Detection of Non-Amplified Mycobacterium tuberculosis Genomic DNA Using Piezoelectric DNA-Based Biosensors
Abstract
:1. Introduction
2. Results and Discussion
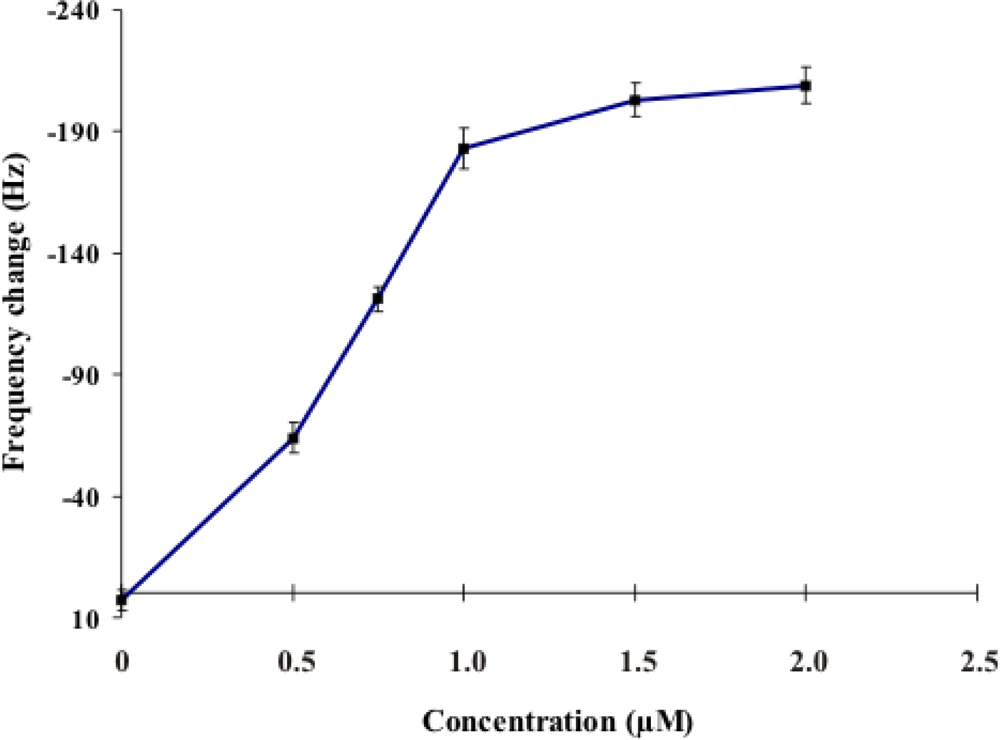
2.1. Investigation of the Optimum Concentration of the DNA Probe for the Performance of the Sensor
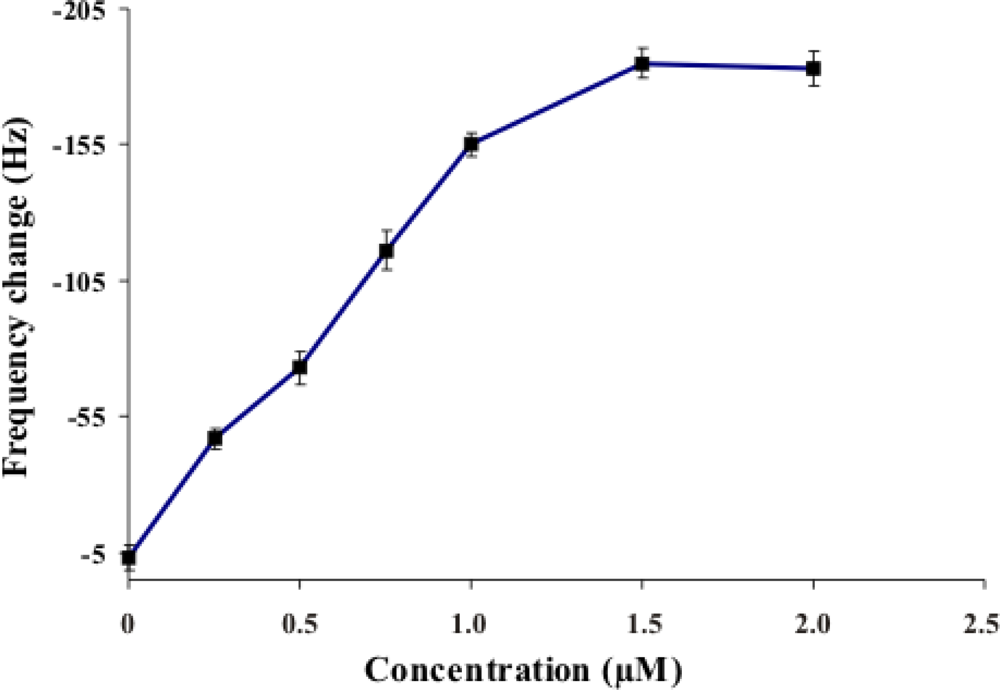
2.2. Investigation of the Optimum Concentration of DNA target for Hybridization
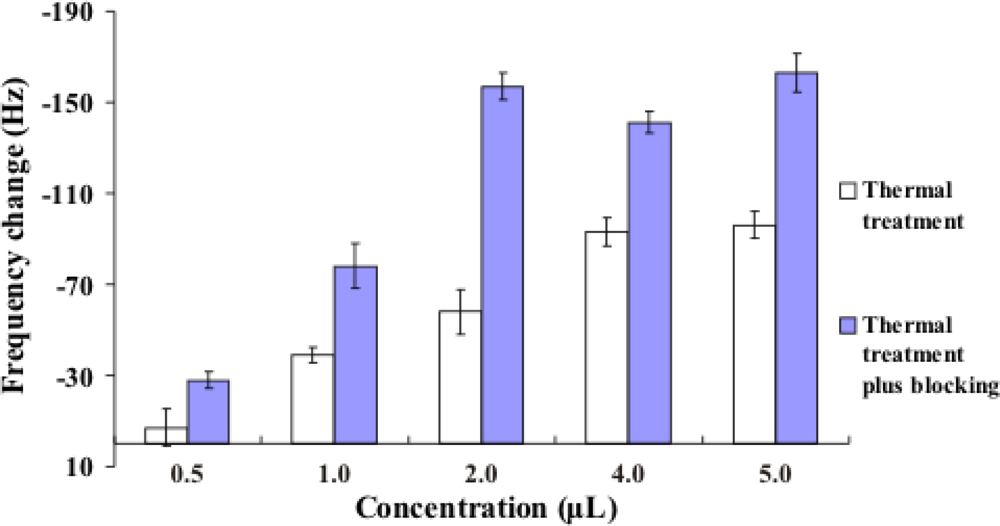
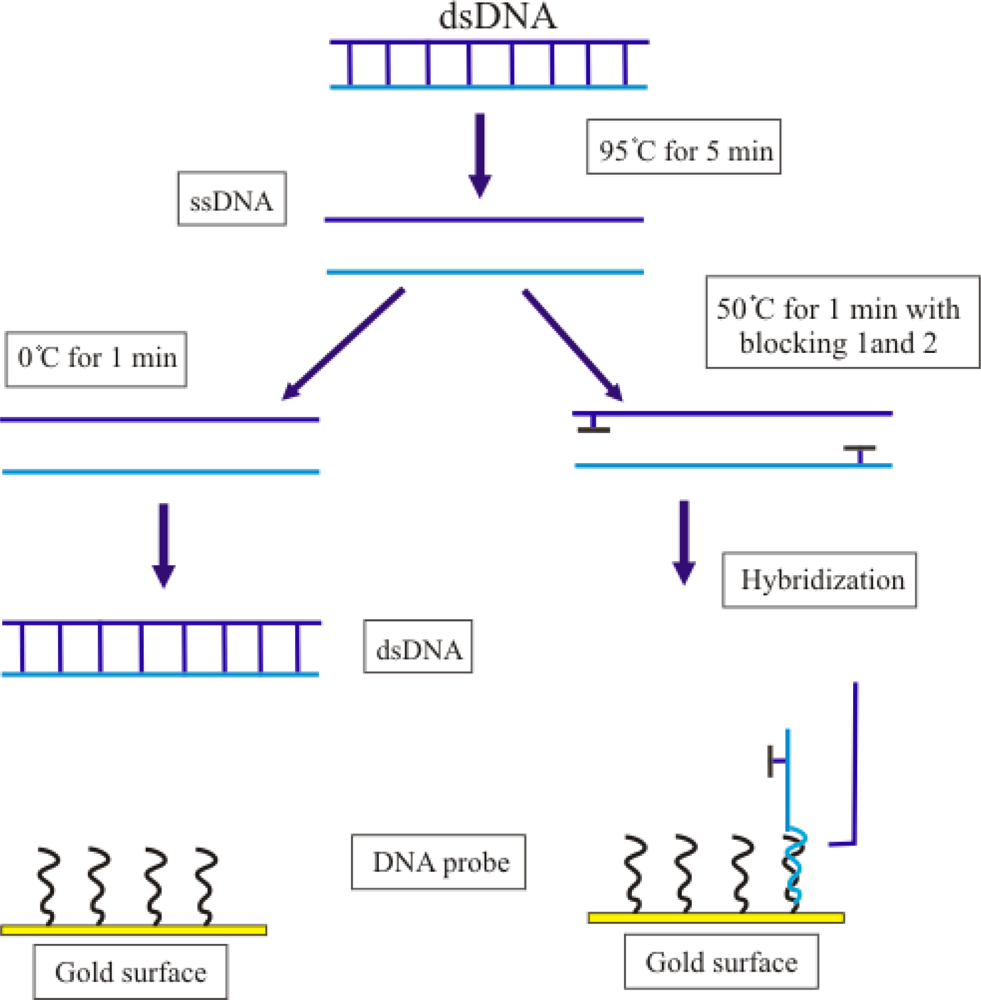
2.3. Improvement in the Hybridization of Bacterial Target DNA
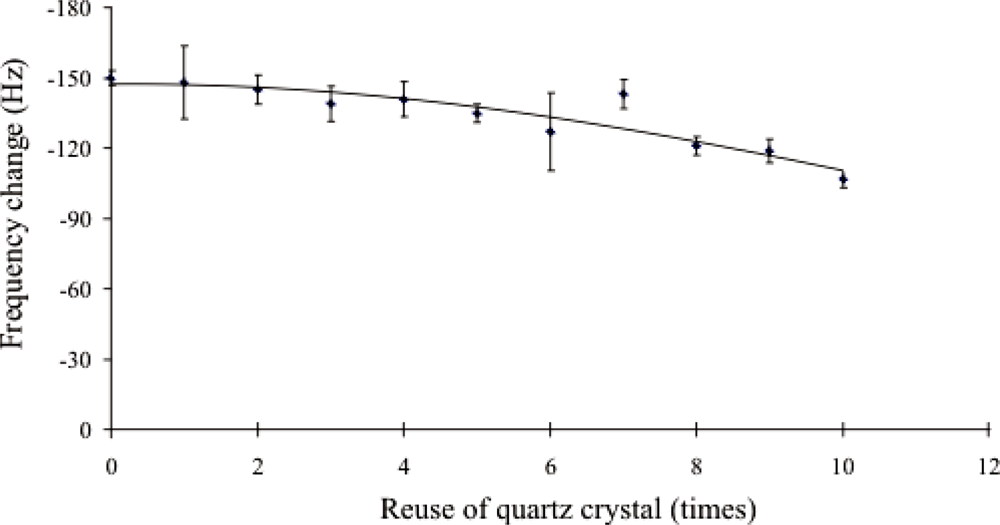
2.4. Evaluation of Piezoelectric DNA-Based Biosensor System
2.5. Bacterial DNA Samples Analysis and Comparison
3. Experimental Section
3.1. Reagents and Oligonucleotides
3.2. Apparatus
3.3. Samples
3.4. Extraction, Fragmentation of Genomic DNA and PCR Amplification
3.5. Preparation of Piezoelectric DNA-Based Biosensor
3.6. DNA Target Hybridization
3.7. Improvement of Hybridization of Bacterial Target DNA
3.8. Bacterial DNA Samples Analysis and Comparison
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
References and Notes
- Tortoli, E.; Lavinia, F.; Simonetti, M.T. Evaluation of a commercial ligase chain reaction kit (Abbott LCx) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. J. Clin. Microbiol 1997, 35, 2424–2426. [Google Scholar]
