Genetic Diversity Assessment of Portuguese Cultivated Vicia faba L. through IRAP Markers
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Material and DNA Extraction
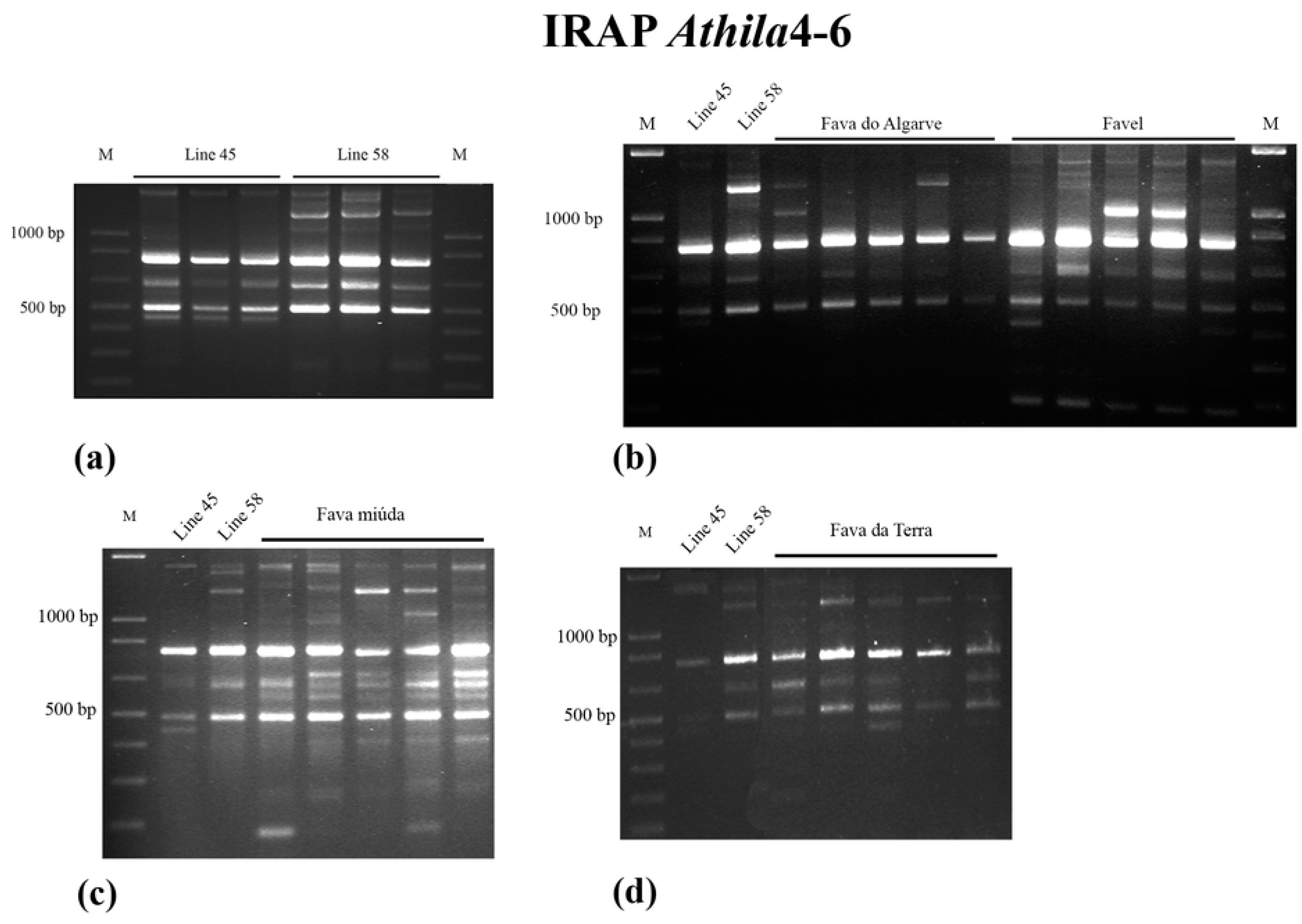
2.2. IRAP
2.3. IRAP Data Analysis
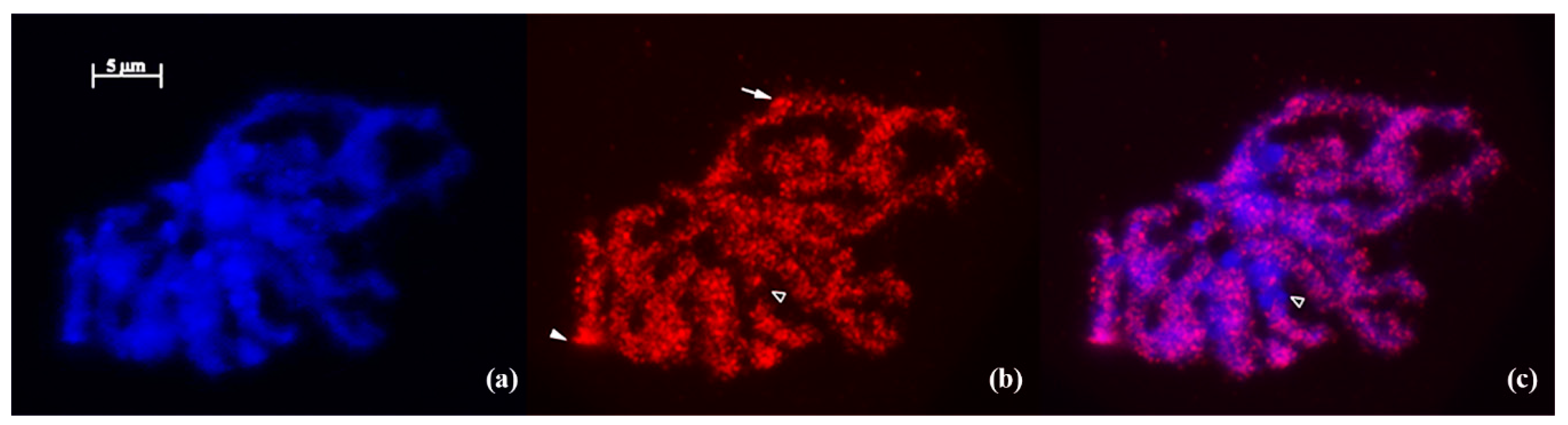
2.4. Fluorescence in Situ Hybridization (FISH)
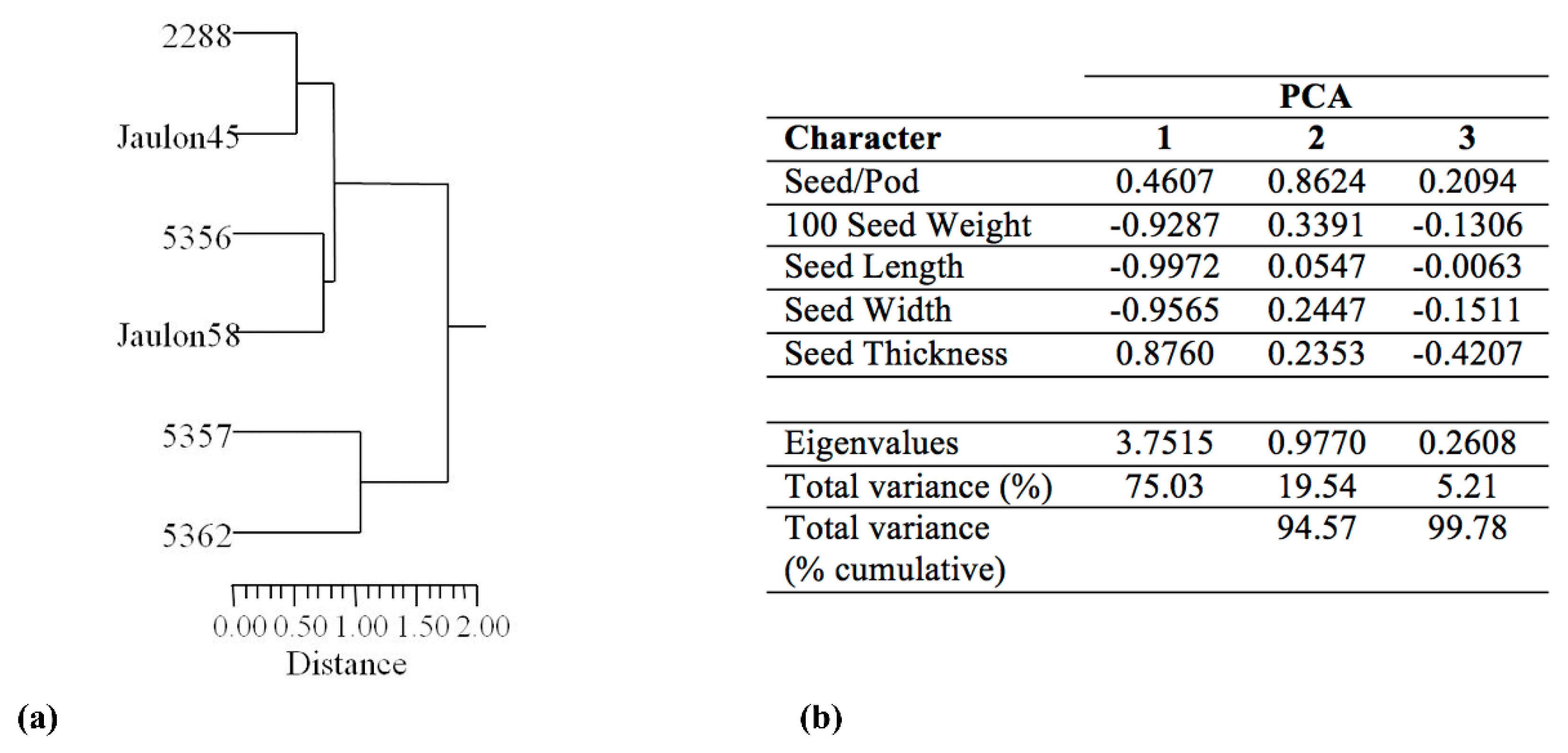
2.5. Morphological Characterization
3. Results and Discussion
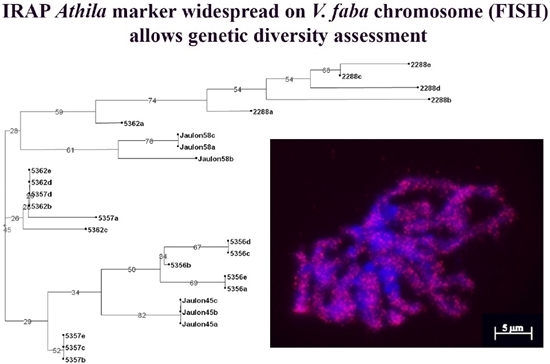
3.1. IRAP Analysis of Genomic Diversity
3.2. Seed Morphological Characterization
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Duc, G.; Bao, S.Y.; Baum, M.; Redden, B.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X.X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crops Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- Suso, M.J.; del Rio, R. A crop-pollinator inter-play approach to assessing seed production patterns in faba bean under two pollination environments. Euphytica 2015, 201, 231–251. [Google Scholar] [CrossRef]
- Cubero, J.I. Evolutionary trends in Vicia faba L. Theor. Appl. Genet. 1973, 43, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Cubero, J.I. On the evolution of Vicia faba L. Theor. Appl. Genet. 1974, 45, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Muratova, V. Common Beans (Vicia faba). Suppl. Bull. Appl. Bot. Genet. Plant Breed 1931, 50, 1–298. [Google Scholar]
- Suso, M.J.; Moreno, M.T.; Cubero, J.I. Variation among spanish faba beans cultivars: Taxonomic and evolutionary implications. Genet. Resour. Crop Evol. 1993, 40, 105–111. [Google Scholar] [CrossRef]
- European Comission. EIP-Focus Group. Available online: http://ec.europa.eu/eip/agriculture/en/content/genetic-resources-cooperation-models (accessed on 2 November 2015).
