Study on a Luminol-based Electrochemiluminescent Sensor for Label-Free DNA Sensing
Abstract
:1. Introduction
2. Experimental Section
2.1. Apparatus
2.2. Chemicals
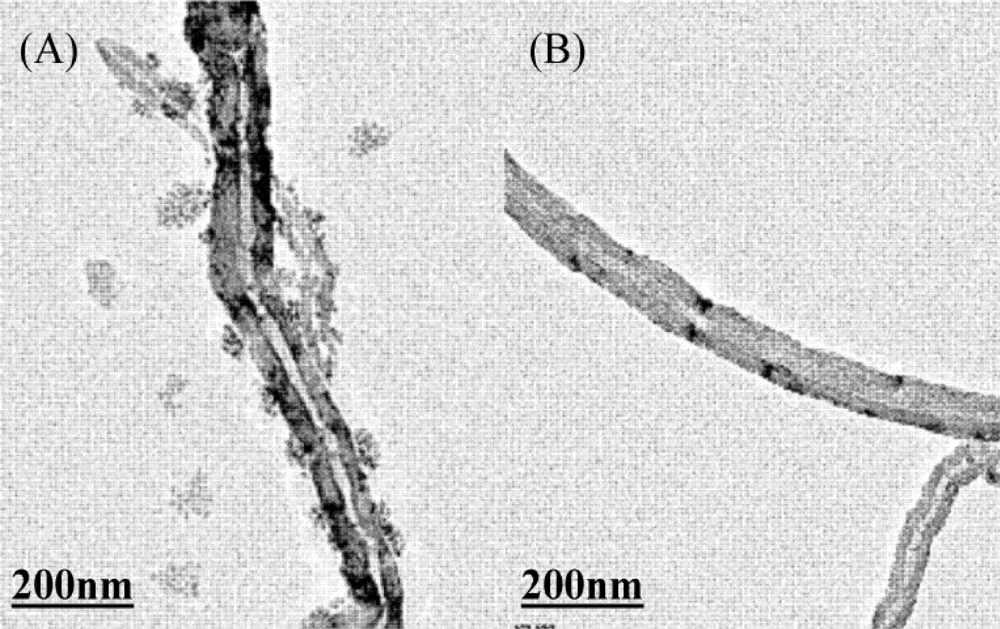
2.3. Preparation of DNA ECL Sensor
2.4. General Procedure
3. Results and Discussion
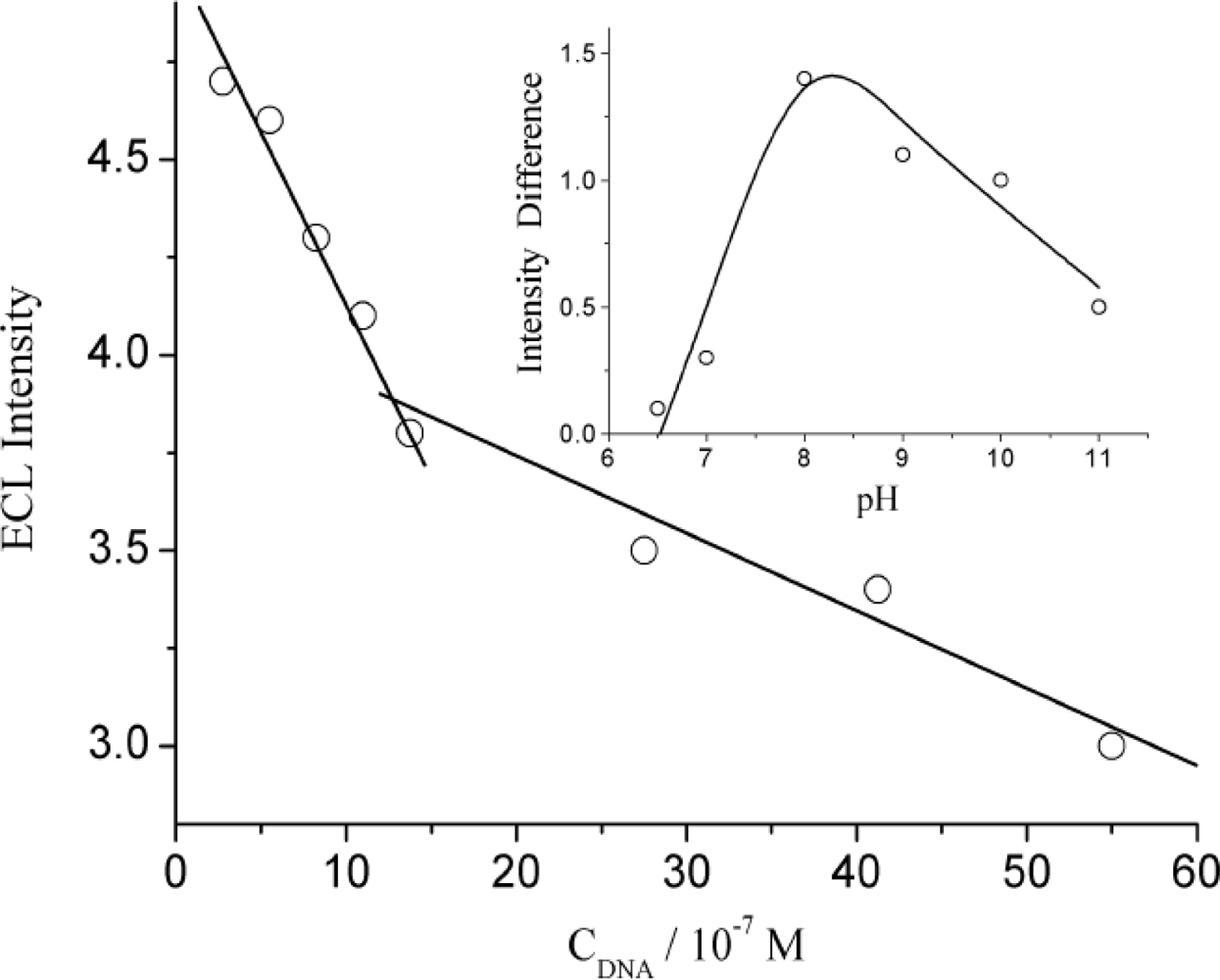
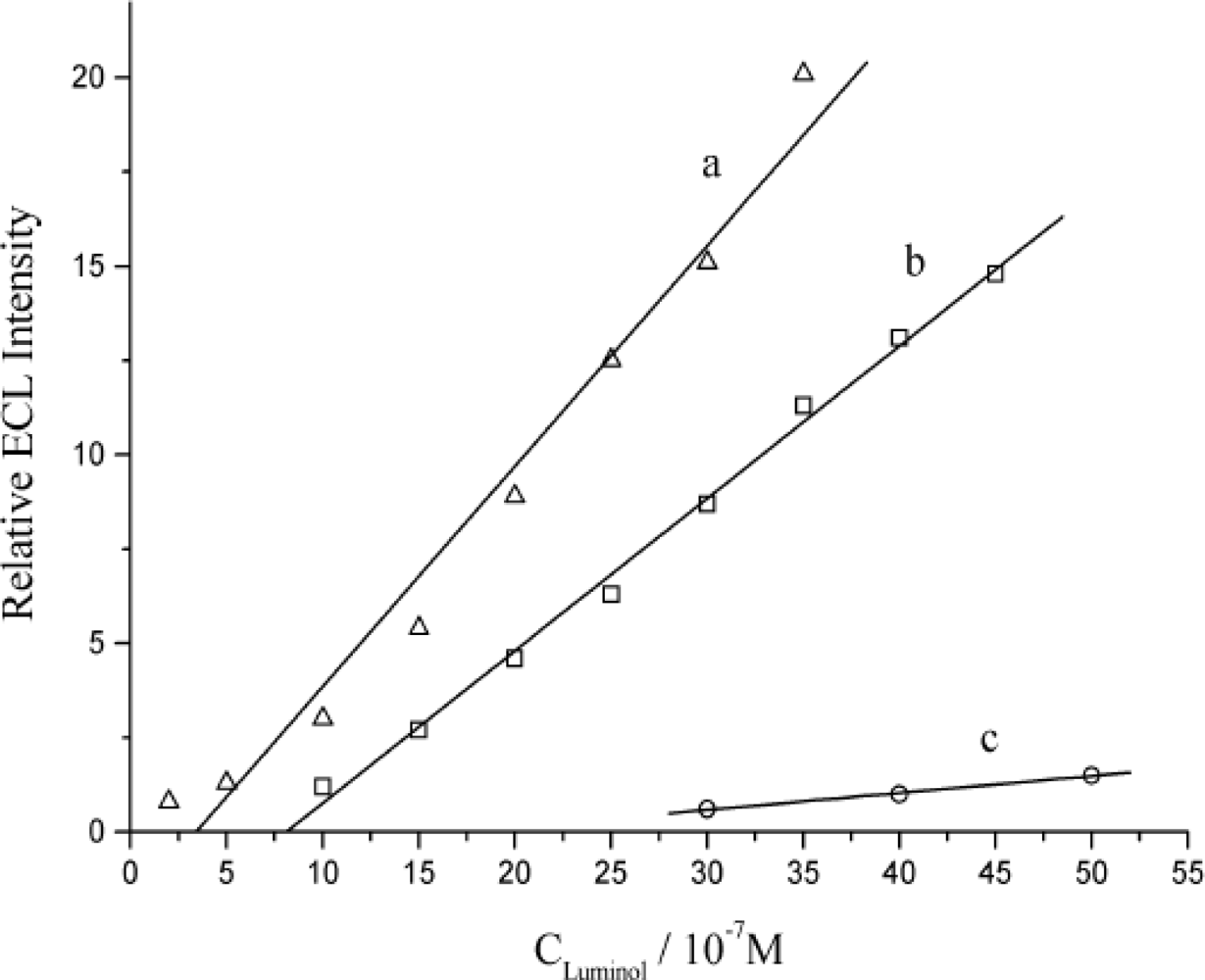
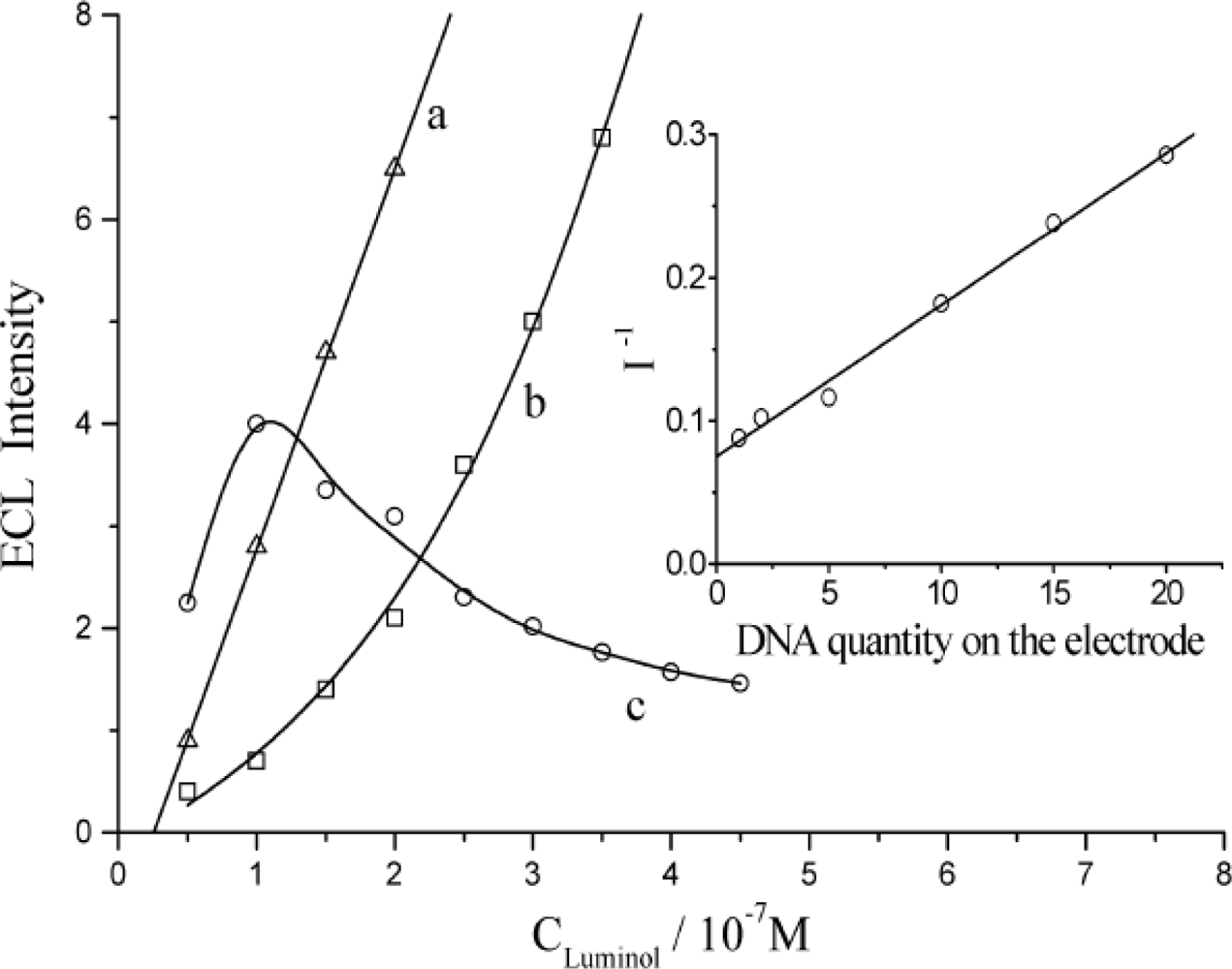
3.1. The Quenching Effect from DNA on ECL of Luminol in Solution
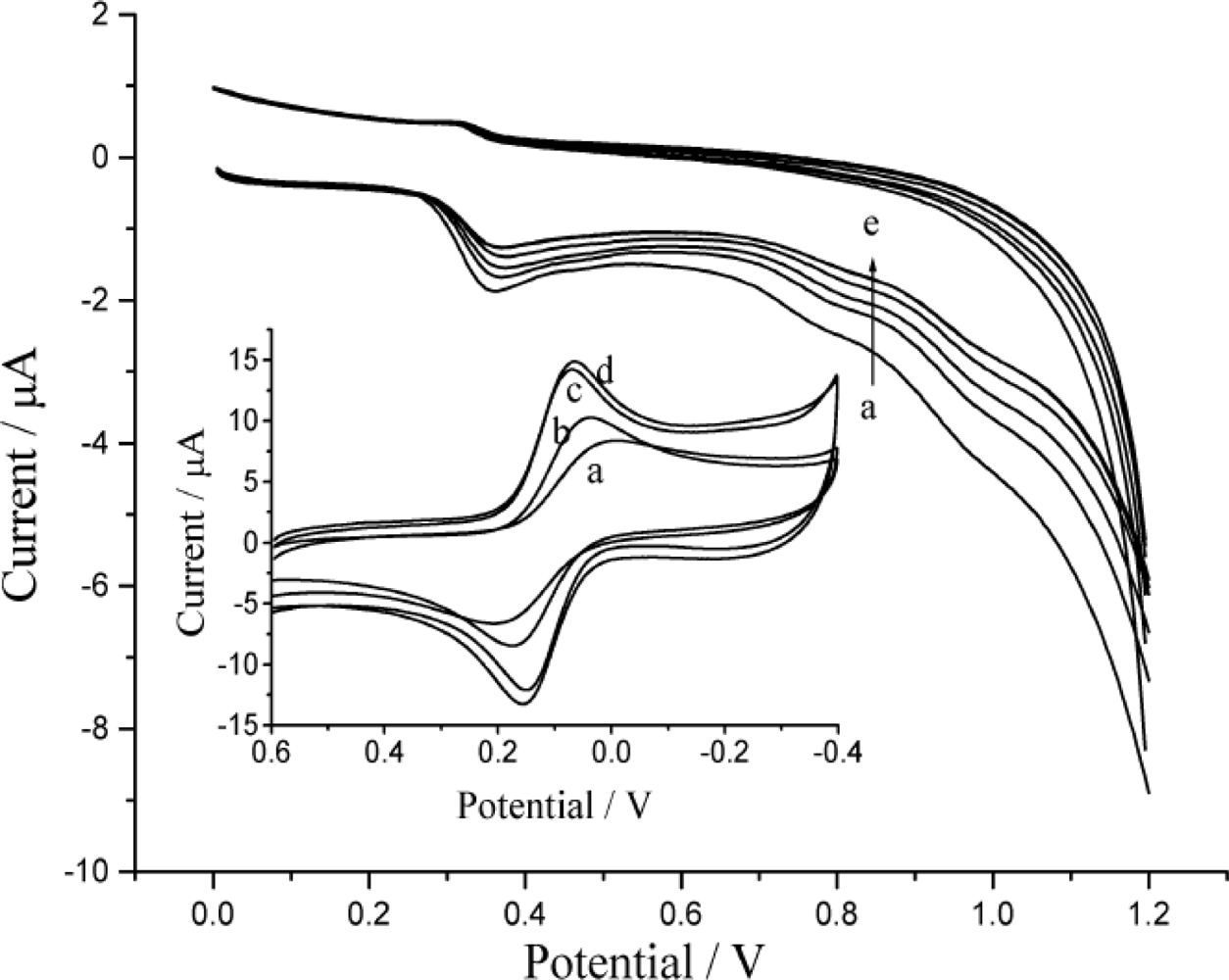
3.2. The Electrochemical Behavior of DNA Modified Electrode
3.3. The Quenching Efficiency of Immobilized DNA for ECL of Luminol
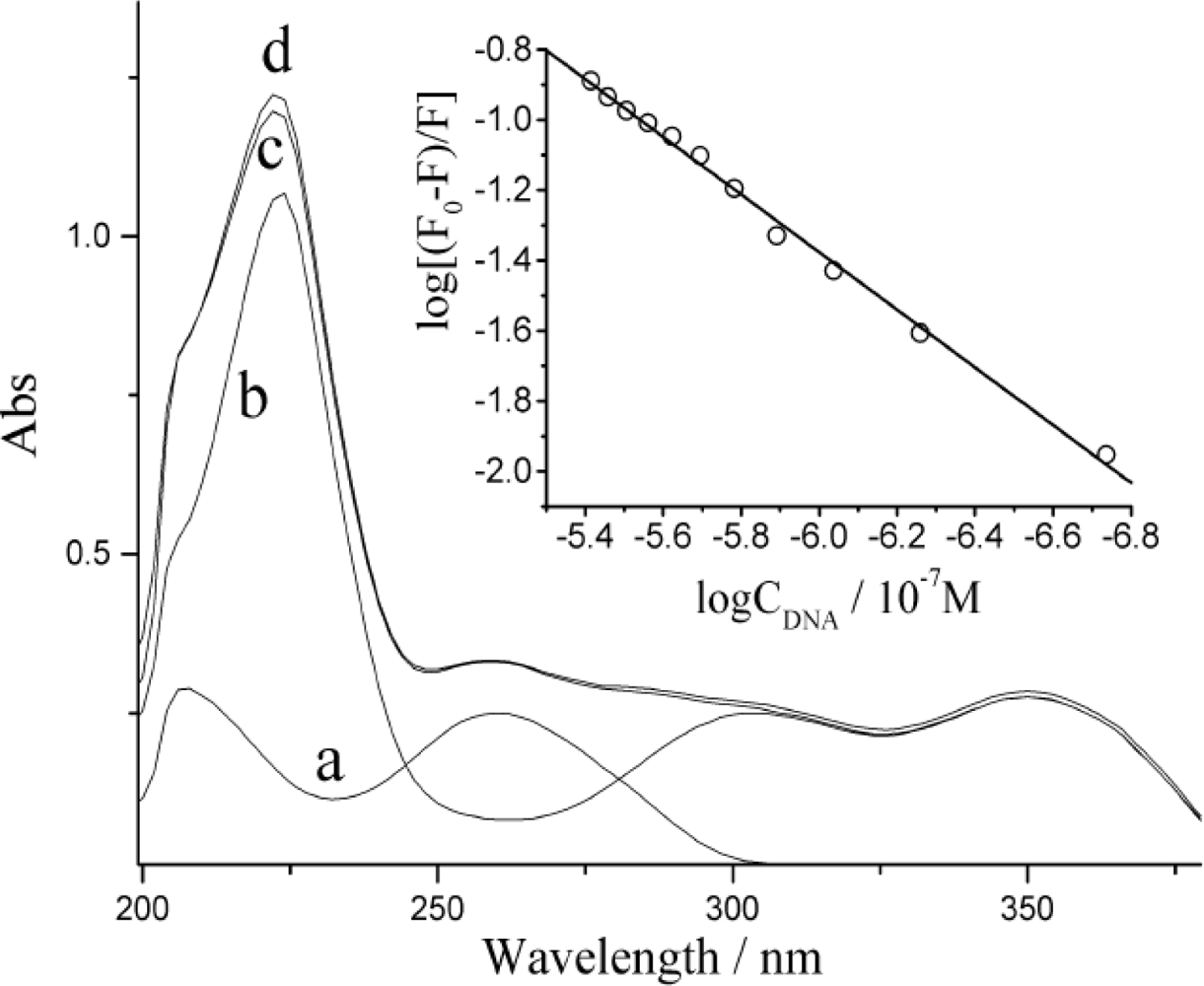
3.4. Discussions on Quenching Mechanism of DNA on ECL Response
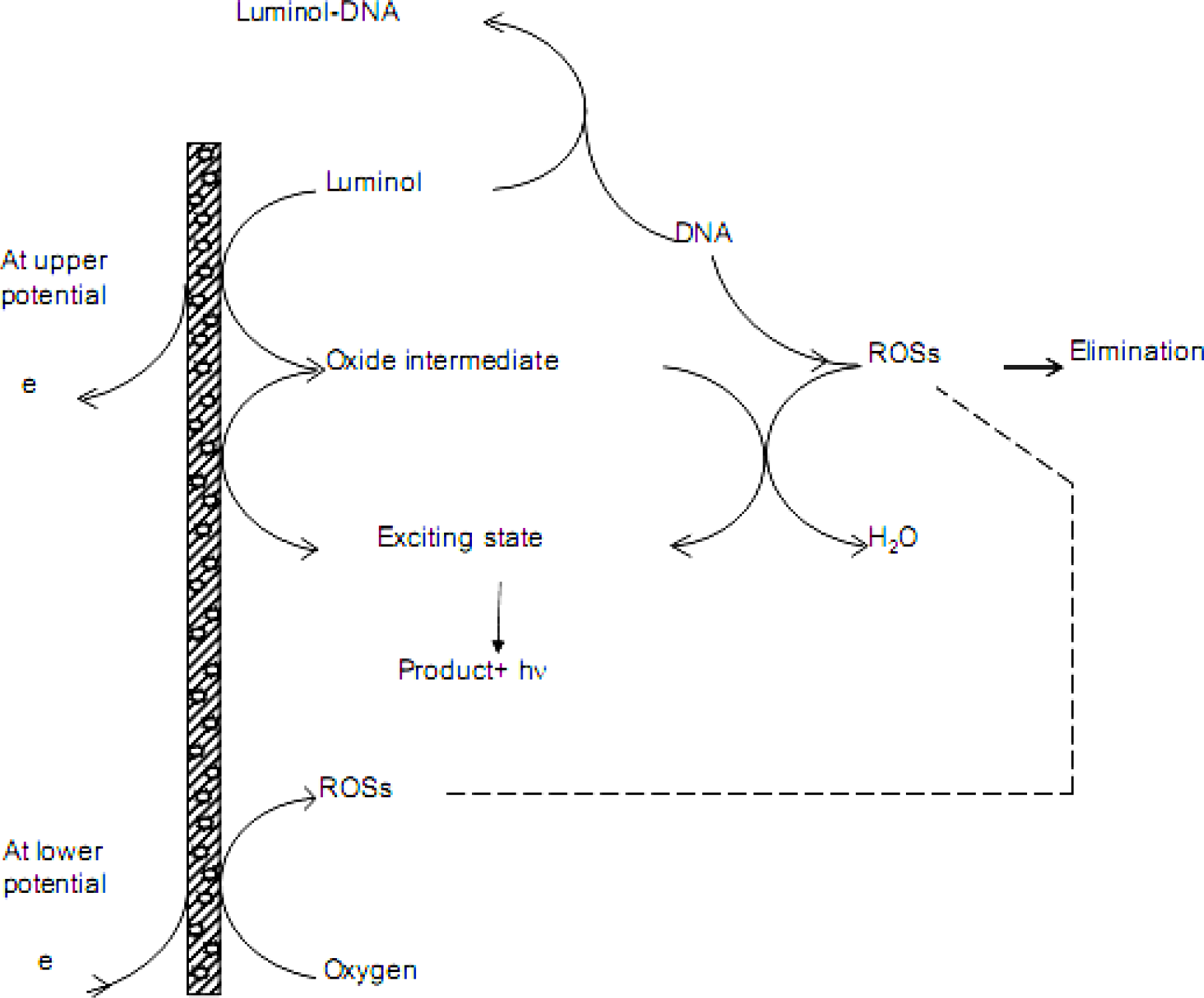
4. Conclusions
Acknowledgments
References
- Qiu, B; Guo, LH; Wang, W; Chen, GN. Synthesis of a Novel Fluorescent Probe Useful for DNA Detection. Biosens. Bioelectron 2007, 22, 2629–2635. [Google Scholar]