- Davies, A.P.; Newport, L.E.; Billington, O.J.; Gillespie, S.H. Length of time to laboratory diagnosis of Mycobacterium tuberculosis infection: comparison of in-house methods with reference laboratory results. J. Infect 1999, 39, 205–208. [Google Scholar]
- Mahaisavariya, P.; Chaiprasert, A.; Manonukul, J.; Khemngern, S.; Tingtoy, N. Detection and identification of Mycobacterium species by polymerase chain reaction (PCR) from paraffin-embedded tissue compare to AFB staining in pathological sections. J. Med. Assoc. Thai 2005, 88, 108–113. [Google Scholar]
- Moore, D.F.; Curry, J.I. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by ligase chain reaction. J. Clin. Microbiol 1998, 36, 1028–1031. [Google Scholar]
- Shah, D.H.; Rishendra, V.; Bakshi, C.S.; Singh, R.K. A multiplex-PCR for the differentiation of Mycobacterium bovis and Mycobacterium tuberculosis. FEMS. Microbiol. Lett 2002, 214, 39–43. [Google Scholar]
- Singh, K.K.; Muralidhar, M.; Kumar, A.; Chattopadhyaya, T.K.; Kapila, K.; Singh, M.K.; Sharma, S.K.; Jain, N.K.; Tyagi, J.S. Comparison of in house polymerase chain reaction with conventional techniques for the detection of Mycobacterium tuberculosis DNA in granulomatous lymphadenopathy. J. Clin. Pathol 2000, 53, 355–361. [Google Scholar]
- Tombelli, S.; Mascini, M.; Sacco, C.; Turner, A.P.F. A DNA piezoelectric biosensor assay coupled with a polymerase chain reaction for bacterial toxicity determination in environmental samples. Anal. Chim. Acta 2000, 418, 1–9. [Google Scholar]
