Aboveground Deadwood Deposition Supports Development of Soil Yeasts
Abstract
:1. Introduction
2. Methods
2.1. Sampling and Isolation of Yeasts
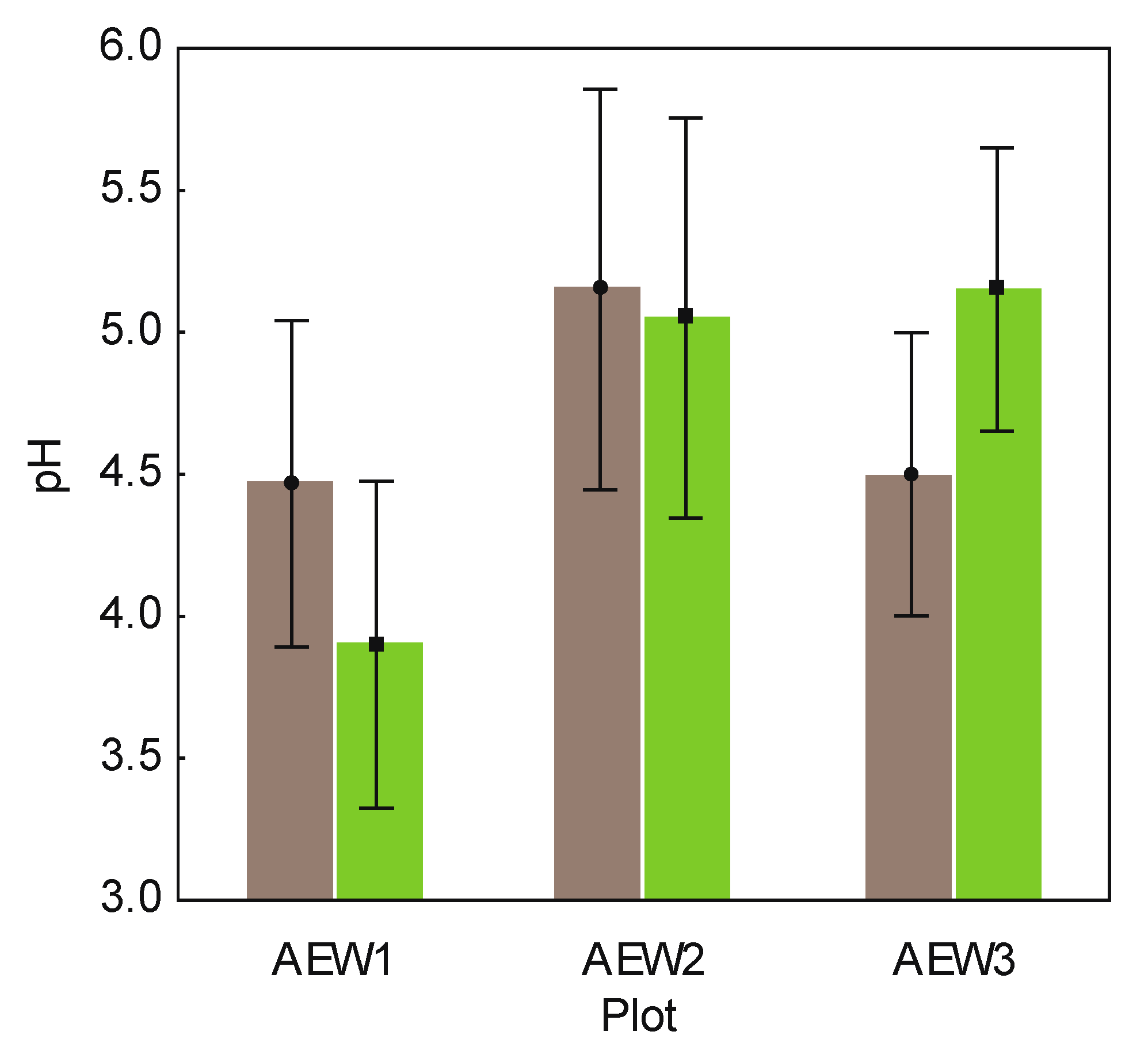
| Sample | Plot ID | Wood Logs | pH | DOC (mg/L) | Corg(%) | Total N (%) | Remaining mass of original stem (%) | Stem length (m) | Stem diam. (foot) (cm) | Brown rot fungi | White rot fungi |
| 8270 | AEW1 | Fagus sylvatica | 4.7 | 251.84 | 6.97 | 0.42 | 65.06 | 3.25 | 22.0 | No | Yes |
| 8271 | AEW1 | Picea abies | 4.6 | 348.35 | 11.57 | 0.62 | 70.46 | 11.25 | 25.0 | Yes | Yes |
| 8273 | AEW1 | Picea abies | 4.1 | 280.27 | 5.48 | 0.31 | 80.81 | 13.25 | 28.0 | Yes | Yes |
| 8820 | AEW2 | Picea abies | 5.3 | 250.49 | 9.29 | 0.55 | 53.13 | 15.30 | 34.0 | No | Yes |
| 8822 | AEW2 | Picea abies | 5.0 | 300.13 | 10.95 | 0.50 | 36.26 | 1.90 | 27.0 | No | Yes |
| 8279 | AEW3 | Picea abies | 4.2 | 247.97 | 5.40 | 0.35 | 64.30 | 13.10 | 21.5 | Yes | Yes |
| 8280 | AEW3 | Fagus sylvatica | 3.7 | 240.00 | 7.04 | 0.51 | 60.03 | 9.80 | 24.0 | Yes | Yes |
| 8281 | AEW3 | Picea abies | 4.8 | 291.20 | 9.96 | 0.57 | 57.96 | 16.10 | 23.0 | Yes | Yes |
| 8282 | AEW3 | Fagus sylvatica | 5.3 | 296.56 | 14.10 | 0.78 | 98.89 | 3.25 | 23.0 | No | Yes |
| 8270 | AEW1 | 4.1 | 216.65 | 6.23 | 0.39 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8271 | AEW1 | 3.7 | 302.65 | 10.40 | 0.53 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8273 | AEW1 | 3.9 | 241.67 | 5.10 | 0.30 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8820 | AEW2 | 5.2 | 240.4 | 8.45 | 0.50 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8822 | AEW2 | 4.9 | 331.24 | 19.41 | 0.78 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8279 | AEW3 | 4.5 | 198.00 | 6.10 | 0.40 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8280 | AEW3 | 5.1 | 168.52 | 6.13 | 0.50 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8281 | AEW3 | 5.5 | 260.43 | 8.01 | 0.52 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| 8282 | AEW3 | 5.5 | 311.3 | 13.7 | 0.80 | n.a. | n.a. | n.a. | n.a. | n.a. |
