Evolutionary Hotspots in the Mojave Desert
Abstract
:1. Introduction
2. Background and Methods
2.1. Study Region

2.2. Compiling Genetic Data
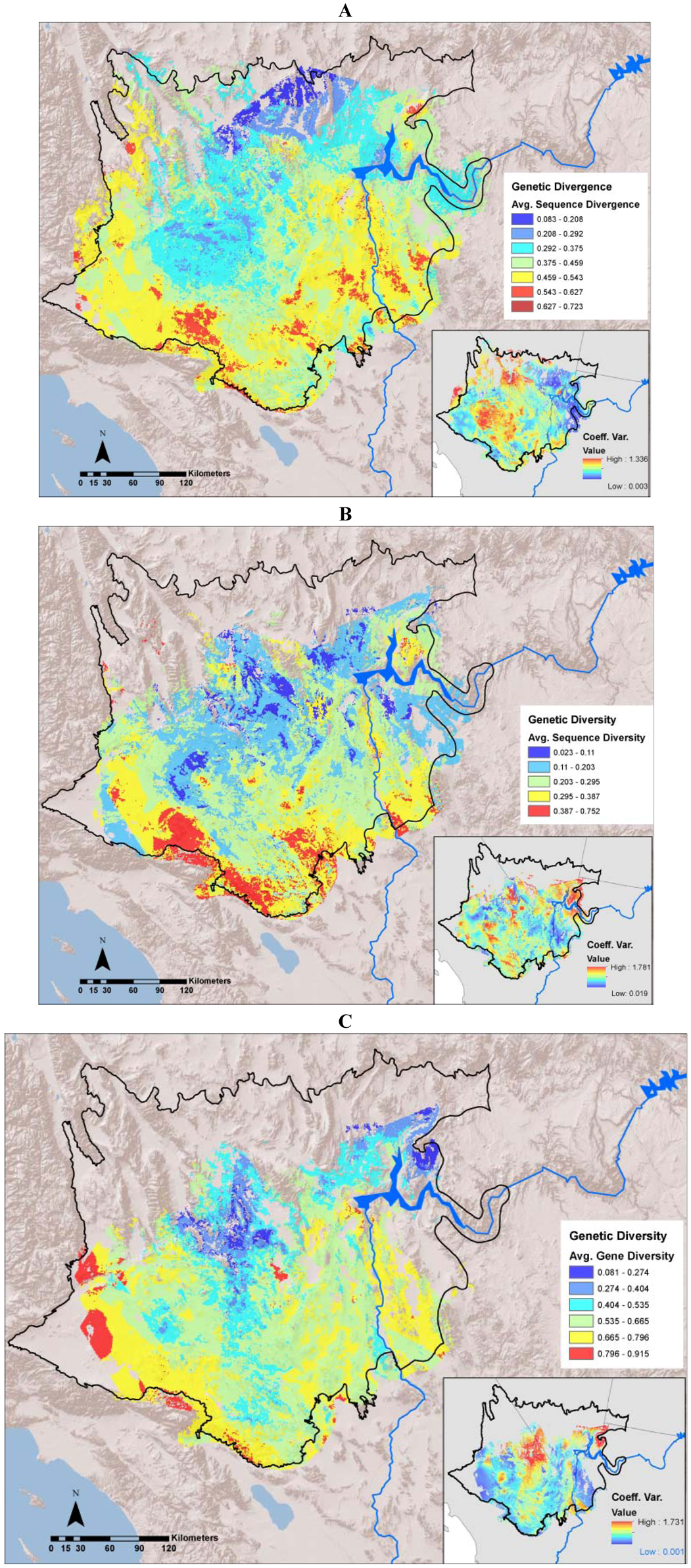
2.3. Mapping Divergence Landscapes
| Species | Genetic Markers | Divergence Sampling Locations | Divergence Mean N | Diversity Sampling Locations | Diversity Mean N |
|---|---|---|---|---|---|
| Herpetofauna | |||||
| Anaxyrus punctatus (red-spotted toad) | mtDNA | 23 | 2.11 | 11 | 4.36 |
| Chionactis occipitalis (shovel-nosed snake) | mtDNA, msats | 33 | 1.12 | 8 (7) | 4.38 (3.71) |
| Crotaphytus bicinctores (collared lizard) | mtDNA | 24 | 1.02 | NI | NI |
| Dipsosaurus dorsalis dorsalis (northern desert iguana) | mtDNA | 8 | 1 | NI | NI |
| Gopherus agassizii (desert tortoise) | mtDNA , msats | 13 | 8.93 | 13 (24) | 8.93 (28.83) |
| Lichanura trivirgata (rosy boa) | mtDNA | 10 | 1.93 | 5 | 4.2 |
| Plestiodon gilberti (Gilbert’s skink) | mtDNA | 42 | 1.11 | 12 | 3.33 |
| Sceloporus magister (desert spiny lizard) | mtDNA, nuDNA | 13 | 1.1 | 7 | 2.14 |
| Uma scoparia (fringe-toed lizard) | mtDNA, nuDNA | 15 | 3.6 | 15 | 3.6 |
| Xantusia vigilis desert night lizard) | mtDNA | 80 | 3.46 | 29 | 8.9 |
| Invertebrates | |||||
| Homalonychus selenopoides (ground-dwelling spider) | mtDNA | 6 | 2.67 | 6 | 2.67 |
| Homalonychus theologus (ground-dwelling spider). | mtDNA | 14 | 2.61 | 10 | 3.3 |
| Mammals | |||||
| Chaetodipus penicillatus (desert pocket mouse) | mtDNA | 22 | 4.34 | 13 | 8.15 |
| Ovis canadensis (bighorn sheep) | mtDNA, msats | 26 | 13.96 | 26 (26) | 13.96 (13.96) |
| Perognathus longimembris (little pocket mouse) | mtDNA | 12 | 2.24 | 5 | 6.2 |
| Thomomys bottae (pocket gopher) | mtDNA | 7 | 1 | NI | NI |
| Xerospermophilus mohavensis (Mohave ground squirrel) | mtDNA, msats | 11 | 4.09 | 11 (12) | 4.09 (21.08) |
2.4. Creating Diversity Landscapes
2.5. Assessing Protected Status and Vulnerability
3. Results and Discussion
3.1. Evolutionary Hotspots
| Map Code | Place Name | Divergence | Sequence Diversity | Gene Diversity | Possible Historical Isolating Factors |
|---|---|---|---|---|---|
| A | Dunmovin - Coso Junction | PLGI, XEMO | XEMO | PLGI, XAVI | Transition from Mojave to Owens Valley |
| B | Sierra -Tehachapi Transition | PLGI, THBO, XAVI | PELO, GOAG, XEMO | CHOC, PLGI, XAVI, XEMO | Transition from Mojave to large mountains in the west |
| C | Antelope Valley - Mojave Desert Transition | PELO, PLGI, THBO, XEMO | XEMO | PELO, PLGI, THBO, XAVI, XEMO | Transition from western grasslands to eastern scrublands |
| D | Ord Mountains - Lucerne Valley | CHPE, PELO, SCMA, THBO, XAVI | PLGI, XAVI | PLGI, XAVI | Historic barrier formed by Mojave River |
| E | Indio Hills - Little San Bernardino Mountains | DIDO, HOTH, LITR, PELO, PLGI, THBO, XAVI | HOTH, LITR, PELO, SCMA, XAVI | CHPE, HOTH, LITR, OVCA, PELO, PLGI | Transition from low Sonoran/Colorado desert (Coachella Valley) into high desert of Little San Bernardino Mountains. |
