1. Introduction
Study of the genomics of falcon species is important for understanding a wide range of aspects of falcon biology, including morphology, ecology, and physiology, as well as being essential for conservation efforts. Whole genome sequencing enables us to understand how the genome relates to phenotype, e.g., to growth, development, maintenance, and disease resistance [
1]. Understanding the genome also helps us to study the regulatory regions and “nonsense” regions, comparing genes across species and identifying genetic variants that lead to certain traits [
2]. In addition to the sequence itself, a key aspect of the genomics of any species is its karyotype [
3].
The karyotype is essentially the organization of the genome expressed as an arrangement of chromosomes (usually smallest to largest). The ultimate aim of any de novo genome sequencing effort therefore is to assign all or most of the sequences to the appropriate chromosomes in the karyotype, with each gene or marker in order–in essence, creating a genomic map [
4]. Making a map of the genome in relation to the karyotype (a so-called chromosome-level assembly) can be useful for genotype-phenotype correlations, followed by marker assisted selection in breeding regimes [
5]. Similarly, with chromosome-level assemblies, we can determine patterns of gross genomic evolution between species [
1]. Despite this, many animals, although sequenced, do not have a chromosome-level genome assembly [
6], and until recently, this also applied to falcons. The purposes of this study are therefore both to report on our very recent results in creating chromosome-level assemblies for two falcon species, as well as presenting hitherto unpublished results on chromosome description and comparative genomics between three of the best-known species.
Avian genomes are usually characterized by the large variation in chromosome size, with most species having ~10 pairs of macrochromosomes and ~30 pairs of microchromosomes. The majority of avian karyotypes typically have large number of chromosomes, more than most other vertebrates, with a diploid count of ~2n = 80 in two-thirds of species [
7]. The
Falco genus is however unusual in having lower diploid chromosome numbers ranging from 40–54, with 7–11 pairs of large and medium-sized chromosomes, and about 13–16 pairs of microchromosomes [
8]. Karyotypes of peregrine falcon and prairie falcons purportedly are similar, and, according to Schmutz and Oliphant [
9], differ from gyr falcons by four chromosomes. However, the karyotypes currently available in the literature are of poor quality. Amaral and Jorge summarized karyotypes of 66 species of
Falconiformes that were analyzed between 1966 to 2001 and their results revealed that the low diploid numbers in
Falco species suggest extensive translocation, as well as the fusion of the microchromosomes into larger chromosomes [
10]. Depending on classification, the
Falco genus comprises 37 or 39 species [
11,
12,
13], from which 10 species have reported karyotype data. They are
Falco columbarius (merlin) (2n = 40),
Falco mexicanus (prairie falcon) (2n = 48),
Falco chicquera (red-necked falcon) (2n = 50),
Falco jugger (laggar falcon) (2n = 50),
Falco sparverius (American kestrel) (2n = 50),
Falco subbuteo (Eurasian hobby) (2n = 50),
Falco peregrinus (peregrine falcon) (2n = 50),
Falco rusticolus (gyr falcon) (2n = 52),
Falco tinunculusi (Common kestrel) (2n = 52), and
Falco biarmicus (lanner falcon) (2n = 52 or 54) [
9,
14,
15,
16,
17].
Among falcon species the most comprehensive account to date of the relationship between their chromosomes and those of other birds is given by Nishida et al. [
17]. In that study, molecular cytogenetic characterization of the chromosomal homologies of three
Falco species, the common kestrel (
F. tinnunculus), Peregrine falcon (
F. peregrinus), and merlin falcon (
F. columbarius) characterization was performed while using chromosome paints derived from chicken chromosomes 1–9 and Z.
F. tinnunculus has a karyotype (2n = 52) consisting of all acrocentric (one arm, centromere at the top) chromosomes, except for the submetacentric (bi-armed) W chromosome.
F. peregrinus has a diploid number of (2n = 50), all acrocentric chromosomes except for the one pair of large sub-metacentric macrochromosomes.
F. columbarius has a lower chromosome number (2n = 40), and, unlike those of other species, has six pairs of large bi-armed (sub-metacentric) chromosomes. Nishida et al. [
17] therefore suggested that the ancestral karyotype of
Falco probably had a diploid number of 2n = 52 or 54, consisting of all acrocentric chromosomes, except for the W chromosome.
F. tinnunculus is considered to have retained the most of the ancestral status of
Falconidae karyotypes [
17]. Until recently, however, comparative studies have been limited to the largest chromosomes (1–9 +Z) using whole chromosome paints.