2.2. Isolation of Cultures
2.3. Identification of the Strains
2.4. Statistical Analysis
3. Results
3.1. Yeast Species Inventory
| Identification results | Taxon example | GenBank | |||
|---|---|---|---|---|---|
| Strain ID | Collection ID | ITS | LSU | Closest match * | |
| Aureobasidium pullulans | TSN-43 | - | FR716139 | - | 0/0 FJ150906 |
| Candida cretensis | TSN-5 | CBS 11766 | - | FN824503 | 10/0 EU011654 |
| C. zeylanoides | TSN-95 | - | HF558653 | FR716579 | 2/0 AY498861 |
| Candida sp. CBS 7170 | TSN-36 | CBS 11767 | - | FN824506 | 0/0 AY574389 |
| Candida sp. CBS 11766 | TSN-95-1 | CBS 11778 | - | FN908211 | 1/0 AY497688 |
| Cryptococcus aerius | TSN-77 TSN-82 | CBS 11764 CBS 12340 | JN942234 JN942257 | FN824504 FR716581 | 2/2 AF181524 1/0 AF181544 |
| Cr. filicatus | TSN-98 | MUCL 52894 | FN908210 | FN908210 | 0/0 EU433984 |
| Cr. musci | TSN-91 | CBS 11765 | FN824491 | FN824491 | 0/0 AB126586 |
| Cr. podzolicus | TSN-16 TSN-41 TSN-23 | CBS 12385 CBS 12341 CBS 12388 | HF558650 HF558651 HF558652 | HF558650 HF558651 HF558652 | 1/0 AF075481 1/0 AF075481 2/0 AF075481 |
| Cr. ramirezgomezianus | TSN-95-14 | - | HF558654 | HF558654 | 0/0 AB126584 |
| Cr. stepposus | TSN-64 | CBS 11763 | FN824490 | FN824490 | 0/0 DQ222456 |
| Cr. tephrensis | TSN-4 | - | - | FR716587 | 0/0 CBS 8934 |
| Cr. terricola | TSN-101 | - | HF558655 | HF558655 | 0/0 AF181520 |
| Cr. victoriae | TSN-53 TSN-84 | CBS 12045 - | HF558648 HF558649 | FR716585 FR716586 | 4/0 AF363647 0/0 AF363647 |
| Cystofilobasidium capitatum | TSN-59 | - | HF558659 | FR716589 | 0/0 AF406889 |
| Cy. macerans | TSN-58 | - | HF558660 | FR716588 | 1/0 AF075477 |
| Kazachstania piceae | TSN-26 | - | HF558661 | FR716594 | 2/0 CBS 7738 |
| Phaffia sp. CBS 11768 | TSN-67 | CBS 11768 | HF558647 | HF558647 | 56/3 AF189871 |
| Rhodotorula colostri | TSN-60 | - | HF558663 | FR716590 | 0/0 AY372177 |
| R. fujisanensis | TSN-63 | - | HF558662 | FR716591 | 0/0 AF189928 |