| F | Pluvial Lakes (Bristol/Cadiz/Danby) | HOTH, OVCA, SCMA, THBO, UMSC, XEMO | HOSE, OVCA, SCMA, UMSC | ANPU, CHOC, CHPE, OVCA, UMSC, XAVI | Historic barrier across low elevation lakes, Mojave/Sonoran Transition |
| G | Colorado River Mountains (Mojave/Black Mountains) | ANPU, CHOC, CRBI, DIDO, HOSE, HOTH, LITR, OVCA, PLGI, SCMA | CHPE | ANPU, CHPE, GOAG | Barrier across river and/or mountains on eastern side of river |
| H | Sacramento-Detrital Valley | DIDO, HOSE | CHPE, HOSE | CHPE, XAVI | Low valley transition between Mojave and Sonoran desert |
| I | Ivanpah Valley | GOAG | CHOC, GOAG, OVCA, XAVI | Historic barrier across low elevation lakes | |
| J | Virgin Mountains | ANPU | ANPU, GOAG | Barrier between Virgin Valley and Shivwits Plateau |
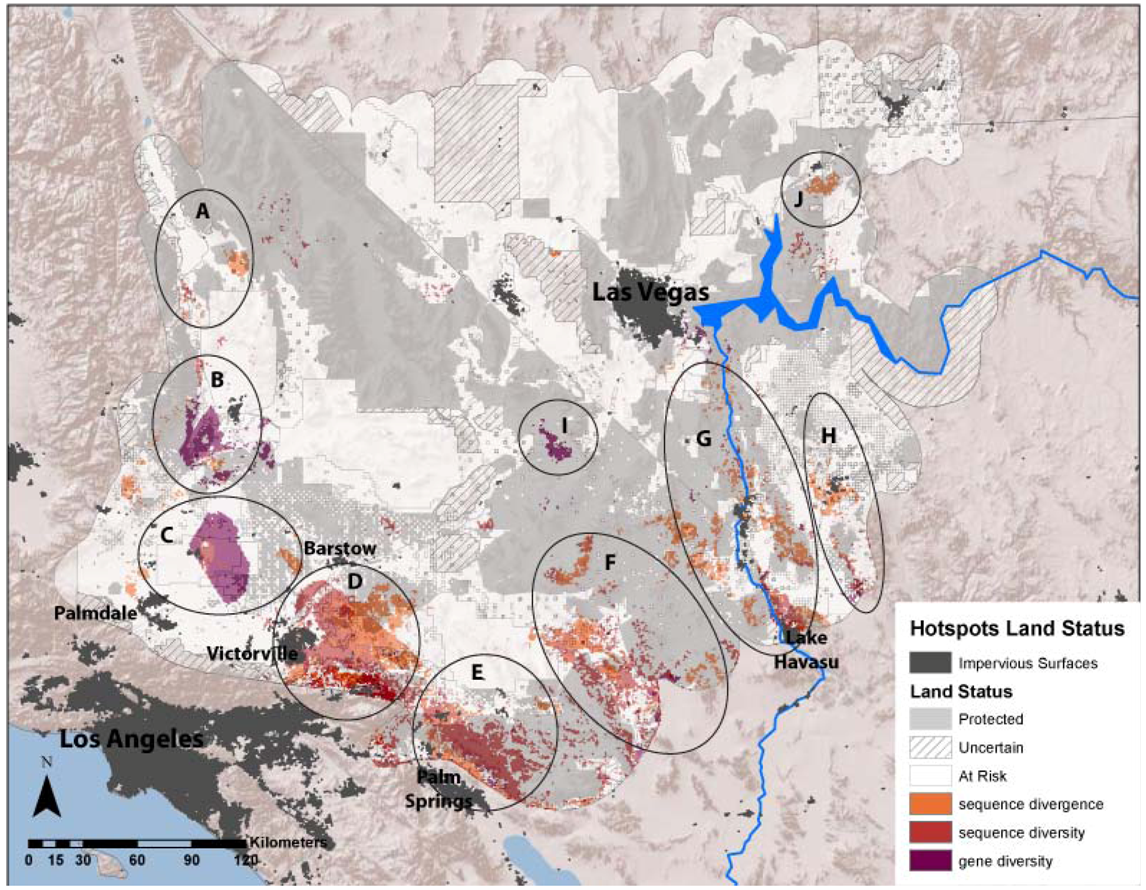
3.2. Protection and Vulnerability of Hotspots

| Layer | Divergence | Sequence Diversity | Gene Diversity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Area (km2) | Mean Score | Score Range | % Hotspot Area | Area (km2) | Mean Score | Score Range | % Hotspot Area | Area (km2) | Mean Score | Score Range | % Hotspot Area | |
| Total Layer | 110,089 | 0.42 | 0.08–0.72 | 5% | 87,982 | 0.25 | 0.02–0.75 | 7% | 71,468 | 0.6 | 0.08–0.91 | 4% |
| “Protected” Lands | 47,428 | 0.42 | 0.19–0.69 | 40% | 37,775 | 0.25 | 0.02–0.58 | 39% | 32,363 | 0.59 | 0.08–0.91 | 29% |
| “Uncertain” Lands | 8,275 | 0.39 | 0.10–0.64 | 5% | 5,213 | 0.24 | 0.05–0.75 | 8% | 3,388 | 0.56 | 0.13–0.87 | 8% |
| “At-Risk” Lands | 54,188 | 0.42 | 0.08–0.72 | 55% | 44,881 | 0.25 | 0.02–0.66 | 53% | 35,709 | 0.61 | 0.10–0.91 | 63% |
| USRED Footprint | 3,209 | 0.44 | 0.08–0.67 | 4% | 2,906 | 0.24 | 0.05–0.65 | 3% | 2,563 | 0.58 | 0.27–0.91 | 7% |
| Transmission Footprint | 10,733 | 0.43 | 0.09–0.66 | 10% | 9,863 | 0.24 | 0.03–0.67 | 8% | 8,503 | 0.6 | 0.18–0.91 | 12% |
| USRED + Transmission | 12,554 | 0.43 | 0.08–0.67 | 13% | 11,477 | 0.24 | 0.03–0.67 | 10% | 9,970 | 0.6 | 0.18–0.91 | 17% |
4. Conclusions
Acknowledgments
References
- Carvalho, S.B.; Brito, J.C.; Crespo, E.J.; Possingham, H.P. Incorporating evolutionary processes into conservation planning using species distribution data: A case study with the western Mediterranean herpetofauna. Divers. Distrib. 2011, 17, 408–421. [Google Scholar] [CrossRef]
- Cowling, R.M.; Pressey, R.L. Rapid plant diversification: Planning for an evolutionary future. Proc. Natl. Acad. Sci. USA 2001, 98, 5452–5457. [Google Scholar] [CrossRef]
- Prendergast, J.R.; Quinn, R.M.; Lawton, J.H. The gaps between theory and practice in selecting natural reserves. Conserv. Biol. 1999, 13, 484–492. [Google Scholar] [CrossRef]
- Davis, E.B.; Koo, M.S.; Conroy, C.; Patton, J.L.; Moritz, C. The California Hotspots Project: Identifying regions of rapid diversification of mammals. Mol. Ecol. 2008, 17, 120–138. [Google Scholar] [CrossRef]
- Kraft, N.J.B.; Baldwin, B.G.; Ackerly, D.D. Range size, taxon age and hotspots of neoendemism in the California flora. Divers. Distrib. 2010, 16, 403–413. [Google Scholar] [CrossRef]