- Minnuni, M.; Tombelli, S.; Fonti, J.; Spiriti, M.M.; Mascini, M.; Bogani, P.; Buiatti, M. Detection of fragmented genomic DNA by PCR-free piezoelectric sensing using a denaturation approach. J. Am. Chem. Soc 2005, 127, 7966–7967. [Google Scholar]
- Yao, C.; Zhu, T.; Tang, J.; Wu, R.; Chen, Q.; Chen, M.; Zhang, B.; Huang, J.; Fu, W. Hybridization assay of hepatitis B virus by QCM peptide nucleic acid biosensor. Biosens. Bioelectron 2008, 23, 879–885. [Google Scholar]
- Zhou, X.C.; Huang, L.Q.; Li, S.F. Microgravimetric DNA sensor based on quartz crystal microbalance: comparison of oligonucleotide immobilization methods and the application in genetic diagnosis. Biosens. Bioelectron 2001, 16, 85–95. [Google Scholar]
- Zhang, B.; Mao, Q.; Zhang, X.; Jiang, T.; Chen, M.; Yu, F.; Fu, W. A novel piezoelectric quartz micro-array immunosensor based on self-assembled monolayer for determination of human chorionic gonadotropin. Biosens. Bioelectron 2004, 19, 711–720. [Google Scholar]
- Lee, J.H.; Hwang, K.S.; Park, J.; Yoon, K.H.; Yoon, D.S.; Kim, T.S. Immunoassay of prostate-specific antigen (PSA) using resonant frequency shift of piezoelectric nanomechanical microcantilever. Biosens. Bioelectron 2005, 20, 2157–2162. [Google Scholar]
- Wang, H.; Zeng, H.; Liu, Z.; Yang, Y.; Deng, T.; Shen, G.; Yu, R. Immunophenotyping of acute leukemia using an integrated piezoelectric immunosensor array. Anal. Chem 2004, 76, 2203–2209. [Google Scholar]
- Junhui, Z.; Hong, C.; Ruifu, Y. DNA based biosensors. Biotechnol. Adv 1997, 15, 43–58. [Google Scholar]