| Schizoblastosporion starkeyi-henricii | TSN-87 | - | HF558658 | FR716592 | 0/0 U40089 |
| Sporobolomyces ruberrimus | TSN-25 | - | HF558664 | FR716593 | 1/0 AF070442 |
| Sp. tsugae | TSN-37 | CBS 11762 | - | FN868152 | 0/0 AF189998 |
| Trichosporon dulcitum | TSN-17 | - | HF558657 | FR716595 | 0/0 AF075517 |
| T. porosum | TSN-24 | - | HF558656 | FR716596 | 0/0 AF189833 |
| Wickerhamomyces anomalus | TSN-95-4 | MUCL 52882 | - | FN868150 | 3/1 EU057562 |
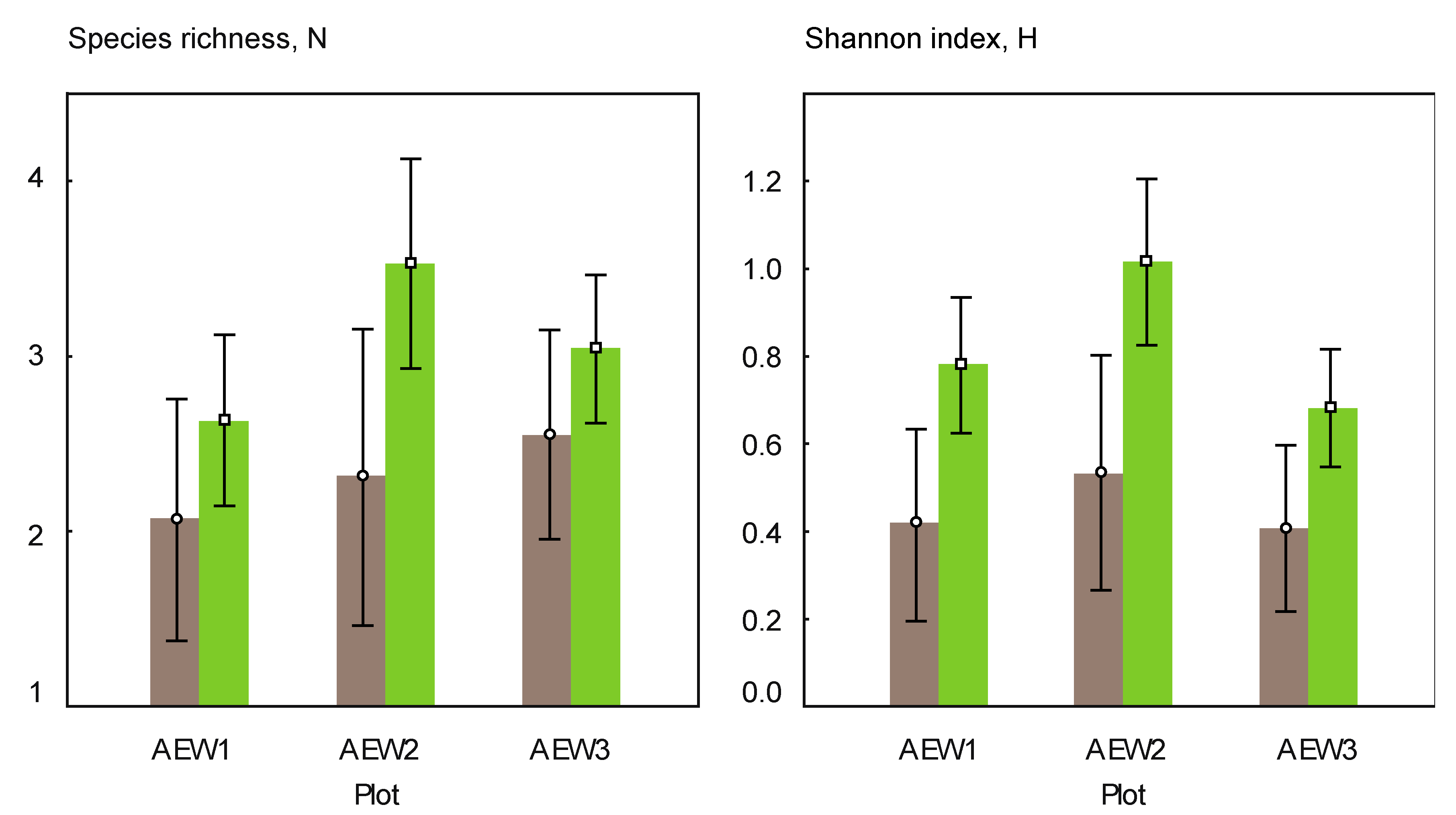
3.2. Yeast Alpha- and Beta-Diversity
| Yeast species | underneath wood logs | in adjacent areas | ||||
|---|---|---|---|---|---|---|
| AEW1 | AEW2 | AEW3 | AEW1 | AEW2 | AEW3 | |
| Aureobasidium pullulans | - | - | 0.003 | - | - | - |
| Candida cretensis | - | - | - | s.i. | - | - |
| C. zeylanoides | - | - | - | s.i. | - | - |
| Candida sp. CBS 7170 | - | - | 0.013 | - | - | - |
| Candida sp. CBS 11766 | 0.167 | - | - | - | - | - |
| Cryptococcus aerius | - | - | - | >0.001 | 0.439 | 0.007 |
| Cr. filicatus | - | - | - | 0.096 | - | - |
| Cr. musci | - | - | - | 0.258 | - | - |
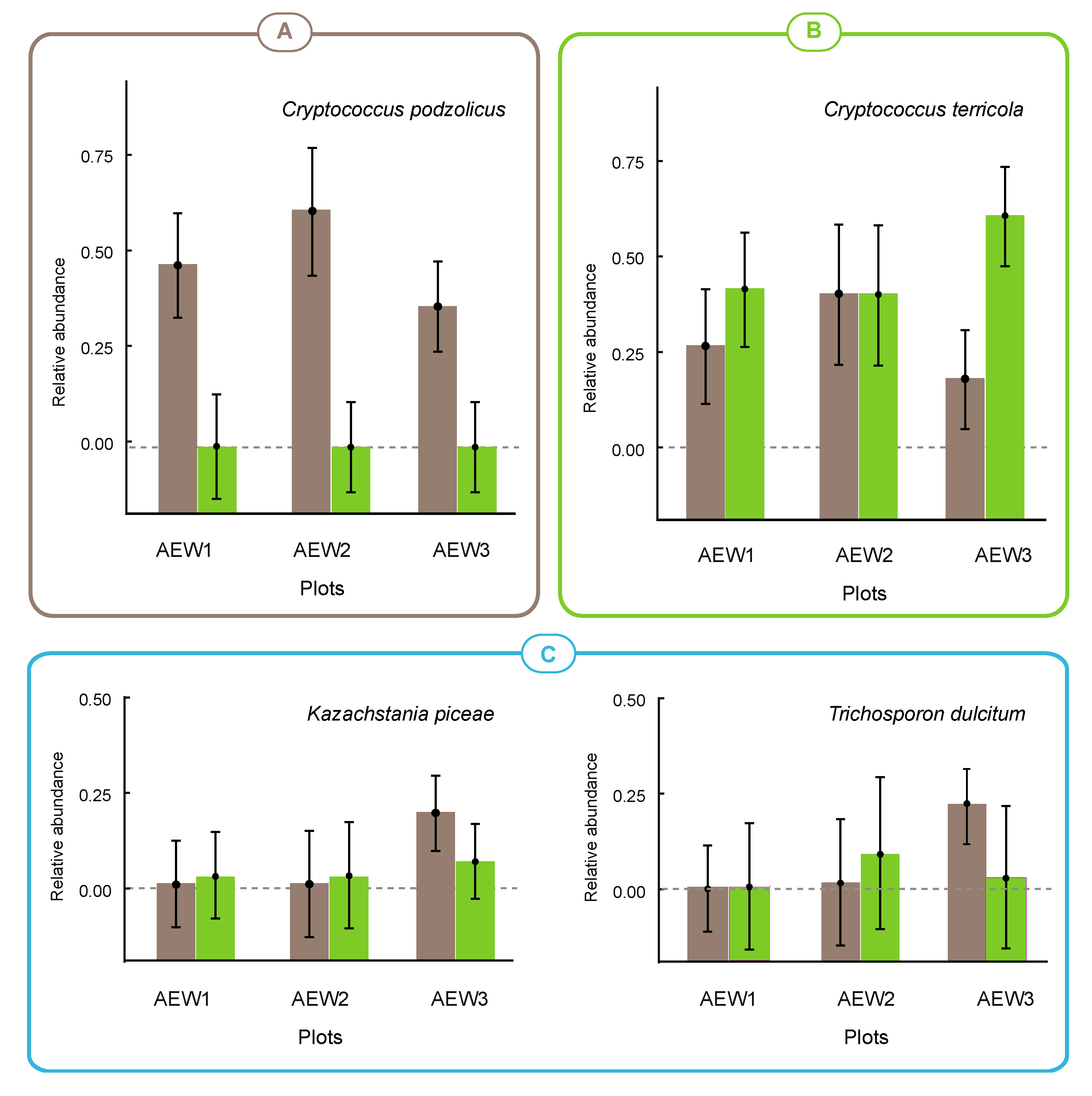
| Cr. podzolicus | 0.449 | 0.590 | 0.363 | - | - | - |
| Cr. ramirezgomezianus | - | - | - | s.i. | - | - |
| Cr. stepposus | - | - | - | 0.096 | - | - |
| Cr. tephrensis | 0.123 | - | - | - | - | - |
| Cr. terricola | 0.252 | 0.387 | 0.176 | 0.381 | 0.367 | 0.573 |
| Cr. victoriae | - | - | - | - | 0.116 | 0.109 |
| Cystofilobasidium capitatum | - | - | - | - | - | 0.017 |
| Cy. macerans | - | - | - | - | - | s.i. |
| Kazachstania piceae | >0.001 | >0.001 | 0.197 | >0.001 | >0.001 | 0.066 |
| Phaffia sp. CBS 11768 | - | - | - | - | - | 0.148 |
| Rhodotorula colostri | 0.003 | - | - | - | 0.002 | 0.022 |
| R. fujisanensis | - | - | - | - | - | 0.039 |
| Schizoblastosporion starkeyi-henricii | 0.006 | - | - | 0.167 | - | - |
| Sporobolomyces ruberrimus | - | - | 0.002 | - | - | - |
| Sp. tsugae | - | - | 0.016 | - | - | - |
| Trichosporon dulcitum | - | 0.010 | 0.220 | - | 0.076 | 0.017 |