- Vandergast, A.G.; Bohonak, A.J.; Hathaway, S.A.; Boys, J.; Fisher, R.N. Are hotspots of evolutionary potential adequately protected in southern California? Biol. Conserv. 2008, 141, 1648–1664. [Google Scholar] [CrossRef]
- Frankham, R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1996, 10, 1500–1508. [Google Scholar]
- Mulligan, C.J.; Kitchen, A.; Miyamoto, M.M. Comment on “Population size does not influence mitochondrial genetic diversity in animals”. Science 2006, 314, 1390a. [Google Scholar] [CrossRef]
- Soulé, M. Allozyme variation: Its determinants in space and time. In Molecular Evolution; Ayala, F.J., Ed.; Sinauer: Sunderland, MA, USA, 1976; pp. 60–77. [Google Scholar]
- Carnaval, A.C.; Hickerson, M.J.; Haddad, C.F.B.; Rodrigues, M.T.; Moritz, C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 2009, 323, 785–789. [Google Scholar] [CrossRef]
- Barton, N.H.; Halliday, R.B.; Hewitt, G.M. Rare electrophoretic variants in a hybrid zone. Heredity 1983, 50, 139–146. [Google Scholar] [CrossRef]
- Nettel, A.; Dood, R.S.; Afzal-Rafii, Z.; Tovilla-Hernandez, C. Genetic diversity enhanced by ancient introgression and secondary contact in East Pacific black mangroves. Mol. Ecol. 2008, 17, 2680–2690. [Google Scholar] [CrossRef]
- O’Brien, S.J.; Roelke, M.E.; Marker, L.; Newman, A.; Winkler, C.A.; Meltzer, D.; Colly, L.; Evermann, J.F.; Bush, M.; Wildt, D.E. Genetic basis for species vulnerability in the cheetah. Science 1985, 227, 1428–1434. [Google Scholar]
- Reed, D.H.; Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular population structure and the biogeographic history of a regional fauna: A case history with lessons for conservation biology. Oikos 1992, 63, 62–76. [Google Scholar] [CrossRef]
- Humphries, C.J.; Williams, P.H.; Vane-Wright, R.I. Measuring biodiversity value for conservation. Ann. Rev. Ecol. Syst. 1995, 26, 93–111. [Google Scholar]
- Moritz, C. Uses of molecular phylogenies for conservation. Philos. Trans. Royal Soc. London B Biol. Sci. 1995, 349, 113–118. [Google Scholar] [CrossRef]
- Moritz, C.; Faith, D.P. Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol. Ecol. 1998, 7, 419–429. [Google Scholar] [CrossRef]
- Riddle, B.R.; Hafner, D.J. A step-wise approach to integrating phylogeographic and phylogenetic biogeographic perspectives on the history of a core North American warm deserts biota. J. Arid Environ. 2006, 66, 435–461. [Google Scholar] [CrossRef]
- Wood, D.A.; Vandergast, A.G.; Barr, K.R.; Inman, R.D.; Esque, T.C.; Nussear, K.E.; Fisher, R.N. Comparative phylogeography reveals deep lineages and regional evolutionary hotspots in the Mojave and Sonoran Deserts. Divers. Distrib. 2012. [Google Scholar] [CrossRef]
- Sgrò, C.M.; Lowe, A.J.; Hoffmann, A.A. Building evolutionary resilience for conserving biodiversity under climate change. Evolut. Appl. 2011, 4, 326–337. [Google Scholar] [CrossRef]
- Reusch, T.B.H.; Ehlers, A.; Hammerli, A.; Worm, B. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl. Acad. Sci. USA 2005, 102, 2826–2831. [Google Scholar] [CrossRef]