- Jiang, X; Shang, L; Wang, ZX; Dong, SJ. Spectrometric and Voltammetric Investigation of Interaction of Neutral Red with Calf Thymus DNA: pH Effect. Biophys. Chem 2005, 118, 42–50. [Google Scholar]
- Palecek, E; Fojta, M; Tomschik, M; Wang, J. Electrochemical Biosensors for DNA Hybridization and DNA Damage. Biosens. Bioelectron 1998, 13, 621–628. [Google Scholar]
- Zhou, YL; Li, YZ. Studies of the Interaction between Poly (Diallyldimethyl Ammonium Chloride) and DNA by Spectroscopic Methods. Colloid. Surf 2004, A233, 129–135. [Google Scholar]
- Andrew, WK; Gillion, MG. Occurrence, Mechanisms and Analytical Applications of Electrogenerated Chemiluminescence. A Review. Analyst 1994, 119, 879–890. [Google Scholar]
- Steven, AS; Isiah, MW; Linda, BM. Molecular Fluorescence, Phosphorescence, and Chemiluminescence Spectrometry. Anal. Chem 1998, 70, 477–494. [Google Scholar]
- Fabian, J; Rosa, IS; Dong, L; Jonathan, KL; David, Y; York, D; Mark, TM. Electrochemiluminescence-Based Quantitation of Classical Clinical Chemistry Analytes. Anal. Chem 1996, 68, 1298–1302. [Google Scholar]