- Zapata, L.; Peña-Chocarro, L.; Pérez-Jordá, G.; Stika, H.-P. Early neolithic agriculture in the iberian peninsula. J. World Prehist. 2004, 18, 283–325. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest asia, Europe, and the Mediterranean Basin; OUP Oxford: New York, NY, USA, 2012. [Google Scholar]
- Moreira, P.M.R.M.; Veloso, M.M. Landraces inventory for portugal. In European Landraces: On Farm Conservation, Management and Use, 1st ed.; Bioversity Technical Bulletin No. 15; Vetelainen, M., Negri, V., Maxted, N., Eds.; Bioversity International: Rome, Italy, 2009; pp. 124–136. [Google Scholar]
- GPPAA. Crop Production Year Book, 1st ed.; Castel Publicações: Lisbon, Portugal, 2005; p. 252. [Google Scholar]
- INE. Estatísticas Agrícolas 2012, 1st ed.; Instituto Nacional de Estatística, IP: Lisbon, Portugal, 2013; p. 178. (In Portuguese) [Google Scholar]
- GPP. Year Book of Agriculture 2014, 1st ed.; Enigma Previsível: Algés, Portugal, 2014; p. 158. (In Portuguese) [Google Scholar]
- INE. Estatísticas Agrícolas 2013., 1st ed.; Instituto Nacional de Estatística, IP: Lisbon, Portugal, 2014; p. 166. (In Portuguese) [Google Scholar]
- Khoury, C.K. Global Strategy for the Ex Situ Conservation of Faba Bean (Vicia faba L.); Global Crop Diversity Trust: Rome, Italy, 2009; p. 74. [Google Scholar]
- Maggioni, L. Regeneration and Safety Duplication of Regionally Prioritized Crop Collections; Global Crop Diversity Trust Technical and Financial Report 2011; Bioversity International: Rome, Italy, 2011; p. 8. [Google Scholar]
- Pereira, M.G.; Tavares de Sousa, M.M.; Meneses, M.; Suso, M.J.; Matos, C.R.; Veloso, M.M. Regeneration and Safety Duplication of Faba Bean Prioritized Collection. In Proceedings of the 28th International Horticultural Congress, Lisbon, Portugal, 22–27 August 2010; p. 570.
- Vitte, C.; Panaud, O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet. Genome Res. 2005, 110, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Bento, M.; Tomas, D.; Viegas, W.; Silva, M. Retrotransposons represent the most labile fraction for genomic rearrangements in polyploid plant species. Cytogenet. Genome Res. 2013, 140, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.R.; Li, D.; Flavell, A.J.; Harrison, G.; Heslop-Harrison, J.S.; Kumar, A. The Ty1-copia group retrotransposons in Vicia species: Copy number, sequence heterogeneity and chromosomal localisation. Mol. Gen. Genet. 1996, 250, 305–315. [Google Scholar] [PubMed]
- Hill, P.; Burford, D.; Martin, D.M.A.; Flavell, A.J. Retrotransposon populations of Vicia species with varying genome size. Mol. Gen. Genom. 2005, 273, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.H.N.; Poyser, S.J.; Knox, M.R.; Vershinin, A.V.; Ambrose, M.J. Ty1-copia class retrotransposon insertion site polymorphism for linkage and diversity analysis in pea. Mol. Gen. Genet. 1998, 260, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Galasso, I.; Harrison, G.E.; Pignone, D.; Brandes, A.; HeslopHarrison, J.S. The distribution and organization of Ty1-copia-like retrotransposable elements in the genome of Vigna unguiculata (L.) Walp. (cowpea) and its relatives. Ann. Bot. 1997, 80, 327–333. [Google Scholar] [CrossRef]
- Porceddu, A.; Albertini, E.; Barcaccia, G.; Marconi, G.; Bertoli, F.B.; Veronesi, F. Development of S-SAP markers based on an LTR-like sequence from Medicago sativa L. Mol. Gen. Genom. 2002, 267, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.M.; Gonzalez, S.G.; Syed, N.H.; Suso, M.J.; Saldana, C.C.; Flavell, A.J. Genetic diversity analysis in Vicia species using retrotransposon-based SSAP markers. Mol. Genet. Genom. 2007, 278, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Flavell, A.J.; Ellis, T.H.N.; Sjakste, T.; Moisy, C.; Schulman, A.H. Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 2011, 106, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Smykal, P. Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. J. Appl. Genet. 2006, 47, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Smykal, P.; Horacek, J.; Dostalova, R.; Hybl, M. Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J. Appl. Genet. 2008, 49, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.S.; Barao, A.; Delgado, M.; Morais-Cecilio, L.; Viegas, W. Genomic analysis of grapevine retrotransposon 1 (Gret1) in Vitis vinifera. Theor. Appl. Genet. 2005, 111, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.C.; Lopez-Ribera, I.; Machado, M.D.P.D.; Vicient, C.M. Genetic diversity of maize germplasm assessed by retrotransposon-based markers. Electrophoresis 2014, 35, 1921–1927. [Google Scholar] [CrossRef] [PubMed]
- Branco, C.J.S.; Vieira, E.A.; Malone, G.; Kopp, M.M.; Malone, E.; Bernardes, A.; Mistura, C.C.; Carvalho, F.I.F.; Oliveira, C.A. IRAP and REMAP assessments of genetic similarity in rice. J. Appl. Genet. 2007, 48, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Bento, M.; Gustafson, J.P.; Viegas, W.; Silva, M. Size matters in triticeae polyploids: Larger genomes have higher remodeling. Genome 2011, 54, 175–183. [Google Scholar] [PubMed]
- Bento, M.; Gustafson, P.; Viegas, W.; Silva, M. Genome merger: From sequence rearrangements in triticale to their elimination in wheat-rye addition lines. Theor. Appl. Genet. 2010, 121, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Tomás, D.; Rodrigues, J.; Varela, A.; Veloso, M.M.; Viegas, W.; Silva, M. Use of repetitive sequences for molecular and cytogenetic characterization of Avena species from Portugal. Int. J. Mol. Sci. 2016, 17, 203. [Google Scholar] [CrossRef] [PubMed]
- Bento, M.; Tomas, D.; Viegas, W.; Silva, M. Unravelling genome dynamics in Arabidopsis synthetic auto and allopolyploid species. Biol. Plantarum. 2015, 59, 661–670. [Google Scholar] [CrossRef]
- Terzopoulos, P.J.; Kaltsikes, P.J.; Bebeli, P.J. Determining the sources of heterogeneity in greek faba bean local populations. Field Crops Res. 2008, 105, 124–130. [Google Scholar] [CrossRef]
- Doyle, J.J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Kalendar, R.; Grob, T.; Regina, M.; Suoniemi, A.; Schulman, A. IRAP and REMAP: Two new retrotransposon-based DNA fingerprinting techniques. Theor. Appl. Genet. 1999, 98, 704–711. [Google Scholar] [CrossRef]
- Leigh, L.; Kalendar, R.; Lea, V.; Lee, D.; Donini, P.; Schulman, A.H. Comparison of the utility of barley retrotransposon families for genetic analysis by molecular marker techniques. Mol. Gen. Genomics 2003, 269, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jacquemoud-Collet, J.P. Darwin Software, CIRAD: Montpellier, France, 2006.
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tomás, D.; Brazao, J.; Viegas, W.; Silva, M. Differential effects of high-temperature stress on nuclear topology and transcription of repetitive noncoding and coding rye sequences. Cytogenet. Genome Res. 2013, 139, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F.J. Ntsys-pc: Numerical taxonomy and multivariate analysis system, version 2.1—Getting started guide. 2000. [Google Scholar]
- Wright, D.A.; Voytas, D.F. Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 1998, 149, 703–715. [Google Scholar] [PubMed]
- Wright, D.A.; Voytas, D.F. Athila 4 of Arabidopsis and calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 2002, 12, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Bento, M.; Pereira, S.; Rocheta, M.; Gustafson, P.; Viegas, W.; Silva, M. Polyploidization as a retraction force in plant genome evolution: Sequence rearrangements in triticale. PLoS ONE 2008, 3, e1402. [Google Scholar] [CrossRef] [PubMed]
- Nadal, S.; Suso, M.J.; Moreno, M.T. Management of Vicia faba genetic resources: Changes associated to the selfing process in the major, equina and minor groups. Genet. Resour. Crop Evol. 2003, 50, 183–192. [Google Scholar] [CrossRef]