- Mo, X.T.; Zhou, Y.P.; Lei, H.; Deng, L. Microbalance-DNA probe method for the detection of specific bacteria in water. Enz. Microbial. Techno 2002, 30, 583–589. [Google Scholar]
- He, F.; Liu, S. Detection of P. aeruginosa using nano-structured electrode separated piezoelectric DNA biosensor. Talanta 2004, 62, 271–277. [Google Scholar]
- Wu, V.C.; Chen, S.H.; Lin, C.S. Real-time detection of Escherichia coli O157:H7 sequences using a circulating-flow system of quartz crystal microbalance. Biosens. Bioelectron 2007, 22, 2967–2975. [Google Scholar]
- Sauerbrey, G. The use of oscillators for weighing thin layer and for microweighing. Z. Phys 1959, 155, 206–212. [Google Scholar]
- Minunni, M.; Mannelli, I.; Spiriti, M.M.; Tombelli, S.; Mascini, M. Detection of highly repeated sequences in non-amplified genomic DNA by bulk acoustic wave (BAW) affinity biosensor. Anal. Chim. Acta 2004, 526, 19–25. [Google Scholar]
- Sekar, B.; Selvaraj, L.; Alexis, A.; Ravi, S.; Arunagiri, K.; Rathinavel, L. The utility of IS6110 sequence based polymerase chain reaction in comparison to conventional methods in the diagnosis of extra-pulmonary tuberculosis. Indian. J. Med. Microbiol 2008, 26, 352–355. [Google Scholar]
- Kent, L.; McHugh, T.D.; Billington, O.; Dale, J.W.; Gillespie, S.H. Demonstration of homology between IS6110 of Mycobacterium tuberculosis and DNAs of other Mycobacterium spp. J. Clin. Microbiol 1995, 33, 2290–2293. [Google Scholar]