| T. porosum | - | 0.012 | 0.010 | 0.002 | - | - |
| Wickerhamomyces anomalus | - | - | s.i. | - | - | |
| underneath wood logs | in adjacent areas | |||||
|---|---|---|---|---|---|---|
| AEW1 | AEW2 | AEW3 | AEW1 | AEW2 | AEW3 | |
| Total yeast quantity, Log10 (CFU/g) | 4.985 | 5.368 | 4.985 | 5.401 | 5.261 | 5.045 |
| Quantity of soil-borne yeasts, Log10 (CFU/g) | 4.968 | 5.386 | 4.972 | 5.309 | 5.206 | 4.865 |
| Species richness | 7 | 5 | 9 | 11 | 6 | 10 |
| Number of soil-borne species | 4 | 5 | 6 | 6 | 4 | 4 |
| Number of transient species | 3 | 0 | 3 | 5 | 2 | 6 |
| Relative abundance of soil-borne species | 0.96 | 1.00 | 0.97 | 0.81 | 0.88 | 0.66 |
| Relative abundance of transient species | 0.04 | - | 0.03 | 0.19 | 0.12 | 0.34 |
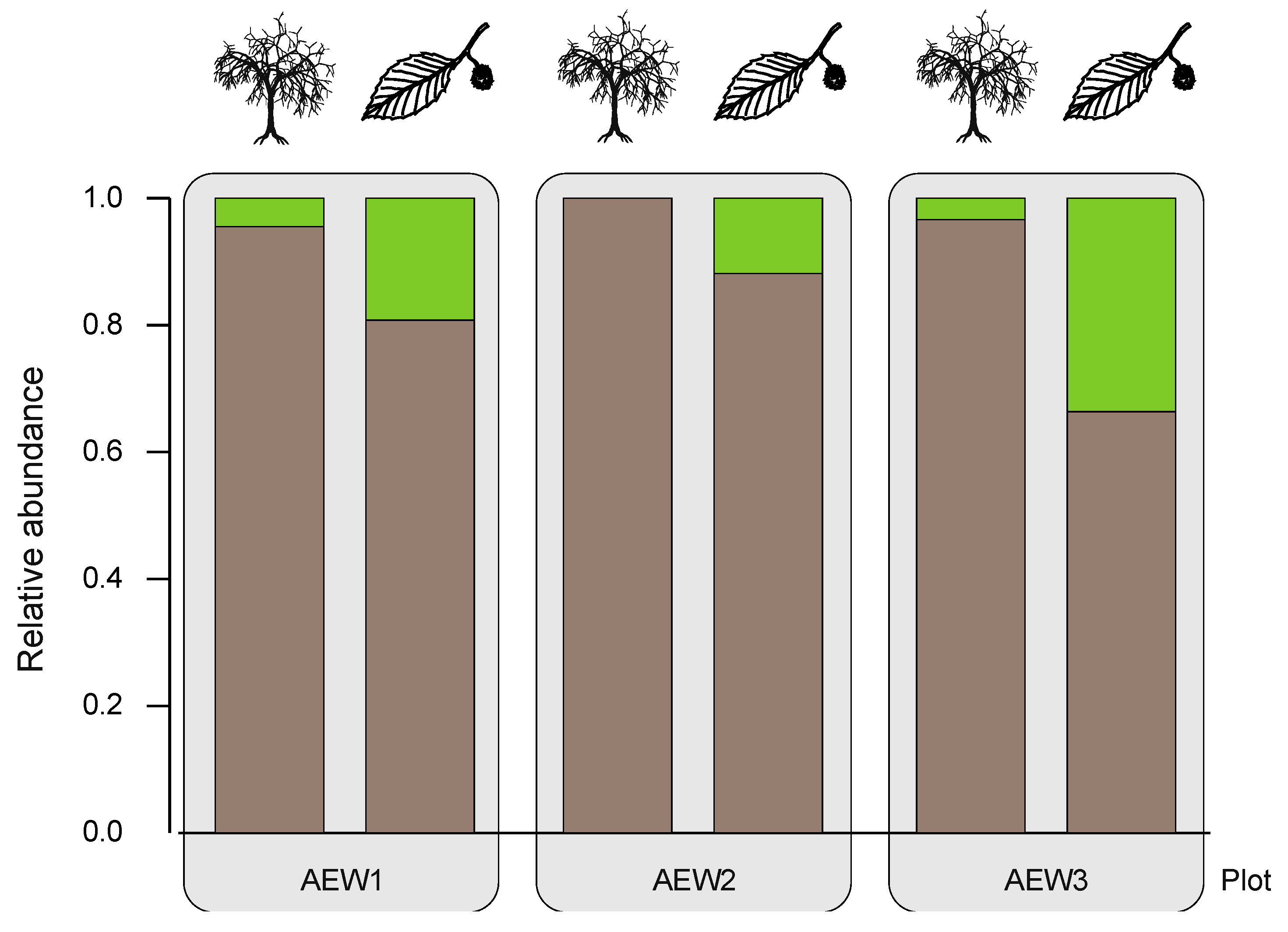


3.3. Yeast Abundance
4. Discussion
4.1. Factors Influencing Distribution of Soil Yeasts
4.2. Autochthonous and Transient Species in Soils
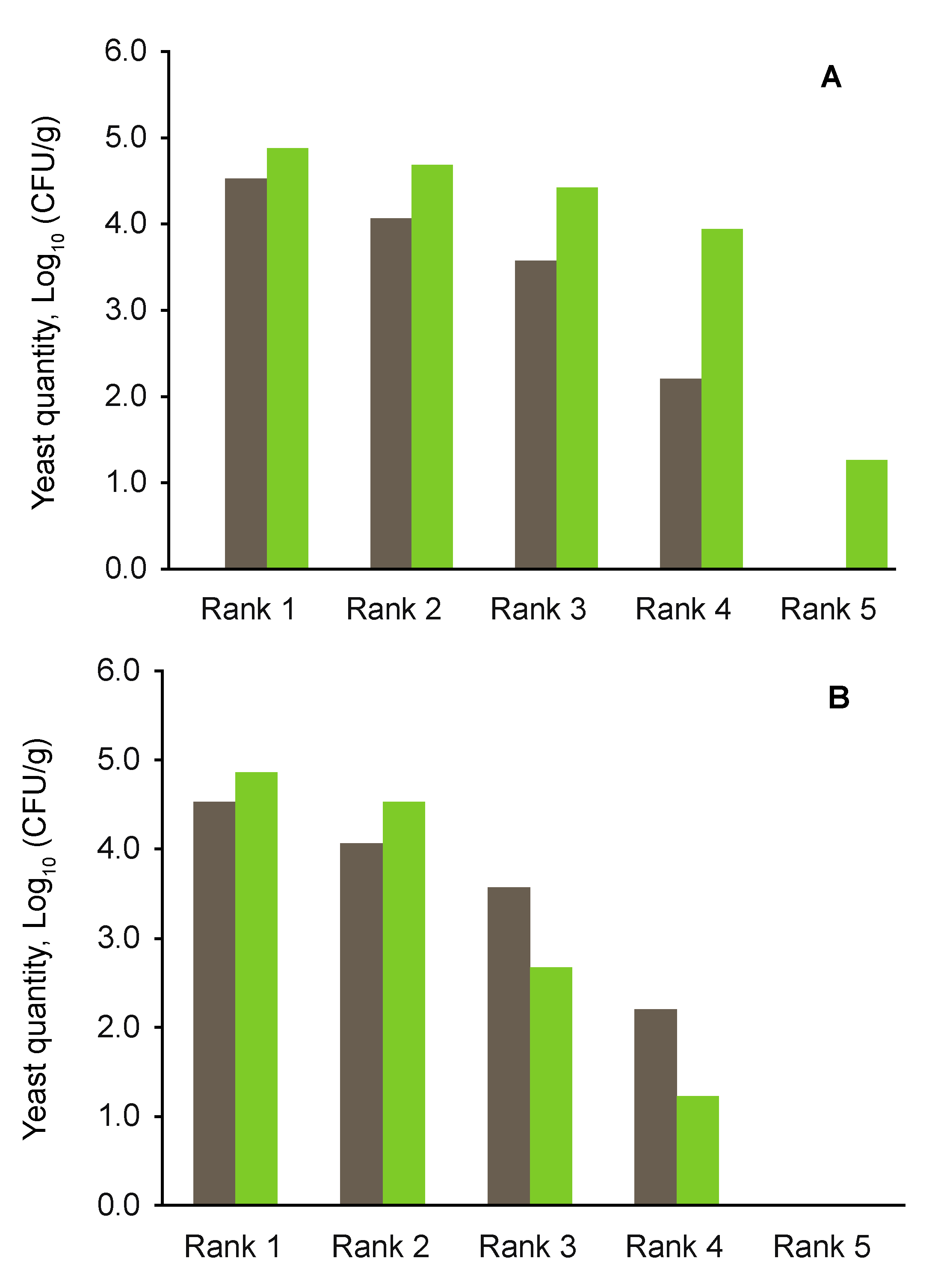
4.3. Effects of Aboveground Deadwood Deposition
5. Conclusions
Supplementary

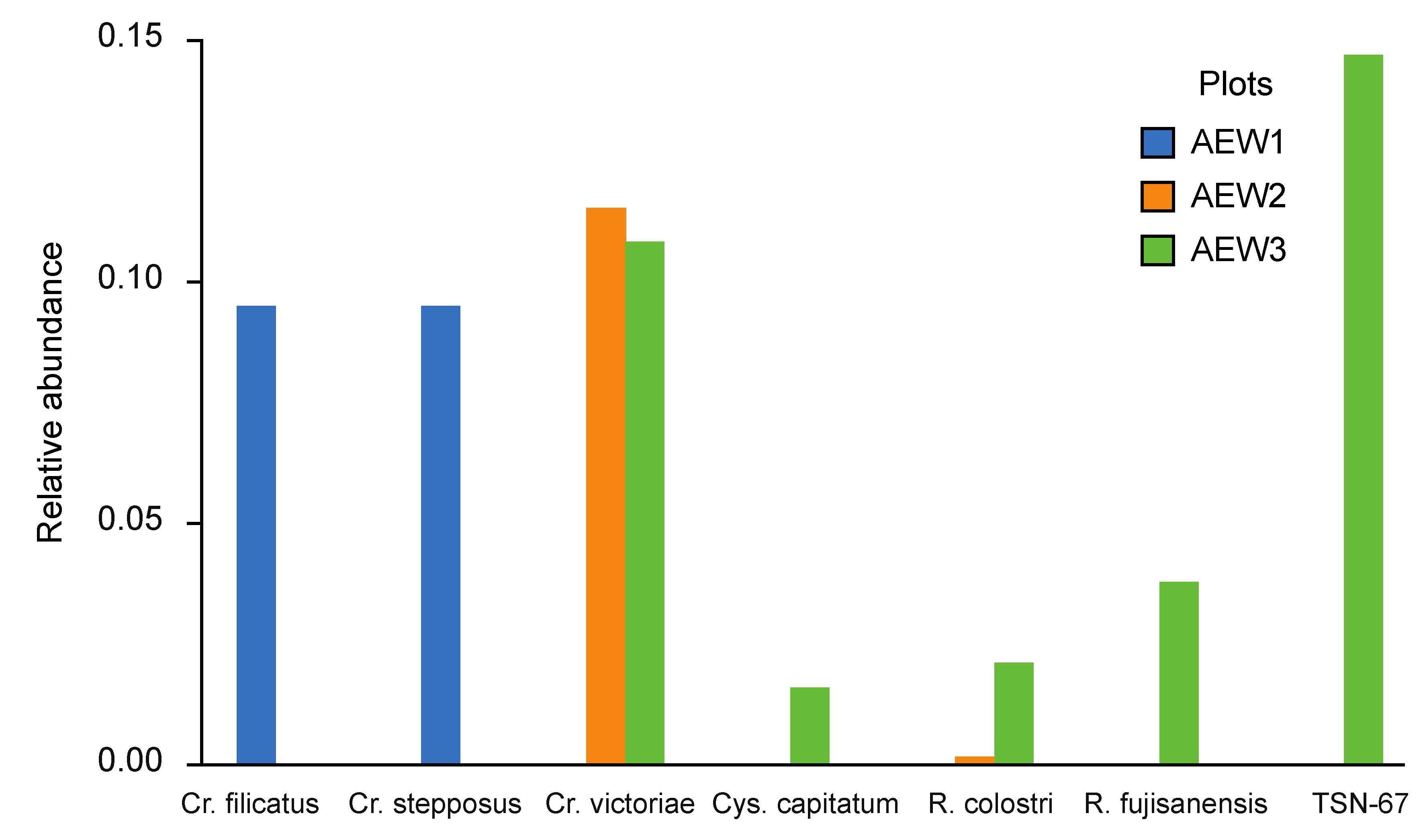
Acknowledgments
References
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar]
- Anderson, J.P.E.; Domsch, K.H. Measurement of bacterial and fungal contributions to respiration of selected agricultural and forest soils. Can. J. Microbiol. 1975, 21, 314–322. [Google Scholar] [CrossRef]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global patterns in belowground communities. Ecol. Lett. 2009, 12, 1238–1249. [Google Scholar] [CrossRef]
- Blackwell, M. The fungi: 1, 2, 3 ... 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Barnett, J.A. A history of research on yeasts 8: taxonomy. Yeast 2004, 21, 1141–1193. [Google Scholar] [CrossRef]
- Botha, A. The importance and ecology of yeasts in soil. Soil Biol. Biochem. 2011, 43, 1–8. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Kemler, M.; Begerow, D. Assessment of yeast diversity in soils under different management regimes. Fungal Ecol. 2012, 5, 24–35. [Google Scholar] [CrossRef]