- Zhang, LL; Zheng, XW. A Novel Electrogenerated Chemiluminescence Sensor for Pyrogallol with Core-Shell Luminol-Doped Silica Nanoparticles Modified Electrode by the Self-Assembled Technique. Anal. Chim. Acta 2006, 570, 207–213. [Google Scholar]
- Miao, WJ. Electrogenerated Chemiluminescence and Its Biorelated Applications. Chem. Rev 2008, 108, 2506–2553. [Google Scholar]
- Zhang, L; Zhou, JM; Hao, YH; He, PG; Fang, YZ. Determination of Co2+ Based on the Cobalt(II)-Catalyzed Electrochemiluminescence of Luminol in Acidic Solution. Electrochim. Acta 2005, 50, 3414–3419. [Google Scholar]
- Sun, XY; He, PG; Liu, SH; Ye, JN; Fang, YZ. Immobilization of Single-Stranded Deoxyribonucleic Acid on Gold Electrode with Self-Assembled Aminoethanethiol Monolayer for DNA Electrochemical Sensor Applications. Talanta 1998, 47, 487–495. [Google Scholar]
- Zhu, NN; Chang, Z; He, PG; Fang, YZ. Electrochemical DNA Biosensors Based on Platinum Nanoparticles Combined Carbon Nanotubes. Anal. Chim. Acta 2005, 545, 21–26. [Google Scholar]