- Cowan, S.T.; Mosher, L.; Diem, L.; Massey, J.P.; Crawford, J.T. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol 2002, 40, 1592–1602. [Google Scholar]

| Number of samples | |||
|---|---|---|---|
| Method | Positive | Negative | Total |
| PCR technique | 150 | 50 | 200 |
| Piezoelectric biosensor | 150 | 50 | 200 |
| Name | Nucleotide sequence 5′ to 3′ |
|---|---|
| Thiol-modified oligonucleotide probe | SH-(CH2)6-TTTTTTGTGGCCATCGTGGAAGCGA |
| Blocking 1 | CCTGCGAGCGTAGGCGTCGG |
| Blocking 2 | ATCGTGGTCCTGCGGGCTTT |
| Synthetic DNA target | TCGCTTCCACGATGGCCAC |
| Primer FMTB | AAAGCCCGCAGGACCACGAT |
| Primer RMTB | GTGGCCATCGTGGAAGCGA |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kaewphinit, T.; Santiwatanakul, S.; Promptmas, C.; Chansiri, K. Detection of Non-Amplified Mycobacterium tuberculosis Genomic DNA Using Piezoelectric DNA-Based Biosensors. Sensors 2010, 10, 1846-1858. https://doi.org/10.3390/s100301846
Kaewphinit T, Santiwatanakul S, Promptmas C, Chansiri K. Detection of Non-Amplified Mycobacterium tuberculosis Genomic DNA Using Piezoelectric DNA-Based Biosensors. Sensors. 2010; 10(3):1846-1858. https://doi.org/10.3390/s100301846
Chicago/Turabian StyleKaewphinit, Thongchai, Somchai Santiwatanakul, Chamras Promptmas, and Kosum Chansiri. 2010. "Detection of Non-Amplified Mycobacterium tuberculosis Genomic DNA Using Piezoelectric DNA-Based Biosensors" Sensors 10, no. 3: 1846-1858. https://doi.org/10.3390/s100301846




