta 2005, 534, 327–334. [Google Scholar]
- Thomas, DH; Rey, M; Jackson, PE. Determination of inorganic cations and ammonium in environmental waters by ion chromatography with a high-capacity cation-exchange column. J. Chromatogr. A 2002, 956, 181–186. [Google Scholar]
- Deyhimi, F; Choobar, BG. Potentiometric determination of activity coefficients for NH4Cl in the ternary NH4Cl/LiCl/H2O mixed electrolyte system. J. Electroanal. Chem 2005, 584, 141–146. [Google Scholar]
- Hassan, SSM; Marei, SA; Badr, IH; Arida, HA. Novel solid-state ammonium ion potentiometric sensor based on zirconium titanium phosphate ion exchanger. Anal. Chim. Acta 2001, 427, 21–28. [Google Scholar]
- Andrew, KN; Worsfold, PJ; Comber, M. On-line flow injection monitoring of ammonia in industrial liquid effluents. Anal. Chim. Acta 1995, 314, 33–43. [Google Scholar]
- Aminot, A; Kerouel, R; Birot, D. A flow injection-fluorometri method for the determination of ammonium in fresh and saline waters with a view to in situ analyses. Wat. Res 2001, 35, 1777–1785. [Google Scholar]
- Haghighi, B; Kurd, SF. Sequential flow injection analysis of ammonium and nitrate using gas phase molecular absorption spectrometry. Talanta 2004, 64, 688–694. [Google Scholar]
- Kerouel, R; Aminot, A. Fluorometric determination of ammonia in sea and estuarine waters by direct segmented flow analysis. Mar. Chem 1997, 57, 265–275. [Google Scholar]
- Oliveira, SM; Lopes, TIMS; Toth, IV; Rangel, AOSS. A multi-commuted flow injection system with a multi-channel propulsion unit placed before detection: Spectrophotometric determination of ammonium. Anal. Chim. Acta 2007, 600, 29–34. [Google Scholar]
- Staden, JFV; Taljaard, RE. Determination of ammonia in water and industrial effluent streams with the indophenol blue method using sequential injection analysis. Anal. Chim. Acta 1997, 344, 281–289. [Google Scholar]
- Mori, M; Tanaka, K; Helaleh, MIH; Xu, Q; Ikedo, M; Ogura, Y; Sato, S; Hu, W; Hasebe, K. Selective determination of ammonium ios by high-speed ion-exclusion chromatography on a weakly baic anion-exchange resin column. J. Chromatogr. A 2003, 997, 191–197. [Google Scholar]
- Kazanskaya, N; Kukhtin, A; Manenkova, M. FET-based sensors with robust photosensitive polymer membranes for determination of ammonium ion and urea. Biosens. Bioelectron 1996, 11, 253–261. [Google Scholar]
- Senillou, A; Renault, NJ; Griffe, CMF. A miniaturized ammonium sensor based on the integration of both ammonium and reference FETs in a sungle chip. Mater. Sci. Eng. C 1998, 6, 59–63. [Google Scholar]
- Dubas, ST; Pimpan, V. Green synthesis of silver nanoparticles for ammonia sensing. Talanta 2008, 76, 29–33. [Google Scholar]
- Li, PQ; Zhang, JZ; Millero, FJ; Hansell, DA. Continuous colorimetric determination of trace ammonium in seawater with a long-path liquid waveguide capillary cell. Mar. Chem 2005, 107, 73–85. [Google Scholar]
- Abass, AK; Hart, JP; Cowell, DC; Chappell, A. Development of an amperometric assay for NH4+ based on a chemically modified screen-printed NADH sensor. Anal. Chim. Acta 1998, 373, 1–8. [Google Scholar]
- Bertocchi, P; Compagnone, D. Amperometric ammonium ion and urea determination with enzyme-based probes. Biosens. Bioelectron 1996, 11, 1–10. [Google Scholar]
- Kwan, RCH; Hon, PYT; Renneberg, R. Amperometric determination of ammonium with bienzyme/poly(carbamoyl) sulfonate hydrogel-based biosensor. Sens. Actuat. B 2005, 107, 616–622. [Google Scholar]
- Agren, D; Stehr, M; Berthold, CL; Kapoor, S; Oehlmann, W; Singh, M; Schneider, G. Three-dimensional structures of apo- and hoho-L-alanine dehydrogenase from mycobacterium tuberculosis reveal conformational changes upon coenzyme binding. J. Mol. Biol 2008, 377, 1161–1173. [Google Scholar]
- Hashimoto, SI; Katsumata, R. l-alanine fermentation by an alanine racemase-deficient mutant of the dl-alanine hyperproducing bacterium Arthrobacter oxydans HAP-1. J. Ferment. Bioeng 1998, 86, 385–390. [Google Scholar]
- Kato, SI; Ohshima, T; Galkin, A; Kulakova, L; Yoshimura, T; Esaki, N. Purification and characterization of alanine dehydrogenase from a marine bacterium, vibrio proteolyticus. J. Mol. Catal. Part B 2003, 23, 373–378. [Google Scholar]
- Ohashima, T; Soda, K. Purification and properties of alanine dehydrogenase from Bacillus spharicus. Eur. J. Biochem 1979, 100, 29–39. [Google Scholar]
- Nassef, HM; Radi, AE; Sullivan, CKO. Electrocatalytic sensing of NADH on a glassy carbon electrode modified with electrografted o-aminophenol film. Electrochem. Commun 2006, 8, 1719–1725. [Google Scholar]
- Behera, S; Retna, CR. Mediatorless voltammetric oxidation of NADH and sensing of ethanol. Biosens. Bioelectron 2007, 21, 949–956. [Google Scholar]
- Retna, CR; Behera, S. Self-assembled monolayers of thio-substituted nucleobases on gold electrode for the electroanalysis of NADH, ethanol and uric acid. Sens. Actuat. B 2005, 128, 31–38. [Google Scholar]
- Rao, TN; Yagi, I; Miwa, T; Tryk, DA; Fujishima, A. Electrochemical oxidation of NADH at highly boron-doped diamond electrodes. Anal. Chem 1999, 71, 2506–2511. [Google Scholar]
- Schroder, I; Vadas, A; Johnson, E; Lim, S; Monbouquette, HG. A novel archeal alanine dehydrogenase homologous to ornithine cyclodeaminase and μ-crystalin. J. Bacteriol 2004, 186, 7680–7689. [Google Scholar]
- Carr, PW; Bowers, LD. Immobilised Enzymes in Analytical and Clinical Chemistry; John Wiley & Sons: New York, NY, USA, 1980. [Google Scholar]
- Kwan, RCH; Hon, PYT; Renneberg, R. Amperometric biosensor for rapid determination of alanine. Anal. Chim. Acta 2004, 523, 81–88. [Google Scholar]
- Renkema, JMS; Gruppen, H; Vliet, TV. Influence of pH and ionic strength on heat-induced formation and rheological properties of soy protein gels in relation to denaturation and their protein compositions. J. Agric. Food Chem 2002, 50, 6064–6071. [Google Scholar]
- Carville, M; Robinson, H. Leachate treatment. Available online: http://www.leachate-treatment.com (accessed on 14 July 2011).