- Fonseca, A. Utilization of tartaric acid and related compounds by yeasts: taxonomic implications. Can. J. Microbiol. 1992, 38, 1242–1251. [Google Scholar] [CrossRef]
- Middelhoven, W.J.; Koorevaar, M.; Schuur, G.W. Degradation of benzene compounds by yeasts in acidic soils. Plant Soil 1992, 145, 37–43. [Google Scholar] [CrossRef]
- Middelhoven, W.J. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeast-like fungi. Antonie Leeuwenhoek 1993, 63, 125–144. [Google Scholar] [CrossRef]
- Middelhoven, W.J. Trichosporon wieringae sp. nov., an anamorphic basidiomycetous yeast from soil, and assimilation of some phenolic compounds, polysaccharides and other nonconventional carbon sources by saprophytic Trichosporon species. Antonie Leeuwenhoek 2004, 86, 329–337. [Google Scholar]
- Middelhoven, W.J. Polysaccharides and phenolic compounds as substrate for yeasts isolated from rotten wood and description of Cryptococcus fagi sp. nov. Antonie Leeuwenhoek 2006, 90, 57–67. [Google Scholar] [CrossRef]
- Sampaio, J.P. Utilization of low molecular weight aromatic compounds by heterobasidiomycetous yeasts: taxonomic implications. Can. J. Microbiol. 1999, 45, 491–512. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; van der Putten, W.H.; Wall, D.H. Ecological linkages between aboveground biota. Science 2004, 304, 1629–1633. [Google Scholar]
- Brockerhoff, E.G.; Jactel, H.; Parrotta, J.A.; Quine, C.P.; Sayer, J.; Hawksworth, D.L. Plantation forests and biodiversity: oxymoron or opportunity? Biodive. Conserv. 2008, 17, 925–951. [Google Scholar] [CrossRef]
- Hjalten, J.; Johansson, T.; Alinvi, O.; Danell, K.; Ball, J.P.; Pettersson, R.; Gibb, H.; Hilszczanski, J. The importance of substrate type, shading and scorching for the attractiveness of dead wood to saproxylic beetles. Basic Appl. Ecol. 2007, 8, 364–376. [Google Scholar] [CrossRef]