- Huang, RF; Zheng, XW; Qu, JJ. Highly Selective Electrogenerated Chemiluminescence (ECL) for Sulfide Ion Determination at Multi-Wall Carbon Nanotubes-modified Graphite Electrode. Anal. Chim. Acta 2007, 582, 267–274. [Google Scholar]
- Willner, I; Willner, B; Katz, E. Biomolecule-Nanoparticle Hybrid Systems for Bioelectronic Applications. Bioelectrochemistry 2007, 70, 2–11. [Google Scholar]
- Zhang, ZY; Berg, A; Levanon, H; Fessenden, RW; Meisel, D. On the Interactions of Free Radicals with Gold Nanoparticles. Am. Chem. Soc 2003, 125, 7959–7963. [Google Scholar]
- Zhou, XM; Xing, D; Zhu, DB; Jia, L. Magnetic Bead and Nanoparticle Based Electrochemiluminescence Amplification Assay for Direct and Sensitive Measuring of Telomerase Activity. Anal. Chem 2009, 81, 255–261. [Google Scholar]
- Will, G; Kudryashov, E; Duggan, E; Fitzmaurice, D; Buckin, V; Waghorne, E; Mukherjee, S. Excited State Complex Formation between 3-Aminophthalhydrazide and DNA: A Fluorescence Quenching Reaction. Spectro. Acta 1999, 55A, 2711–2717. [Google Scholar]