- Turner, R.M. Mohave Desertscrub. In Biotic Communities Southwestern United States and Northwestern Mexico; Brown, D.E., Ed.; University of Utah Press: Salt Lake City, UT, USA, 1994; pp. 157–168. [Google Scholar]
- Keeler-Wolf, T. Mojave Desert Scrub Vegetation. In Terrestrial Vegetation of California, 3rd; Barbour, M.G., Keeler-Wolf, T., Schoenherr, A.A., Eds.; University of California Press: Berkeley, CA, USA, 2007; pp. 609–656. [Google Scholar]
- Thorne, R.F.; Schoenherr, A.A.; Clements, C.D.; Young, J.A. Transmontane Coniferous Vegetation. In Terrestrial Vegetation of California, 3rd; Barbour, M.G., Keeler-Wolf, T., Schoenherr, A.A., Eds.; University of California Press: Berkeley, CA, USA, 2007; pp. 574–586. [Google Scholar]
- Axelrod, D.I. History of the Mediterranean Ecosystem in California. In Mediterranean Type Ecosystems: Origin and Structure; di Castri, F., Mooney, H.A., Eds.; Springer-Verlag: New York, NY, USA, 1973; pp. 225–277. [Google Scholar]
- Betancourt, J.L.; van Devender, T.R.; Martin, P.S. Packrat Middens: The Last 40,000 Years of Biotic Change; University of Arizona Press: Tucson, AZ, USA, 1990. [Google Scholar]
- Ivo, L. Late Cenozoic uplift of the southwestern Colorado Plateau and adjacent lower Colorado River region. Tectonophysics 1979, 61, 63–95. [Google Scholar] [CrossRef]
- McDougall, K.; Poore, R.; Matti, J. Age and paleoenvironment of the Imperial Formation near San Gorgonio Pass, southern California. J. Foraminiferal Res. 1999, 29, 4–25. [Google Scholar]
- Smith, G.I.; Street-Perrott, F.A. Pluvial lakes of the western United States. In Late-Quaternary Environments of the United States; Wright, H.E.J., Ed.; Longman Group, Limited: London, UK, 1983; Volume 1, pp. 190–212. [Google Scholar]
- Jezkova, T.; Jaeger, J.; Marshall, Z.; Riddle, B. Pleistocene impacts on the phylogeography of the desert pocket mouse (Chaetodipus penicillatus). J. Mammal. 2009, 90, 306–320. [Google Scholar] [CrossRef]
- Axelrod, D.I. Paleobotanical History of the Western Deserts. In Origin and Evolution of the Deserts; Wells, S.G., Haragan, D.R., Eds.; University of New Mexico Press: Albuquerque, NM, USA, 1983; pp. 113–129. [Google Scholar]
- Leu, M.; Hanser, S.E.; Knick, S.T. The human footprint in the west: A large-scale analysis of anthropogenic impacts. Ecol. Appl. 2008, 18, 1119–1139. [Google Scholar] [CrossRef]
- Lovich, J.E.; Bainbridge, D. Anthropogenic degradation of the southern California desert ecosystem and prospects for natural recovery and restoration. Environ. Manag. 1999, 24, 309–326. [Google Scholar] [CrossRef]
- Webb, R.H.; Fenstermaker, L.F.; Heaton, J.S.; Hughson, D.L.; Mcdonald, E.V.; Miller, D.M. The Mojave Desert Ecosystem Processes and Sustainability; University of Nevada Press: Reno, NV, USA, 2009; p. 481. [Google Scholar]
- Commission for Environmental Cooperation of North America, Ecological Regions of North America: Toward a Common Perspective. 1997; Unpublished report.