- Gresta, F.; Avola, G.; Albertini, E.; Raggi, L.; Abbate, V. A study of variability in the sicilian faba bean landrace ‘Larga di Leonforte’. Genet. Resour. Crop Evol. 2010, 57, 523–531. [Google Scholar] [CrossRef]
- Ouji, A.; El Bok, S.; Syed, N.H.; Abdellaoui, R.; Rouaissi, M.; Flavell, A.J.; El Gazzah, M. Genetic diversity of faba bean (Vicia faba L.) populations revealed by sequence specific amplified polymorphism (SSAP) markers. Afr. J. Biotechnol. 2012, 11, 2162–2168. [Google Scholar]
- Terzopoulos, P.J.; Bebeli, P.J. Genetic diversity analysis of mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crops Res. 2008, 108, 39–44. [Google Scholar] [CrossRef]
- Kwon, S.J.; Hu, J.G.; Coyne, C.J. Genetic diversity and relationship among faba bean (Vicia faba L.) germplasm entries as revealed by TRAP markers. Plant Genet. Resour.-Charact. Util. 2010, 8, 204–213. [Google Scholar] [CrossRef]
- Suso, M.J.; Hunady, I.; Solis, I.; Mondragão-Rodrigues, F.; Winkler, J. Germplasm management of Vicia faba L.: Comparative study of the mating system of local and common cultivars growing under different agro-ecological conditions. Plant Genet. Resour. Newsl. 2008, 155, 46–51. [Google Scholar]
- Kumar, A.; Bennetzen, J.L. Plant retrotransposons. Ann. Rev. Genet. 1999, 33, 479–532. [Google Scholar] [CrossRef] [PubMed]
- Choulet, F.; Wicker, T.; Rustenholz, C.; Paux, E.; Salse, J.; Leroy, P.; Schlub, S.; Le Paslier, M.C.; Magdelenat, G.; Gonthier, C.; et al. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 2010, 22, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, J. Triticeae genome structure and evolution. In Genetics and Genomics of the Triticeae; Muehlbauer, G.J., Feuillet, C., Eds.; Springer: New York, NY, USA, 2009; Volume 7, pp. 685–711. [Google Scholar]
- Peterson-Burch, B.D.; Nettleton, D.; Voytas, D.F. Genomic neighborhoods for Arabidopsis retrotransposons: A role for targeted integration in the distribution of the metaviridae. Genome Biol. 2004, 5, R78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polignano, G.B.; Zeuli, P.L.S. Variation and covariation in Vicia faba L. populations of mediterranean origins. Euphytica 1985, 34, 659–668. [Google Scholar] [CrossRef]
- Zeid, M.; Schon, C.C.; Link, W. Genetic diversity in recent elite faba bean lines using AFLP markers. Theor. Appl. Genet. 2003, 107, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Negri, V. Landraces in central italy: Where and why they are conserved and perspectives for their on-farm conservation. Genet. Resour. Crop Evol. 2003, 50, 871–885. [Google Scholar] [CrossRef]

| Population Common Name/Accession Number | Origin/Pedigree | Botanical Group |
|---|---|---|
| “Fava Miuda”/2288 | Madeira Island (Portugal)/locally adapted population | Equina |
| “Fava da Terra”/5362 | Azores Island (Portugal)/locally adapted population | Major |
| “Fava Do Algarve”/5357 | Algarve (Portugal)/locally adapted population | Major |
| “Favel”/5356 | Elvas (Portugal)/locally adapted population | Equina |
| “Jaulon 45” | CSIC (Spain)/Inbred line | Equina |
| “Jaulon 58” | CSIC (Spain)/Inbred line | Equina |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomás, D.; Dias, A.L.; Silva, M.; Oliveira, H.R.; Suso, M.J.; Viegas, W.; Veloso, M.M. Genetic Diversity Assessment of Portuguese Cultivated Vicia faba L. through IRAP Markers. Diversity 2016, 8, 8. https://doi.org/10.3390/d8020008
Tomás D, Dias AL, Silva M, Oliveira HR, Suso MJ, Viegas W, Veloso MM. Genetic Diversity Assessment of Portuguese Cultivated Vicia faba L. through IRAP Markers. Diversity. 2016; 8(2):8. https://doi.org/10.3390/d8020008
Chicago/Turabian StyleTomás, Diana, Ana Luisa Dias, Manuela Silva, Hugo R. Oliveira, Maria José Suso, Wanda Viegas, and Maria Manuela Veloso. 2016. "Genetic Diversity Assessment of Portuguese Cultivated Vicia faba L. through IRAP Markers" Diversity 8, no. 2: 8. https://doi.org/10.3390/d8020008