- Lee, JG; Yun, K; Lim, GS; Lee, SE; Kim, S; Park, J. DNA Biosensor Based on the Electrochemiluminescence of Ru(bpy)32+ with DNA-Binding Intercalators. Bioelectrochemistry 2007, 70, 228–234. [Google Scholar]
- Pérez-Ruiz, T; Martínez-Lozano, C; Tomás, V; Mart, J. Flow-Injection Chemiluminescence Determination of DNA Based on a Photochemical Reaction. Anal. Chim. Acta 1999, 402, 13–19. [Google Scholar]
- Miao, WJ; Bard, A. Electrogenerated Chemiluminescence. 77. DNA Hybridization Detection at High Amplification with [Ru(bpy)3]2+-Containing Microspheres. Anal. Chem 2004, 76, 5379–5386. [Google Scholar]
- Zhu, DB; Tang, YB; Xing, D; Chen, WR. PCR-Free Quantitative Detection of Genetically Modified Organism from Raw Materials. An Electrochemiluminescence-Based Bio Bar Code Method. Anal. Chem 2008, 80, 3566–3571. [Google Scholar]
- Bertoncello, P; Forster, RJ. Nanostructured Materials for Electrochemiluminescence (ECL)—Based Detection Methods: Recent Advances and Future Perspectives. Biosens. Bioelectron 2009, 24, 3191–3200. [Google Scholar]