- Jaeger, J.R.; Riddle, B.R.; Bradford, D.F. Cryptic Neogene vicariance and Quaternary dispersal of the red-spotted toad (Bufo punctatus): Insights on the evolution of North American warm desert biotas. Mol. Ecol. 2005, 14, 3033–3048. [Google Scholar] [CrossRef]
- Wood, D.; Meik, J.; Holycross, A.; Fisher, R.; Vandergast, A. Molecular and phenotypic diversity in Chionactis occipitalis (Western shovel-nosed snake), with emphasis on the status of C. o. klauberi (Tucson shovel-nosed snake). Conserv. Genet. 2008, 9, 1489–1507. [Google Scholar] [CrossRef]
- McGuire, J.A.; Linkem, C.W.; Koo, M.S.; Hutchison, D.W.; Lappin, A.K.; Orange, D.I.; Lemos-Espinal, J.; Riddle, B.R.; Jaeger, J.R. Mitochondrial introgression and incomplete lineage sorting through space and time: phylogenetics of crotaphytid lizards. Evolution 2007, 61, 2879–2897. [Google Scholar] [CrossRef]
- Hagerty, B.E.; Nussear, K.E.; Esque, T.C.; Tracy, C.R. Making molehills out of mountains: landscape genetics of the Mojave Desert tortoise. Landscape Ecol. 2011, 26, 267–280. [Google Scholar] [CrossRef]
- Murphy, R.W.; Berry, K.H.; Edwards, T.; McLuckie, A.M. A genetic assessment of the recovery units for the Mojave population of the desert tortoise, Gopherus agassizii. Chelonian Conserv. Biol. 2007, 6, 229–251. [Google Scholar] [CrossRef]
- Wood, D.; Fisher, R.; Reeder, T. Novel patterns of historical isolation, dispersal, and secondary contact across Baja California in the rosy boa (Lichanura trivirgata). Mol. Phylogenet. Evol. 2008, 46, 484–502. [Google Scholar] [CrossRef]
- Richmond, J.; Jockusch, E. Body size evolution simultaneously creates and collapses species boundaries in a clade of scincid lizards. Proc. Royal Soc. B Biol. Sci. 2007, 274, 1701–1708. [Google Scholar] [CrossRef]
- Leaché, A.D.; Mulcahy, D.G. Phylogeny, divergence times and species limits of spiny lizards (Sceloporus magister species group) in western North American deserts and Baja California. Mol. Ecol. 2007, 16, 5216–5233. [Google Scholar] [CrossRef]
- Murphy, R.; Trepanier, T.; Morafka, D. Conservation genetics, evolution and distinct population segments of the Mojave fringe-toed lizard, Uma scoparia. J. Arid Environ. 2006, 67, 226–247. [Google Scholar] [CrossRef]
- Leavitt, D.H.; Bezy, R.L.; Crandall, K.A.; Sites, J.W., Jr. Multi-locus DNA sequence data reveal a history of deep cryptic vicariance and habitat-driven convergence in the desert night lizard Xantusia vigilis species complex (Squamata: Xantusiidae). Mol. Ecol. 2007, 16, 4455–4481. [Google Scholar] [CrossRef]
- Epps, C.W.; Palsboll, P.J.; Wehausen, J.D.; Roderick, G.K.; Ramey, R.R., II; McCullough, D.R. Highways block gene flow and cause a rapid decline in genetic diversity of desert bighorn sheep. Ecol. Lett. 2005, 8, 1029–1038. [Google Scholar] [CrossRef]
- Swei, A.; Brylski, P.V.; Spencer, W.D.; Dodd, S.C.; Patton, J.L. Hierarchical genetic structure in fragmented populations of the little pocket mouse (Perognathus longimembris) in southern California. Conserv. Genet. 2003, 4, 501–514. [Google Scholar] [CrossRef]
- lvarez-Castaneda, S. Phylogenetic structure of the Thomomys bottae-umbrinus complex in North America. Mol. Phylogenet. Evol. 2010, 54, 671–679. [Google Scholar] [CrossRef]
- Bell, K.C.; Hafner, D.J.; Leitner, P.; Matocq, M.D. Phylogeography of the ground squirrel subgenus Xerospermophilus and assembly of the Mojave Desert biota. J. Biogeogr. 2010, 37, 363–378. [Google Scholar] [CrossRef]
- Bell, K.C.; Matocq, M.D. Regional genetic subdivision in the Mohave ground squirrel: Evidence of historic isolation and ongoing connectivity in a Mojave Desert endemic. Anim. Conserv. 2010, 14, 371–381. [Google Scholar] [CrossRef]
- Crews, S.; Hedin, M. Studies of morphological and molecular phylogenetic divergence in spiders (Araneae: Homalonychus) from the American southwest, including divergence along the Baja California Peninsula. Mol. Phylogenet. Evol. 2006, 38, 470–487. [Google Scholar] [CrossRef]
- Murphy, R.W.; Berry, K.H.; Edwards, T.; Leviton, A.E.; Lathrop, A.; Riedle, J.D. The dazed and confused identity of Agassiz’s land tortoise, Gopherus agassizii (Testudines: Testudinidae) with the description of a new species and its consequences for conservation. ZooKeys 2011, 113, 39–71. [Google Scholar] [CrossRef]
- Higgins, D.G.; Bleasby, A.J.; Fuchs, R. CLUSTAL-V - Improved software for multiple sequence alignment. Comp. Appl. Biosciences 1992, 8, 189–191. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Dieringer, D.; Schlotterer, C. MICROSATELLITE ANALYSER (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Edwards, S.V.; Beerli, P. Perspective: Gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 2000, 54, 1839–1854. [Google Scholar]
- Nei, M.; Li, W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Jensen, J.L.; Bohonak, A.J.; Kelley, S.T. Isolation by distance, web service. BMC Genet. 2005, 6, 13. [Google Scholar] [CrossRef]
- Vandergast, A.G.; Perry, W.M.; Lugo, R.V.; Hathaway, S.A. Genetic landscapes GIS Toolbox: tools to map patterns of genetic divergence and diversity. Mol. Ecol. Resour. 2011, 11, 158–161. [Google Scholar] [CrossRef]
- Manni, F.; Guerard, E.; Heyer, E. Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by using Monmonier's algorithm. Hum. Biol. 2004, 76, 173–190. [Google Scholar] [CrossRef]
- Miller, M.P. Alleles In Space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. J. Hered. 2005, 96, 722–724. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Nussear, K.E.; Esque, T.C.; Inman, R.D.; Gass, L.; Thomas, K.A.; Wallace, C.S.A.; Blainey, J.B.; Miller, D.M.; Webb, R.H. Modeling habitat of the desert tortoise (Gopherus agassizii) in the Mojave and parts of the Sonoran Deserts of California, Nevada, Utah, and Arizona: U.S. Geological Survey Open-File Report 2009–1102. 2009, p. 18. Available online: http://pubs.usgs.gov/of/2009/1102/ (accessed on 1 April 2013).