Peregrine (
F. peregrinus) and saker (
F. cherrug) genomes were sequenced around five years ago in an attempt to understand evolutionary aspects of predatory adaptations of falcons [
18]. Sequencing of males of both species was achieved by a next-generation genome sequencing platform, generating genome sizes of both species estimated at 1.2 Gb with a genome coverage of 106.72× for
F. peregrinus and 113.51× for
F. cherrug [
18]. Protein-coding genes were predicted while using homology and de novo methods and RNA sequencing data was used to process gene structure. As a result of this combined effort, 16,263 genes were predicted for
F. peregrinus and 16,204 were predicted for
F. cherrug [
18].
Genome sequences available for
F. peregrinus and
F. cherrug were however not chromosome-level assemblies, but in the form of sub-chromosomal sized scaffolds [
18]. The purpose of this study is first to report on (and review) our recent published findings [
6,
19] in generating chromosome level genome assemblies for
F. peregrinus and
F. cherrug, second to report novel data on the generation of analyzable karyotypes and ideograms of
F. peregrinus,
F. cherrug, and
F. rusticolis, third to generate a novel, low coverage de-novo genome sequence of
F. rusticolis falcon, and finally to perform comparative genomics between all three species (two of which are nested under the subgenus hierofalco (
F. cherrug and
F. rusticolis) and chicken (representing the ancestral avian karyotype).
3. Results
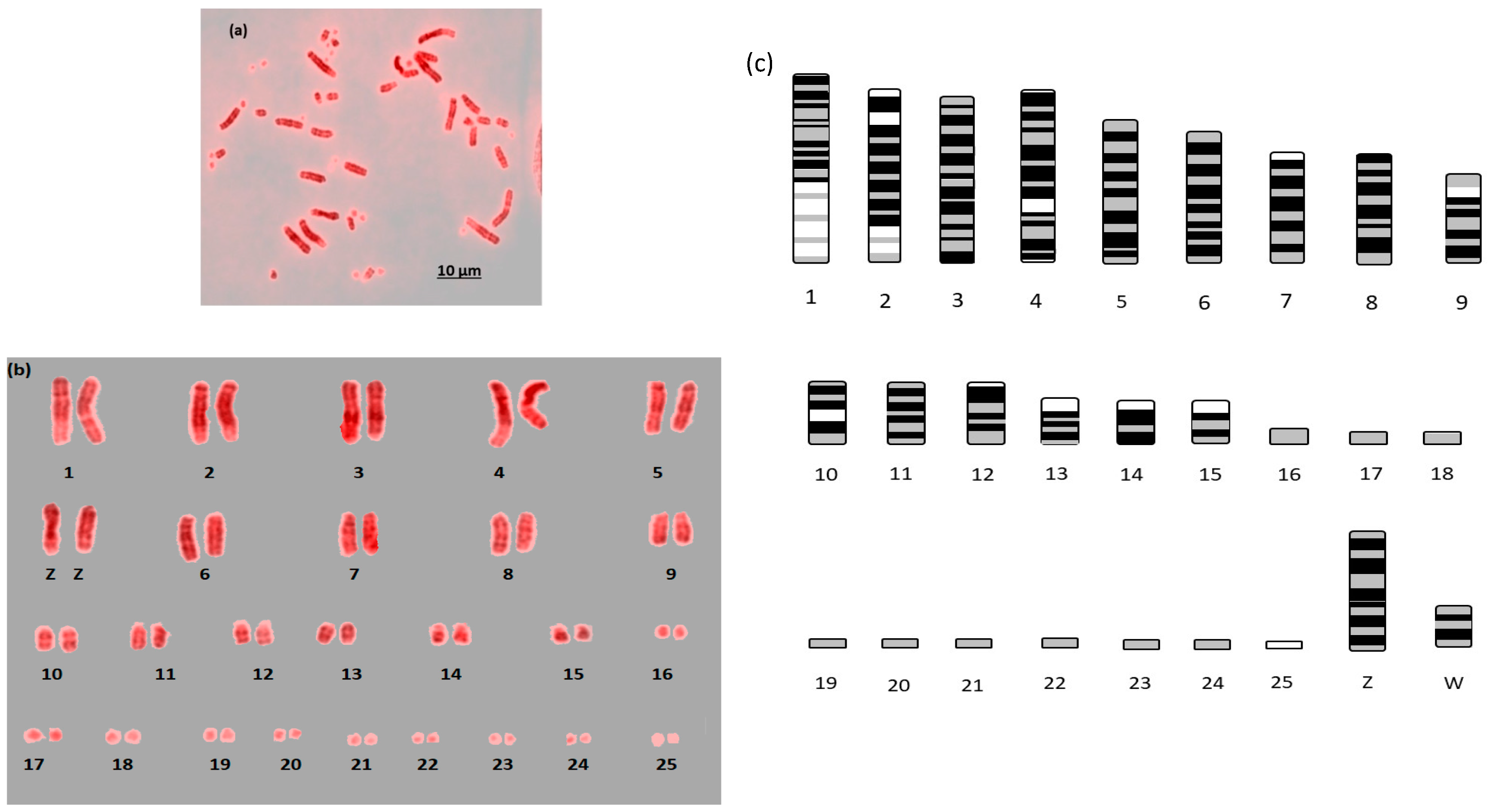
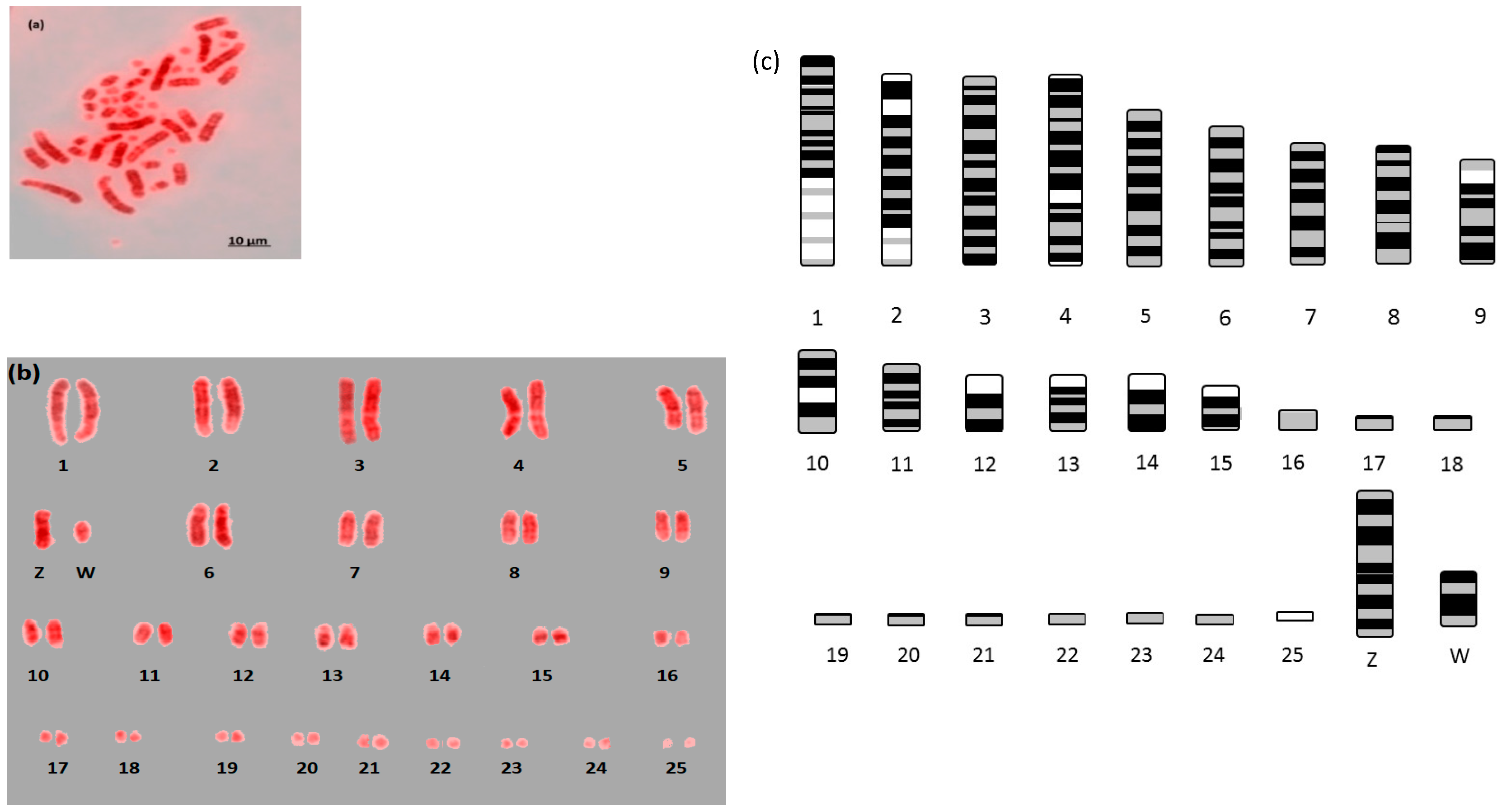
Here we report the first example of near fully analyzable metaphases of three falcon species (
F. peregrinus,
F. cherrug and
F. rusticolis) with diploid numbers of 2n = 50 for
F. peregrinus and 2n = 52 for
F. cherrug and
F. rusticolis (
Figure 1,
Figure 2 and
Figure 3). After some experimentation, a combination of DAPI and propidium iodide gave the sharpest and most distinct banding. Using simple measurement and visual inspection we generated standard ideograms (
Figure 1,
Figure 2 and
Figure 3). In addition, the smallest chromosome using DAPI staining was disproportionally bright on propidium iodide, and it was thus named chromosome 24 in