- Chu, HH; Guo, WY; Di, JW; Wu, Y; Tu, YF. Study on Sensitization from Reactive Oxygen Species for Electrochemiluminescence of Luminol in Neutral Medium. Electroanalysis 2009, 21, 1630–1635. [Google Scholar]
- Yu, HX; Cui, H. Comparative Studies on the Electrochemiluminescence of the Luminol System at a Copper Electrode and a Gold Electrode under Different Transient-State Electrochemical Techniques. J. Electroanal. Chem 2005, 580, 1–8. [Google Scholar]
- Chiorcea, PAM; Diculescu, VC; Oretskaya, TS; Oliveira-Brett, AM. AFM and Electroanalytical Studies of Synthetic Oligonucleotide Hybridization. Biosens. Bioelectron 2004, 20, 933–944. [Google Scholar]
- Piedade, JAP; Oliveira, PSC; Lopes, MC; Oliveira-Brett, AM. Voltammetric Determination of γ Radiation-Induced DNA Damage. Anal. Biochem 2006, 355, 39–49. [Google Scholar]
- Cui, H; Xu, Y; Zhang, ZF. Multichannel Electrochemiluminescence of Luminol in Neutral and Alkaline Aqueous Solutions on a Gold Nanoparticle Self-Assembled Electrode. Anal. Chem 2004, 76, 4002–4010. [Google Scholar]