- Inman, R.D.; Esque, T.C.; Nussear, K.E.; Lieitner, P.; Matocq, M.D.; Weisberg, P.J.; Dilts, T.E.; Vandergast, A.G. Is there room for all of us? Renewable energy and Xerospermophilus mohavensis. Endanger. Species Res. 2013, 20, 1–18. [Google Scholar] [CrossRef]
- Inman, R.D.; Nussear, K.E.; Esque, T.C.; Vandergast, A.G.; Hathaway, S.A.; Barr, K.R.; Fisher, R.N. Mapping Habitat in the Desert Southwest. U.S. Geological Survey Open-File Report. 2013; in preparation. [Google Scholar]
- Cumming, G.S. Using between-model comparisons to fine-tune linear models of species ranges. J. Biogeogr. 2000, 27, 441–455. [Google Scholar] [CrossRef]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Boyce, M.S.; Vernier, P.R.; Nielsen, S.E.; Schmiegelow, F.K.A. Evaluating resource selection functions. Ecol. Model. 2002, 157, 281–300. [Google Scholar] [CrossRef]
- Anderson, D.R.; Burnham, K.P.; Thompson, W.L. Null hypothesis testing: Problems, prevalence, and an alternative. J. Wildlife Manag. 2000, 64, 912–923. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Multimodel inference - understanding AIC and BIC in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Liu, C.; Berry, P.; Dawson, T.; Pearson, R. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 2005, 28, 385–393. [Google Scholar] [CrossRef]
- Penrod, K.; Beier, P.; Garding, E.; Cabañero, C. A Linkage Network for the California Deserts; Prepared for the Bureau of Land Management and The Wildlands Conservancy. Science and Collaboration for Connected Wildlands and Northern Arizona University: Fair Oaks, CA, USA, 2012; p. 361. Available online: http://www.scwildlands.org/reports/Default.aspx/ (accessed on 14 December 2012).
- Fu, Y.-X. Statistical properties of segregating sites. Theor. Popul. Biol. 1995, 48, 172–197. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- USDI-BLM Programmatic Environmental Impact Statement for Solar Energy Development in Six Southwestern States. Available online: http://solareis.anl.gov/documents/fpeis/Solar_FPEIS_ExecutiveSummary.pdf (accessed on 16 July 2012).
- USDI-BLM California Desert Conservation Area Plan. Available online: http://www.blm.gov/pgdata/etc/medialib//blm/ca/pdf/pdfs/cdd_pdfs.Par.aa6ec747.File.pdf/CA_Desert_.pdf (accessed on 18 July 2012).
- USDI-BLM Approved Resource Management Plan Amendments/Record of Decision (ROD) for Designation of Energy Corridors on Bureau of Land Management-Administered Lands in the 11 Western States. Available online: http://corridoreis.anl.gov/documents/docs/Energy_Corridors_final_signed_ROD_1_14_2009.pdf (accessed on 18 July 2012).
- Remington, C.L. Suture-Zones of Hybrid Interaction between Recently Joined Biotas. In Evolutionary Biology; Dobzhansky, T., Hecht, M.K., Steere, W.C., Eds.; Appleton-Century-Crofts: New York, NY, USA, 1968; Volume 2. [Google Scholar]
- Stebbins, G.L.; Major, J. Endemism and speciation in the California flora. Ecol. Monogr. 1965, 35, 1–35. [Google Scholar] [CrossRef]
- Epps, C.W.; Palsbøll, P.J.; Wehausen, J.D.; Roderick, G.K.; McCullough, D.R. Elevation and connectivity define genetic refugia for mountain sheep as climate warms. Mol. Ecol. 2006, 15, 4295–4302. [Google Scholar] [CrossRef]
- Markert, J.A.; Champlin, D.M.; Gutjahr-Gobell, R.; Grear, J.S.; Kuhn, A.; McGreevy, T.J.; Roth, A.; Bagley, M.J.; Nacci, D.E. Population genetic diversity and fitness in multiple environments. BMC Evolut. Biol. 2010, 10, 205. [Google Scholar]
- McKay, J.K.; Latta, R.G. Adaptive population divergence: Markers, QTL and traits. Trends Ecol. Evol. 2002, 17, 285–291. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 2001, 55, 1095–1103. [Google Scholar]
- Barton, N.H.; Hewitt, G.M. Analysis of hybrid zones. Ann. Rev. Ecol. Syst. 1985, 16, 113–148. [Google Scholar]
- Ebert, D.; Haag, C.; Kirkpatrick, M.; Riek, M.; Hottinger, J.W.; Pajunen, V.I. A selective advantage to immigrant genes in a Daphnia metapopulation. Science 2002, 295, 485–488. [Google Scholar] [CrossRef]
- Lavergne, S.; Molofsky, J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl. Acad. Sci. USA 2007, 104, 3883–3888. [Google Scholar] [CrossRef]
- Tallmon, D.A.; Luikart, G.; Waples, R.S. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 2004, 19, 489–496. [Google Scholar] [CrossRef]
- Turgeon, J.; Estoup, A.; Bernatchez, L. Species flock in the North American Great Lakes: Molecular ecology of Lake Nipigon Ciscoes (Teleostei: Coregonidae: Coregonus). Evolution 1999, 53, 1857–1871. [Google Scholar] [CrossRef]