- Nur Ellina, A; Jaafar, A; Musa, A; Heng, LY; Hamidah, S; Nadarajah, K. Biosensor based on glutamate dehydrogenase immobilized in chitosan for the determination of ammonium in water samples. Anal. Biochem 2009, 388, 28–32. [Google Scholar]


| Time (day) | Current Difference (×10−7 A) | Standard deviation | Relative response (%) |
|---|---|---|---|
| 1 | 0.9541 | 0.0695 | 100.0 |
| 4 | 0.6354 | 0.0555 | 66.6 |
| 19 | 0.5754 | 0.0062 | 60.3 |
| 23 | 0.5099 | 0.0418 | 53.4 |
| 26 | 0.4697 | 0.0059 | 49.2 |
| 30 | 0.4490 | 0.0378 | 47.1 |
| Interferent | Current difference (×10−6 A) at different molar ratios of interferent: NH4+ ion | ||||
|---|---|---|---|---|---|
| 0.00:1 | 0.01:1 | 0.10:1 | 0.50:1 | 1.00:1 | |
| Na+ | 0.887 ± 0.013 | 0.850 ± 0.033 | 0.843 ± 0.016 | 0.831 ± 0.036 | 0.844 ± 0.039 |
| K+ | 0.887 ± 0.013 | 0.863 ± 0.113 | 0.866 ± 0.086 | 0.901 ± 0.049 | 0.797 ± 0.036 * |
| CH3NH2 | 0.887 ± 0.013 | 0.877 ± 0.033 | 1.013 ± 0.001 * | 1.005 ± 0.007 * | 1.778 ± 0.061 * |
| C2H5NH2 | 0.887 ± 0.013 | 0.851 ± 0.042 | 0.825 ± 0.024 * | 1.114 ± 0.017 * | 1.599 ± 0.110 * |
*t value > t critical value at 95% confident level with 4 degrees of freedom.
| Sample | Concentration of NH4+ ion spiked in each sample (mM) | Concentration of NH4+ ion determined by biosensor (n = 3), (mM) | Concentration of NH4+ ion by Nessler method (n = 3), mM | Recovery (%) |
|---|---|---|---|---|
| Sample 1 | 10.00 | 10.29 ± 0.95 | 10.41 ± 0.79 | 102.9 |
| Sample 2 | 18.00 | 18.63 ± 1.05 | 18.20 ± 0.91 | 103.5 |
| Sample 3 | 22.00 | 21.56 ± 1.52 | 21.90 ± 0.76 | 98.0 |
| Sample 4 | 26.00 | 26.13 ± 0.32 | 26.10 ± 0.15 | 100.5 |
| Sample 5 | 30.00 | 29.01 ± 2.33 | 30.38 ± 1.20 | 96.7 |
| Parameters | Present study | Abass et al. [17] | Bertocchi et al. [18] | Kwan et al. [19] |
|---|---|---|---|---|
| Enzyme | AlaDH | GLDH | GLDH | GLDH & glutamate oxidase |
| Dynamic linear range (mM) | 10–100 | 0.0017–0.25 | 0.01–0.3 | 0.01–0.30 |
| Detection limit (mM) | 0.18 | 0.0017 | 0.01 | 0.002 |
| Response time (min) | <3 | – | – | 4 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tan, L.L.; Musa, A.; Lee, Y.H. Determination of Ammonium Ion Using a Reagentless Amperometric Biosensor Based on Immobilized Alanine Dehydrogenase. Sensors 2011, 11, 9344-9360. https://doi.org/10.3390/s111009344
Tan LL, Musa A, Lee YH. Determination of Ammonium Ion Using a Reagentless Amperometric Biosensor Based on Immobilized Alanine Dehydrogenase. Sensors. 2011; 11(10):9344-9360. https://doi.org/10.3390/s111009344
Chicago/Turabian StyleTan, Ling Ling, Ahmad Musa, and Yook Heng Lee. 2011. "Determination of Ammonium Ion Using a Reagentless Amperometric Biosensor Based on Immobilized Alanine Dehydrogenase" Sensors 11, no. 10: 9344-9360. https://doi.org/10.3390/s111009344