F. peregrinus and 25 in
F. cherrug and
F. rusticolis.
For F. rusticolis and F. cherrug: Chromosome 1 has a large pale band near the base, there is a similar but smaller pale band for chromosome 2, and chromosome 4 is easy to distinguish because of its pale band in the centre. Chromosomes 10–16 mostly have two dark bands top and bottom (not dissimilar from a human 14 or 18) and they could be distinguished from one another (looking at subtle differences) with some degree of confidence. Chromosomes 17–25 are generally indistinguishable microchromosomes with the exception of chromosome 25, which is much brighter under propidium iodide. Indeed, chromosome 25 was only visible and distinguishable using propidium iodide, similarly the bright portion of the p-arm of chromosome 12 was much brighter. F. peregrinus is similar, with the two fused chromosomes making up chromosome 1 and the inversions taken into account.
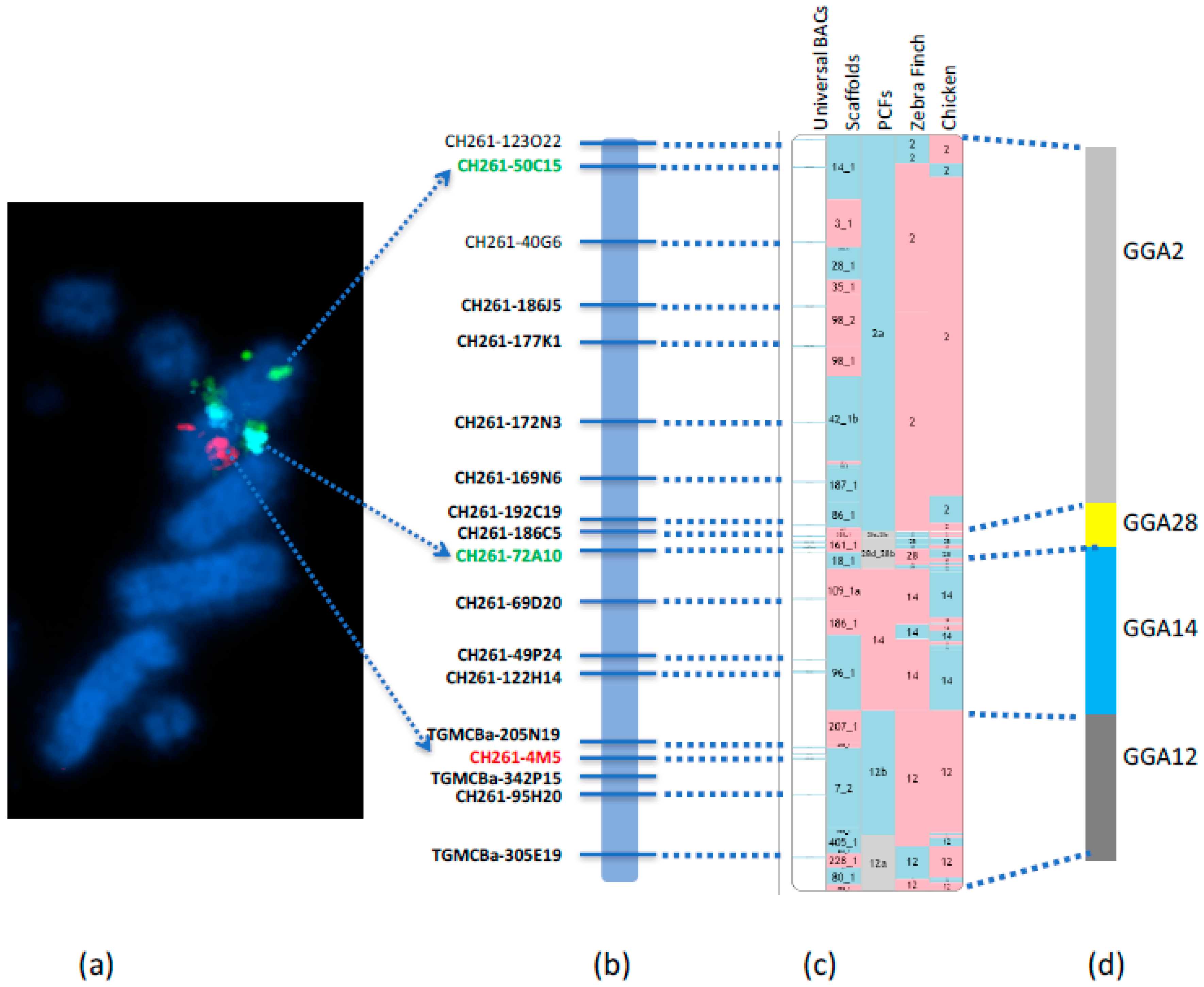
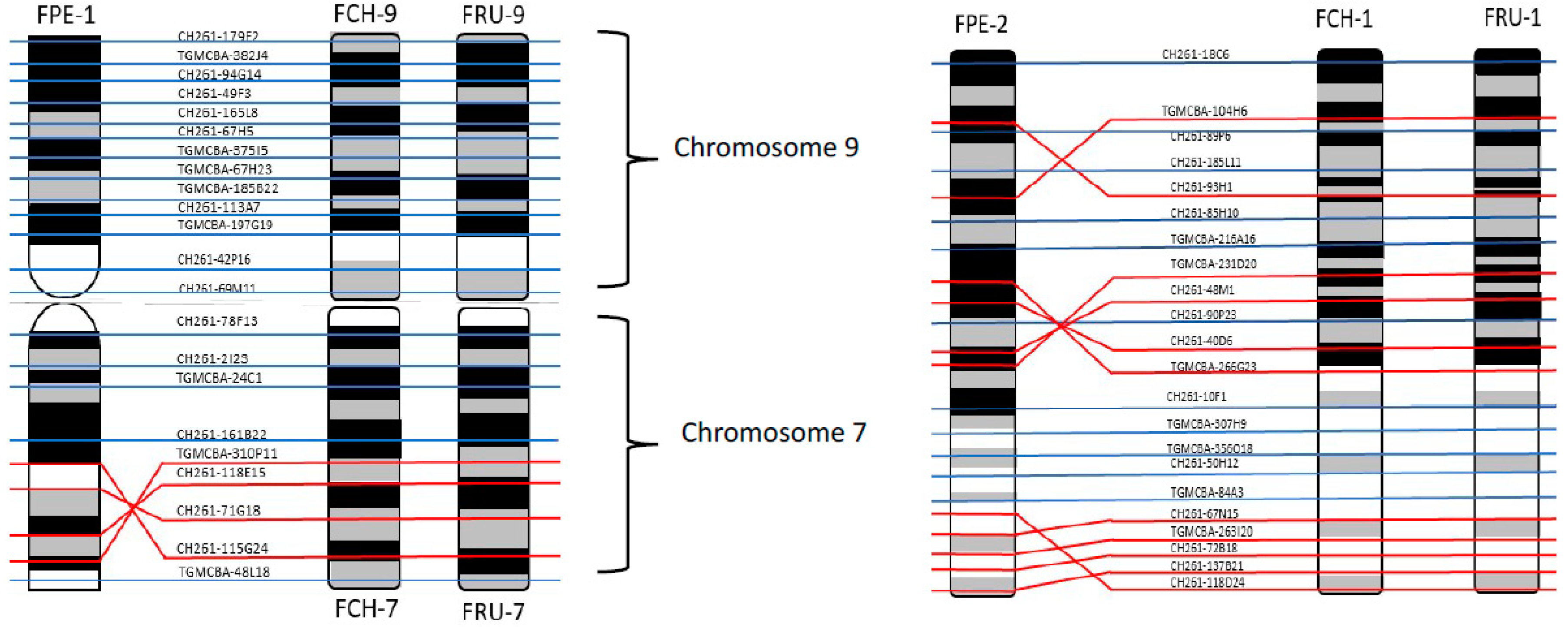
We recently reported the first falcon chromosome level genome assembly [
6] through the development of a new approach to upgrade the scaffold-based
F. peregrinus genome to chromosome level. This was achieved by using RACA to generate PCFs, combined with the verification of scaffolds by PCR and the physical mapping to chromosome by hybridizing with a universal set of chicken BAC probes by FISH (
Figure 4). The Damas et al. [
6] study successfully generated a cytogenetically anchored genome map of the
F. peregrinus, and it was subsequently repeated for the
F. cherrug in 2018 by O’Connor et al. [
19].