- Freedman, A.H.; Thomassen, H.A.; Buermann, W.; Smith, T.B. Genomic signals of diversification along ecological gradients in a tropical lizard. Mol. Ecol. 2010, 19, 3773–3788. [Google Scholar] [CrossRef]
- Smith, T.B.; Kark, S.; Schneider, C.J.; Wayne, R.K.; Moritz, C. Biodiversity hotspots and beyond: the need for preserving environmental transitions. Trends Ecol. Evol. 2001, 16, 431. [Google Scholar]
- Thomassen, H.A.; Cheviron, Z.A.; Freedman, A.H.; Harrigan, R.J.; Wayne, R.K.; Smith, T.B. Spatial modelling and landscape-level approaches for visualizing intra-specific variation. Mol. Ecol. 2010, 19, 3532–3548. [Google Scholar] [CrossRef]
- Patton, J.L.; Huckaby, D.G.; Alvarez-Castaneda, S.T. The Systematic and Evolutionary History of Woodrats of the Neotoma lepida Complex; University of California Press: Berkeley, CA, USA, 2008. [Google Scholar]
- Epling, C.; Lewis, H.; Ball, F.M. The breeding group and seed storage: A study in population dynamics. Evolution 1960, 14, 238–255. [Google Scholar] [CrossRef]
- Schemske, D.W.; Bierzychudek, P. Evolution of flower color in the desert annual Linanthus parryae: Wright revisited. Evolution 2001, 55, 1269–1282. [Google Scholar]
- Schemske, D.W.; Bierzychudek, P. Spatial differentiation for flower color in the desert annual Linanthus parryae: was Wright right? Evolution 2007, 61, 2528–2543. [Google Scholar] [CrossRef]
- Etterson, J.R. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. II. Genetic architecture of three populations reciprocally planted along an environmental gradient in the Great Plains. Evolution 2004, 58, 1459–1471. [Google Scholar]
- Hellmann, J.J.; Pineda-Krch, M. Constraints and reinforcement on adaptation under climate change: Selection of genetically correlated traits. Biol. Conserv. 2007, 137, 599–609. [Google Scholar] [CrossRef]
- Kuvlesky, W.P.; Brennan, L.A.; Morrison, M.L.; Boydston, K.K.; Ballard, B.M.; Bryant, F.C. Wind energy development and wildlife conservation: Challenges and opportunities. J. Wildl. Manag. 2007, 71, 2487–2498. [Google Scholar] [CrossRef]
- Kunz, T.H.; Arnett, E.B.; Cooper, B.M.; Erickson, W.P.; Larkin, R.P.; Mabee, T.; Morrison, M.L.; Strickland, M.D.; Szewczak, J.M. Assessing impacts of wind-energy development on nocturnally active birds and bats: A guidance document. J. Wildl. Manag. 2007, 71, 2449–2486. [Google Scholar] [CrossRef]
- Baerwald, E.F.; Barclay, R.M.R. Patterns of activity and fatality of migratory bats at a wind energy facility in Alberta, Canada. J. Wildl. Manag. 2011, 75, 1103–1114. [Google Scholar] [CrossRef]
- Smallwood, K.S.; Rugge, L.; Morrison, M.L. Influence of behavior on bird mortality in wind energy developments. J. Wildl. Manag. 2009, 73, 1082–1098. [Google Scholar] [CrossRef]
- Drewitt, A.L.; Langston, R.H.W. Assessing the impacts of wind farms on birds. Ibis 2006, 148, 29–42. [Google Scholar] [CrossRef]
- Rabin, L.A.; Coss, R.G.; Owings, D.H. The effects of wind turbines on antipredator behavior in California ground squirrels (Spermophilus beecheyi). Biol. Conserv. 2006, 131, 410–420. [Google Scholar] [CrossRef]
- Lovich, J.E.; Ennen, J.R.; Madrak, S.; Meyer, K.; Loughran, C.; Bjrulin, C.; Arundel, T.; Turner, W.; Jones, C.; Groenendaal, G.M. Effects of wind energy production on growth, demography and survivorship of a desert tortoise (Gopherus agassizii) population in southern California with comparisons to natural populations. Herpetol. Conserv. Biol. 2011, 6, 161–174. [Google Scholar]
- Lovich, J.E.; Ennen, J.R. Wildlife conservation and solar energy development in the desert southwest, United States. Bioscience 2011, 61, 982–992. [Google Scholar] [CrossRef]
- Holderegger, R.; Di Giulio, M. The genetic effects of roads: A review of empirical evidence. Basic Appl. Ecol. 2010, 11, 522–531. [Google Scholar] [CrossRef]
- Trombulak, S.C.; Frissell, C.A. Review of ecological effects of roads on terrestrial and aquatic communities. Conserv. Biol. 2000, 14, 18–30. [Google Scholar] [CrossRef]
- Keyghobadi, N.; Roland, J.; Matter, S.F.; Strobeck, C. Among- and within-patch components of genetic diversity respond at different rates to habitat fragmentation: an empirical demonstration. Proc. R. Soc. B-Biol. Sci. 2005, 272, 553–560. [Google Scholar] [CrossRef]
- Richmond, J.Q.; Reid, D.T.; Ashton, K.G.; Zamudio, K.R. Delayed genetic effects of habitat fragmentation on the ecologically specialized Florida sand skink (Plestiodon reynoldsi). Conserv. Genet. 2009, 10, 1281–1297. [Google Scholar] [CrossRef]