- Tian, B; Xu, ZJ; Sun, ZT; Lin, J; Hua, YJ. Evaluation of the Antioxidant Effects of Carotenoids from Deinococcus Radiodurans through Targeted Mutagenesis, Chemiluminescence, and DNA Damage Analyses. Biochim. Biophys. Acta 2007, 1770, 902–911. [Google Scholar]
- Chu, HH; Wu, Y; Di, JW; Tu, YF. Determination of SOD by Electrochemiluminescence Quenching Analysis. J. Instru. Anal 2006, 25, 125–126. [Google Scholar]
- Huang, BQ; Guo, WY; Xu, Y; Tu, YF. Study on the Enhancement of Dissolved Oxygen for Electrochemiluminescence of Luminol. Spectros. Spec. Anal 2003, 23, 849–851. [Google Scholar]
- Guo, SJ; Dong, SJ. Biomolecule-Nanoparticle Hybrids for Electrochemical Biosensors. Trends Anal. Chem 2009, 28, 96–109. [Google Scholar]
- Liu, SN; Wu, P; Zhou, B; Zhou, YM; Cai, CX. Immobilization, Characterization of DNA on the Surface of Carbon Nanotube and Electrochemical Detection of DNA Damage. Acta Chimica Sinica 2008, 66, 424–432. [Google Scholar]
- Lim, SH; Wei, J; Lin, JY. Electrochemical Genosensing Properties of Gold Nanoparticle-Carbon Nanotube Hybrid. Chem. Phys. Lett 2004, 400, 578–582. [Google Scholar]