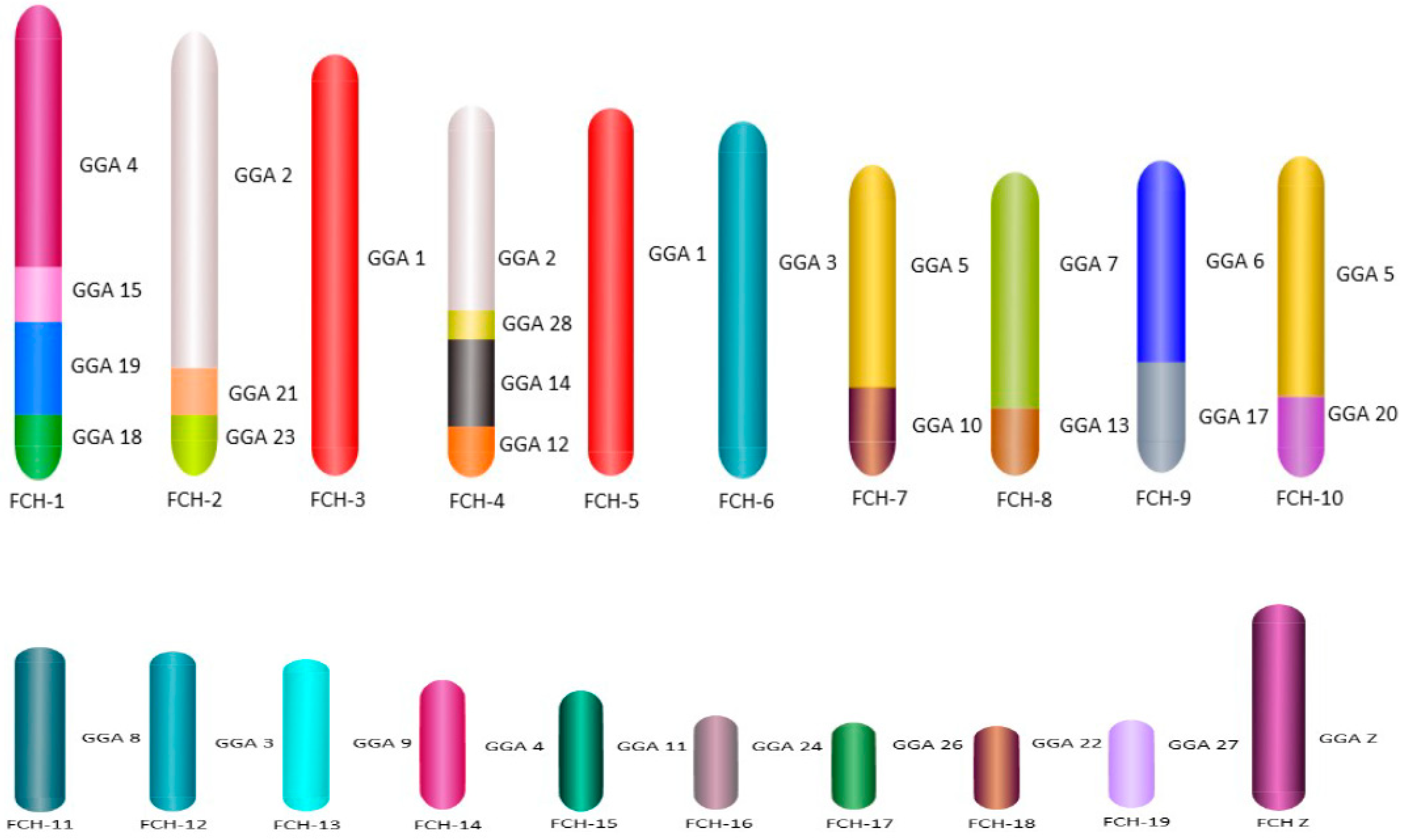
For the first time we carried out an extensive homology study between the three falcon species (
F. peregrinus,
F. cherrug and
F. rusticolis) and chicken (
Figure 5 and
Figure 6). FISH was performed with a selected set of BAC clones developed by Damas et al. [