- dor, P.; Heilmann-Clausen, J.; Christensen, M.; Aude, E.; van Dort, K.W.; Piltaver, A.; Siller, I.; Veerkamp, M.T.; Walleyn, R.; Standovár, T.; et al. Diversity of dead wood inhabiting fungi and bryophytes in semi-natural beech forests in Europe. Biol. Conserv. 2006, 131, 58–71. [Google Scholar] [CrossRef]
- Paillet, Y.; Bergès, L.; Hjältén, J.; Ódor, P.; Avon, C.; Bernhardt-Römermann, M.; Bijlsma, R.J.; De Bruyn, L.; Fuhr, M.; Grandin, U.; et al. Biodiversity differences between managed and unmanaged forests: meta-analysis of species richness in Europe. Conserv. Biol. 2010, 24, 101–112. [Google Scholar] [CrossRef]
- Christensen, M.; Emborg, J. Biodiversity in Natural Versus Managed Forest in Denmark. Forest Ecol. Manag. 1996, 85, 47–51. [Google Scholar]
- Di Menna, M.E. Yeasts in New Zealand soils. New Zeal. J. Bot. 1965, 3, 194–203. [Google Scholar] [CrossRef]
- Sláviková, E.; Vadkertiová, R. The occurrence of yeasts in the forest soils. J. Basic Microbiol. 2000, 40, 207–212. [Google Scholar] [CrossRef]
- Maksimova, I.A.; Chernov, I.Yu. Community structure of yeast fungi in forest biogeocenoses. Microbiology 2004, 73, 474–481. [Google Scholar] [CrossRef]
- Mestre, M.C.; Rosa, C.A.; Safar, S.V.; Libkind, D.; Fontenla, S.B. Yeast communities associated with the bulk-soil, rhizosphere and ectomycorrhizosphere of a Nothofagus pumilio forest in northwestern Patagonia, Argentina. FEMS Microbiol. Ecol. 2011, 78, 531–541. [Google Scholar] [CrossRef]
- Wuczkowski, M.; Prillinger, H. Molecular identification of yeasts from soils of the alluvial forest national park along the river Danube downstream of Vienna, Austria (“Nationalpark Donauauen”). Microbiol. Res. 2004, 159, 263–275. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Kemler, M.; Begerow, D. Species accumulation curves and incidence-based species richness estimators to appraise the diversity of cultivable yeasts from beech forest soils. PLoS ONE 2011, 6, e23671. [Google Scholar]
- Chernov, I.Yu. The latitude-zonal and spatial-successional trends in the distribution of yeasts. Zhurnal obshchei biologii 2005, 66, 123–135. [Google Scholar]