- Carthew, S.M.; Horner, B.; Jones, K.M.W. Do utility corridors affect movements of small terrestrial fauna? Wildl. Res. 2009, 36, 488–495. [Google Scholar] [CrossRef]
- Fahrig, L.; Rytwinski, T. Effects of roads on animal abundance: An empirical review and synthesis. Ecol. Soc. 2009, 14, 21. [Google Scholar]
- Schreiber, R.K.; Graves, J.H. Powerline corridors as possible barriers to the movements of small mammals. Am. Midl. Nat. 1977, 97, 504–508. [Google Scholar] [CrossRef]
- Forman, R.T.T.; Alexander, L.E. Roads and their major ecological effects. Ann. Rev. Ecol. Syst. 1998, 29, 207–231. [Google Scholar] [CrossRef]
- Gelbard, J.L.; Belnap, J. Roads as conduits for exotic plant invasions in a semiarid landscape. Conserv. Biol. 2003, 17, 420–432. [Google Scholar] [CrossRef]
- Stiles, J.H.; Jones, R.H. Distribution of the red imported fire ant, Solenopsis invicta, in road and powerline habitats. Landsc. Ecol. 1998, 13, 335–346. [Google Scholar] [CrossRef]
- Coates, P.S.; Connelly, J.W.; Delehanty, D.J. Predators of Greater Sage-Grouse nests identified by video monitoring. J. Field Ornithol. 2008, 79, 421–428. [Google Scholar] [CrossRef]
- Kristan, W.B.; Boarman, W.I. Spatial pattern of risk of common raven predation on desert tortoises. Ecology 2003, 84, 2432–2443. [Google Scholar] [CrossRef]
- Peery, M.Z.; Henry, R.W. Recovering marbled murrelets via corvid management: A population viability analysis approach. Biol. Conserv. 2010, 143, 2414–2424. [Google Scholar] [CrossRef]
- Boarman, W. Threats to Desert Tortoise Populations: A Critical Review of the Literature. 2002. Available online: http://www.werc.usgs.gov/ProductDetails.aspx?ID=2574/ (accessed on 14 December 2012). Prepared for West Mojave Planning Team, Bureau of Land Management.
- Hawlena, D.; Bouskila, A. Land management practices for combating desertification cause species replacement of desert lizards. J. Appl. Ecol. 2006, 43, 701–709. [Google Scholar] [CrossRef]
- Loarie, S.R.; Carter, B.E.; Hayhoe, K.; McMahon, S.; Moe, R.; Knight, C.A.; Ackerly, D.D. Climate change and the future of California’s endemic flora. Plos One 2008, 3, e2502. [Google Scholar] [CrossRef]
- Notaro, M.; Mauss, A.; Williams, J.W. Projected vegetation changes for the American Southwest: combined dynamic modeling and bioclimatic-envelope approach. Ecol. Appl. 2012, 22, 1365–1388. [Google Scholar] [CrossRef]
- Spencer, W.; Abella, S.R.; Barrows, C.; Berry, K.; Esque, T.C.; Garrett, K.; Howell, C.A.; Kobaly, R.; Noss, R.F.; Redak, R.A.; et al. Recommendations of Independent Science Advisors for the California Desert Renewable Energy Conservation Plans (DRECP); Prepared for: Renewable Energy Action Team. Conservation Biology Institute: Corvallis, OR, 2010. Available online: http://www.energy.ca.gov/2010publications/DRECP-1000–2010–008/DRECP-1000–2010–008-F.PDF.
- Salazar, K. Addressing the impacts of climate change on America’s water, land, and other natural and cultural resources. Order No. 3289 from the Director of the Department of the Interior. 2009. Available online: http://www.doi.gov/whatwedo/climate/cop15/upload/SecOrder3289.pdf (accessed on 14 December 2012).
- Abby, R.V. The Bureau of Land Management’s Landscape Approach for Managing the Public Lands. Information Bulletin No. 2012-058. Available online: http://www.blm.gov/wo/st/en/info/regulations/Instruction_Memos_and_Bulletins/national_information/2012/IB_2012–058.html/ (accessed on 14 December 2012).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vandergast, A.G.; Inman, R.D.; Barr, K.R.; Nussear, K.E.; Esque, T.C.; Hathaway, S.A.; Wood, D.A.; Medica, P.A.; Breinholt, J.W.; Stephen, C.L.; et al. Evolutionary Hotspots in the Mojave Desert. Diversity 2013, 5, 293-319. https://doi.org/10.3390/d5020293
Vandergast AG, Inman RD, Barr KR, Nussear KE, Esque TC, Hathaway SA, Wood DA, Medica PA, Breinholt JW, Stephen CL, et al. Evolutionary Hotspots in the Mojave Desert. Diversity. 2013; 5(2):293-319. https://doi.org/10.3390/d5020293
Chicago/Turabian StyleVandergast, Amy G., Richard D. Inman, Kelly R. Barr, Kenneth E. Nussear, Todd C. Esque, Stacie A. Hathaway, Dustin A. Wood, Philip A. Medica, Jesse W. Breinholt, Catherine L. Stephen, and et al. 2013. "Evolutionary Hotspots in the Mojave Desert" Diversity 5, no. 2: 293-319. https://doi.org/10.3390/d5020293