6] and identified 13
F. peregrinus specific fusions and five fissions when compared to chicken.
F. peregrinus has undergone approximately 38 intrachromosomal rearrangements during the evolution of avian lineages. Comparing homology between the chicken and
F. cherrug showed that in total, 12 fusions, five fissions, and 36 inversions occurred during evolution from their common ancestor. Moreover, out of 17 mapped chicken microchromosomes, 12 were found to be fused with other chromosomes in both species.
Comparative mapping of BAC clones between the three species displayed no inter nor intrachromosomal rearrangements between
F. rusticolis and
F. cherrug. A total of nine intrachromosomal and one interchromosomal changes were identified between
F. peregrinus and the two other species (
Figure 7 and
Figure S1). Finally, we report here, for the first time, the genome sequencing of
F. rusticolus (
Supplementary Materials).
4. Discussion
This study significantly contributed to the understanding of genome organization and evolution in the genus Falco. Specifically, it provides the first example of standard ideograms, analyzable metaphases, chromosome level genome assemblies, and comparative genomics between three species. Our approach combines classical cytogenetics, molecular cytogenetics, and computational algorithms to merge scaffolds into chromosomal fragments. To our knowledge is the only example of genome sequencing in F. rusticolus, and, although a chromosome level assembly was not generated, the evidence shows no difference between the genome organization of F. rusticolus and F. cherrug.
Only partial karyotypes were generated until now for species of the genus Falco. Reasons for a significant improvement in banding patterns are possibly due to the combination of DAPI (which preferentially recognizes AT rich regions of the genome) and propidium iodide (preferentially intercalating between bases), which produced a banding pattern that made karyotyping much clearer. Interestingly, although we have not done an exhaustive study, such a defined banding pattern is not something that we have seen in the chromosomes of other avian species. Whether this is a technical issue borne of the fact that our falcon preparations are fresher than most, or a function of a greater differentiation of AT and GC rich regions in falcons remains to be established.
There are few reports of comparative molecular cytogenetic studies for falcons and only one prior comprehensive comparative FISH study [
17]. That study was limited however in that it compared only the largest chromosomes (using chicken macrochromosome paints). It nonetheless provided a baseline for our current data set. The molecular karyotype that is generated in this study and by Damas et al. [
6] and O’Connor et al. [
19] largely correlates to the preliminary homology study results between chicken and peregrine falcon [
17], but it fills in more of the gaps, in particular for the microchromosomes. The chromosome painting data (not shown) and the BAC data generated in these studies suggests a large degree of similarity in the overall genome organization of
F. cherrug and
F. rusticolus when compared with
F. tinnunculus. Nishida et al. [
17] suggested that
F. tinnunculus had the ancestral falcon karyotype, and, if this is the case, the same would therefore apply to
F. cherrug and
F. rusticolus falcons. The lower diploid number of 2n = 50 found in the
F. peregrinus therefore probably originated from the centric fusion of
F. cherrug/
F. rusticolus chromosomes 7 and 9, forming the metacentric chromosome 1 of the peregrine.
With the genome sequencing of the
F. peregrinus and
F. cherrug, interest in falcon biology has gained tremendous momentum [
18]. Here, we summarize and review our recent efforts to upgrade these scaffold-based genome assemblies to those of the chromosomally assembled genomes for
F. peregrinus and
F. cherrug, with
F. rusticolus implied by its similarity with
F. cherrug. The overall strategy used for scaffold assembly by RACA, and physical mapping using a panel of universal BACs [
6,
20], provides proof of principle for an approach that could be applied to any animal genome. Furthermore, by uploading the chromosomally-assembled genomes to Evolution Highway (
F. cherrug and
F. peregrinus), users will be able to compare multiple species, with falcons, in order to identify evolutionary breakpoint regions and homologous synteny blocks.