- Vishniac, H.S. A multivariate analysis of soil yeasts isolated from a latitudinal gradient. Microbial Ecol. 2006, 52, 90–103. [Google Scholar]
- Birkhofer, K.; Schöning, I.; Alt, F.; Herold, N.; Klarner, B.; Maraun, M.; Marhan, S.; Oelmann, Y.; Wubet, T.; Yurkov, A.; et al. General relationships between abiotic soil properties and soil biota across spatial scales and different land-use types. PLoS ONE 2012, 7, e43292. [Google Scholar]
- Begerow, D.; Nilsson, H.; Unterseher, M.; Maier, W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 2010, 87, 99–108. [Google Scholar]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar]
- Biodiversity Exploratories. Available online: http://www.biodiversity-exploratories.de (accessed on 6 December 2012).
- Fischer, M.; Bossdorf, O.; Gockel, S.; Hansel, F.; Hemp, A.; Hessenmöller, D.; Korte, G.; Nieschulze, J.; Pfeiffer, S.; Prati, D.; et al. Implementing large-scale and long-term functional biodiversity research: The Biodiversity Exploratories. Basic Appl. Ecol. 2010, 11, 473–485. [Google Scholar]
- IUSS Working Group WRB, World Reference Base for Soil Resources 2006, 2nd ed; FAO: Rome, Italy, 2006; p. 128, World Soil Resources Reports No. 103.
- Alvarez, C.V. Influence of dead wood on soil organic carbon along a gradient of management intensity.
- The National Center for Biotechnology Information. Available online: http://www.ncbi.nih.gov (accessed on 6 December 2012).
- MycoBank. Available online: http://www.mycobank.org (accessed on 6 December 2012).
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform (describes the FFT-NS-1, FFT-NS-2 and FFT-NS-i strategies). Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Chao, A.; Chazdon, R.L.; Colwell, R.K.; Shen, T.J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 2005, 8, 148–159. [Google Scholar]
- EstimateS: Biodiversity Estimations. Available online: http://purl.oclc.org/estimates (accessed on 22 August 2012).