- Ding, DY; Hu, JH. UV Study on the Ways of Interactions of Drugs with DNA-Effects of CDDP, Cp2TiCl2 and DHE on Alkali Denaturation of DNA. Spectros. Spec. Anal 1991, 11, 7–10. [Google Scholar]
- Qi, HL; Zhang, CX. Electrogenerated Chemiluminescence Determination of Procaine Hydrochloride. Chin. J. Anal. Lab 2004, 23, 20–22. [Google Scholar]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chu, H.-H.; Yan, J.-L.; Tu, Y.-F. Study on a Luminol-based Electrochemiluminescent Sensor for Label-Free DNA Sensing. Sensors 2010, 10, 9481-9492. https://doi.org/10.3390/s101009481
Chu H-H, Yan J-L, Tu Y-F. Study on a Luminol-based Electrochemiluminescent Sensor for Label-Free DNA Sensing. Sensors. 2010; 10(10):9481-9492. https://doi.org/10.3390/s101009481
Chicago/Turabian StyleChu, Hai-Hong, Ji-Lin Yan, and Yi-Feng Tu. 2010. "Study on a Luminol-based Electrochemiluminescent Sensor for Label-Free DNA Sensing" Sensors 10, no. 10: 9481-9492. https://doi.org/10.3390/s101009481