One of the primary benefits of whole genome sequences (particularly chromosome-level assemblies) is to provide a better understanding of evolutionary history of genome organization and chromosome structural variation that is caused by chromosome rearrangements [
1]. Multiple projects, including the Bird 10K programme [
21], are working to generate draft genome sequences of thousands of extant bird species over the next ten years using next generation sequencing (NGS) technologies to produce de novo assemblies, some of which will be to chromosome-level. Chromosome-level assemblies are also essential for agricultural species where an established order of DNA markers is required to establish phenotype-to-genotype associations for gene-assisted selection and breeding [
5]. With this information, high-resolution SNP genotyping is very effective for association studies among different species, which in turn facilitated the mapping of Mendelian disorders, accurate identification of (e.g., cryptic) chromosome translocations [
22], discovery of quantitative trait nucleotides (QTNs) and expression quantitative trait loci (eQTLs), and studies of long-range regulatory interactions [
23]. This has resulted in significant economic improvement, more efficient food production, and improved global food security in farm animals [
23]. The same principles could be applied to establish genomic selection and genome-assisted breeding and/or conservation regimes for falcons.
With these assemblies, comparative genomics also becomes possible in silico [
24], particularly when such assemblies are available for multiple species. The comparative genomic maps that are generated here demonstrate the similarities between the three species, with complete synteny between
F. cherrug and
F. rusticolus. Lack of apparent chromosome rearrangements between these two species raises the question of whether they could be considered the same species. Helbig et al. [
25] conducted a phylogenetic relationship study among Falcon species based on cytochrome-b gene variations reporting that the
F. cherrug mtDNA haplotypes are almost identical to those of
F. rusticolus. Further studies by Nittinger and colleagues used control region and microsatellite markers to elucidate the evolutionary patterns within the hierofalco complex [
26,
27]. Moreover,
F. cherrug and
F. rusticolus falcons can produce fertile hybrids in the wild as well as in captivity with extended viability over indefinite generations [
28,
29]. The detection of hybrid falcons is becoming increasingly important in falcon racing. The nine intrachromosomal differences that were identified between
F. peregrinus and
F. cherrug/
F. rusticolus could theoretically form the basis for establishing a testing device (FISH based) that could detect hybrids. Such a device could have eight spatially separated hybridization chambers, each of which carries specific DNA FISH probes that are labelled and designed to identify
F. peregrinus and
F. cherrug/
F. rusticolus chromosomes (one for the fusion and seven for the inversions). In recent studies, we have performed multiple (up to 24) hybridizations on single slides for chromosome translocation screening in domestic animals [
22], suggesting that a falcon hybrid detection device would be possible to manufacture. Developing such a testing tool to identify
F. rusticolus ×
F. cherrug falcon hybrids would not be possible as a result of there being no apparent intrachromosomal differences between them.
Falconidae and
Accipitridae, together with
Psittaciformes members, are recognized as avian species with ‘atypical’ karyotypes and previous studies have shown that these avian species have the highest numbers of rearrangements occurring on their macrochromosomes [
17,
30,
31,
32]. Recent studies conducted on such ‘atypical’ karyotype species collectively highlight the substantial amount of rearrangements of macrochromosomes that have occurred throughout their evolutionary history. Until now, the only information available was that of their homology to chicken chromosomes GGA 1–9 and Z, while knowledge of their microchromosomal rearrangements has been limited until this and our other recent studies [
6,
20]. By performing microchromosome analysis and thus evaluating the inter-and intrachromosomal relationship between more falcon species,
Gyps species and
Psittaciformes our understanding of avian chromosome evolution will improve. Moreover, further analysis of the recent availability of several sequenced bird genomes, such as the White-tailed eagle (
Accipitriformes), Kea (
Psittaciformes), and Turkey vulture (
Cathartiformes) [
33], provide additional opportunities for insight into the role of evolutionarily conserved synteny blocks in avian evolution. Romanov et al. [
34] suggested that chicken was most similar to the common ancestor when compared to ostrich, turkey, duck, budgerigar, and zebra finch. That is, the chicken lineage underwent the fewest number of chromosomal rearrangements. The comparison therefore of falcon interchromosomal and intrachromosomal rearrangements against chicken in this study gives an indication of the number of changes that have occurred in the lineage of the falcons.
In conclusion, the results that are presented here represent the most comprehensive account of chromosomally related genome mapping in falcon species to date. The ideograms presented in the text, the fact that the comparative genomic information is presented on an interactive browser (
http://eh-demo.ncsa.uiuc.edu/birds/#/SynBlocks) and the freely available
F. rusticolus data provide a useful resource for the scientific community.