- McGill, B.J.; Etienne, R.S.; Gray, J.S.; Alonso, D.; Anderson, M.J.; Benecha, H.K.; Dornelas, M.; Enquist, B.J.; Green, J.L.; He, F.; et al. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecol. Lett. 2007, 10, 995–1015. [Google Scholar] [CrossRef]
- Suh, S.O.; McHugh, J.V.; Blackwell, M. Expansion of the Candida tanzawaensis yeast clade: 16 novel Candida species from basidiocarp-feeding beetles. Int. J. Syst. Evol. Micr. 2004, 54, 2409–2429. [Google Scholar] [CrossRef]
- Fonseca, A.; Inacio, J. Phylloplane yeasts. In Biodiversity and Ecophysiology of Yeasts; Peter, G., Rosa, C., Eds.; Springer-Verlag: Berlin, Germany, 2006; pp. 263–301. [Google Scholar]
- Golubev, W.I.; Sampaio, J.P.; Golubeva, E.W. Cryptococcus stepposus, a new filobasidiaceous yeast species found in the Prioksko-terrasny biosphere reserve in Russia. Mycol. Res. 2006, 110, 957–961. [Google Scholar] [CrossRef]
- Golubev, W.I.; Sampaio, J.P. New filobasidiaceous yeasts found in the phylloplane of a fern. J. Gen. Appl. Microbiol. 2009, 55, 441–446. [Google Scholar]
- Herzberg, M.; Fischer, R.; Titze, A. Conflicting results obtained by RAPD-PCR and large-subunit rDNA sequences in determining and comparing yeast strains isolated from flowers: a comparison of two methods. Int. J. Syst. Evol. Micr. 2002, 52, 1423–1433. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Chernov, I.Yu.; Tiunov, A.V. Influence of Lumbricus terrestris earthworms on the structure of the yeast community of forest litter. Microbiology 2008, 77, 107–111. [Google Scholar] [CrossRef]
- Babjeva, I.P.; Chernov, I.Yu. Geographical aspects of yeast ecology. Physiol. Gen. Biol. Rev. 1995, 9, 1–54. [Google Scholar]
- Yurkov, A.M.; Krüger, D.; Begerow, D.; Arnold, N.; Tarkka, M.T. Basidiomycetous yeasts from Boletales fruiting bodies and their interactions with the mycoparasite Sepedonium chrysospermum and the host fungus Paxillus. Microbial Ecol. 2012, 63, 295–303. [Google Scholar] [CrossRef]
- Babjeva, I.P.; Blagodatskaya, V.M. Physiological properties and ecology of the yeast Schizoblastosporion starkeyi-henricii Ciferri. Microbiology 1972, 41, 99–104. [Google Scholar]
- Botha, A. Yeasts in soil. In Biodiversity and Ecophysiology of Yeasts; Peter, G., Rosa, C., Eds.; Springer-Verlag: Berlin, Germany, 2006; pp. 221–240. [Google Scholar]
- Di Menna, M.E. Schizoblastosporion starkeyi-henricii Ciferri. Mycopathologia 1964, 25, 205–212. [Google Scholar]
- Babjeva, I.P.; Rheshetova, I.S. A new yeast species Candida podzolica sp. nov. isolated from the soil. Microbiology 1975, 44, 333–338. [Google Scholar]
- Chesworth, W. Encyclopedia of Soil Science, Encyclopedia of Earth Sciences Series; Springer: Dordrecht, Netherlands, 2008; p. 902. [Google Scholar]
- Giller, K.E.; Witter, E.; McGrath, S.P. Heavy metals and soil microbes. Soil Biol. Biochem. 2009, 41, 2031–2037. [Google Scholar] [CrossRef]
- Raspor, P.; Zupan, J. Yeasts in Extreme Environments. In Biodiversity and Ecophysiology of Yeasts; Peter, G., Rosa, C., Eds.; Springer-Verlag: Berlin, Germany, 2006; pp. 371–417. [Google Scholar]
- Kachalkin, A.V.; Glushakova, A.M.; Yurkov, A.M.; Chernov, I.Yu. Characterization of yeast groupings in the phyllosphere of Sphagnum mosses. Microbiology 2008, 77, 474–481. [Google Scholar] [CrossRef]
- Golubev, W.I. Taxonomic evaluation of mycocins produced by basidiomycetous yeast Cryptococcus podzolicus. Mikrobiology 1991, 60, 115–121. [Google Scholar]
- Middelhoven, W.J.; Kurtzman, C.P. Four novel yeasts from decaying organic matter: Blastobotrys robertii sp. nov., Candida cretensis sp. nov., Candida scorzettiae sp. nov. and Candida vadensis sp. nov. Antonie Leeuwenhoek 2007, 92, 233–244. [Google Scholar] [CrossRef]
- Grinbergs, J.; Yarrow, D. Two new Candida species: Candida chilensis sp. n. and Candida valdiviana sp. n. Antonie Leeuwenhoek 1970, 36, 143–148. [Google Scholar] [CrossRef]
- Sampaio, A.; Sampaio, J.P.; Leão, C. Dynamics of yeast populations recovered from decaying leaves in a nonpolluted stream: a 2-year study on the effects of leaf litter type and decomposition time. FEMS Yeast Res. 2007, 7, 595–603. [Google Scholar] [CrossRef]
- Amend, A.S.; Seifert, K.A.; Bruns, T.D. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol. Ecol. 2010, 19, 5555–5565. [Google Scholar] [CrossRef]
- Unterseher, M.; Jumpponen, A.; Opik, M.; Tedersoo, L.; Moora, M.; Dormann, C.F.; Schnittler, M. Species abundance distributions and richness estimations in fungal metagenomics - lessons learned from community ecology. Mol. Ecol. 2011, 10, 275–285. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yurkov, A.; Wehde, T.; Kahl, T.; Begerow, D. Aboveground Deadwood Deposition Supports Development of Soil Yeasts. Diversity 2012, 4, 453-474. https://doi.org/10.3390/d4040453
Yurkov A, Wehde T, Kahl T, Begerow D. Aboveground Deadwood Deposition Supports Development of Soil Yeasts. Diversity. 2012; 4(4):453-474. https://doi.org/10.3390/d4040453
Chicago/Turabian StyleYurkov, Andrey, Thorsten Wehde, Tiemo Kahl, and Dominik Begerow. 2012. "Aboveground Deadwood Deposition Supports Development of Soil Yeasts" Diversity 4, no. 4: 453-474. https://doi.org/10.3390/d4040453





