Olive Tree (Olea europaea L.) Diversity in Traditional Small Farms of Ficalho, Portugal
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphological Study
2.2. DNA Extraction, SSR Markers, PCR Amplification and Fragment Sizing
2.3. Data Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- International Olive Council (IOC). Market Newsletter; International Olive Council: Madrid, Spain, 2017; Volume 121, p. 6. [Google Scholar]
- Instituto Nacional de Estatística (INE). EstatísticasAgrícolas 2016, 1st ed.; Instituto Nacional de Estatística: Lisboa, Portugal, 2017; p. 171. [Google Scholar]
- Breton, C.; Pinatel, C.; Médail, F.; Bonhomme, F.; Bervillé, A. Comparison between classical and Baysean methods to investigate the history of olive cultivars using SSR-polymorphisms. Plant Sci. 2008, 175, 524–532. [Google Scholar] [CrossRef]
- Moreira, P.M.R.M.; Veloso, M.M. Landraces inventory for Portugal. In European Landraces: On Farm Conservation, Management and Use, 1st ed.; Vetelainen, M., Negri, V., Maxted, N., Eds.; Bioversity Technical Bulletin No. 15; Bioversity International: Rome, Italy, 2009; pp. 124–136. ISBN 978-92-9043-805-2. [Google Scholar]
- Matos, M. As oliviculturas nacionais: Uma nova realidade em Portugal. In Olival Tradicional: Contextos, Realidades e Sustentabilidade, 1st ed.; Rota do Guadiana, Ed.; Rota do Guadiana: Serpa, Portugal, 2014; pp. 47–53. ISBN 978-989-98484-1-2. [Google Scholar]
- Duarte, F.; Jones, N.; Fleskens, L. Traditional olive orchards on sloping land: Sustainability or abandonment? J. Environ. Manag. 2008, 89, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Gemas, V.J.; Rijo-Johansen, M.J.; Tenreiro, R.; Fevereiro, P. Inter and intra-varietal analysis of three Olea europaea L. cultivars using RAPD technique. J. Hortic. Sci. Biotechnol. 2000, 75, 312–319. [Google Scholar] [CrossRef]
- Cidraes, F.G. Estudo das variedades de oliveira do Baixo Alentejo, Região de Serpa. In Brigada Técnica da XVI Região Beja; Série II, Numero 5; Direcção Geral dos Serviços Agrícolas: Beja, Portugal, 1939. [Google Scholar]
- Veloso, M.M. Os agroecosistemas tradicionais na conservação da diversidadegenética da oliveira (Olea europaea) em Vila Verde de Ficalho. In OlivalTradicional: Contextos, Realidades e Sustentabilidade, 1st ed.; Rota do Guadiana, Ed.; Rota do Guadiana: Serpa, Portugal, 2014; pp. 147–153. ISBN 978-989-98484-1-2. [Google Scholar]
- Veloso, M.M.; Reis, P.; Machado, D. On farm conservation of the olive tree (Olea europaea) landraces at Vila Verde de Ficalho (Portugal). Landraces 2015, 3, 16–17. [Google Scholar]
- Fendri, M.; Trujillo, I.; Trigui, A.; Rodriguez-Garcia, I.M.; Ramirez, J.D.A. Simple Sequence Repeat identification and endocarp characterization of olive accessions in a Tunisian germplasm collection. Hortscience 2010, 45, 1429–1436. [Google Scholar]
- Trujillo, I.; Ojeda, M.A.; Urdiroz, N.M.; Potter, D.; Barranco, D.; Rallo, L.; Diez, C.M. Identification of the worlwide olive germplasm bank of Córdoba (Spain) using SSR and morphological markers. Tree Genet. Genomes 2014, 10, 141–155. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lopes, M.S.; Mendonça, D.; Rodrigues Dos Santos, M.; Da Câmara Machado, M.L.; Da Câmara Machado, A. Identification of microsatellite loci in olive (Olea europaea L.) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, R.; James, C.; Tobutt, K.R. Isolation and characterization of polymorphic microsatellite in Olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol. Ecol. Notes 2002, 2, 265–267. [Google Scholar] [CrossRef]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojda, M.; et al. Consensus list of microsatellite markers for olive genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Poljuha, D.; Sladonja, B.; Setic, E.; Milotic, A.; Bandelj, D.; Jakse, J.; Javornik, B. DNA fingerprinting of olive varieties in Istria (Croatia) by microsatellite markers. Sci. Hortic. 2008, 115, 223–230. [Google Scholar] [CrossRef]
- Bracci, T.; Sebastiani, L.; Busconi, M.; Fogher, C.; Belaj, A.; Trujillo, I. SSR markers reveal the uniqueness of olive cultivars from the Italian region of Liguria. Sci. Hortic. 2009, 122, 209–215. [Google Scholar] [CrossRef]
- Delgado-Martinez, F.J.; Amaya, I.; Sánchez-Sevilla, J.F.; Gomez-Jimenez, M.C. Microsatellite marker-based identification and genetic relationships of olive cultivars from the Extremadura region of Spain. Genet. Mol. Res. 2012, 11, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Roubos, K.; Moustakas, M.; Aravanopoulos, F.A. Molecular identification of greek olive (Olea europaea) cultivars based on microsatellite loci. Genet. Mol. Res. 2010, 9, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Bari, A.; Martin, A.; Boulouha, B.; Gonzalez-Andujar, J.L.; Barranco, D.; Ayad, G.; Padulosi, S. Use of fractals and moments to describe olive cultivars. J. Agric. Sci. 2003, 141, 63–71. [Google Scholar] [CrossRef]
- Navero, D.B.; Cimato, A.; Fiorino, P.; Romero, L.R.; Touzani, A.; Castañeda, C.; Serafin, F.; Trujillo, I. World Catalogue of Olive Varieties, 1st ed.; International Olive Oil Council: Madrid, Spain, 2000; pp. 15–21. ISBN 84-931663-3-2. [Google Scholar]
- Rohlf, J.F. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.1; Exeter Software: Setauket, NY, USA, 2000. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Minch, E.; Ruiz-Linares, A.; Goldstein, D.; Feldman, M.; Cavalli-Sforza, L.L. MICROSAT: A Computer Program for Calculating Various Statistics on Microsatellite Allele Data (Version 1.5d); Stanford University: Stanford, CA, USA, 1997. Available online: http://hpgl.stanford. edu/projects/microsat (accessed on 15 April 2016).
- Perrier, X.; Jacquemoud-Collet, J.P.D. ARwin Software. 2006. Available online: http://darwin.cirad.fr/ (accessed on 20 June 2016).
- Cantini, C.; Cimato, A.; Sani, G. Morphological evaluation of olive germplasm present in Tuscany region. Euphytica 1999, 109, 173–181. [Google Scholar] [CrossRef]
- Lopes, S.M.; Mendonça, D.; Sefc, K.M.; Gil, S.F.; Câmara-Machado, A. Genetic evidence of intra cultivar variability within Iberian olive cultivars. HortScience 2004, 39, 1562–1565. [Google Scholar]
- Gomes, S.; Martins-Lopes, P.; Lopes, J.; Guedes-Pinto, H. Assessing genetic diversity in Oleaeuropaea L. using ISSR and SSR markers. Plant Mol. Biol. Report. 2009, 27, 365–373. [Google Scholar] [CrossRef]
- Erre, P.; Chessa, I.; Muñoz-Diez, C.; Belaj, A. Genetic diversity and relationships between wild and cultivated olives in Sardinia as assessed by SSR markers. Genet. Resour. Crop Evol. 2010, 57, 41–54. [Google Scholar] [CrossRef]
- Gemas, V.J.V.; Almadanim, M.C.; Tenreiro, R.; Martins, A.; Fevereiro, P. Genetic diversity in the Olive tree (Olea europaea L. subsp. europaea) cultivated in Portugal revealed by RAPD and ISSR markers. Genet. Resour. Crop Evol. 2004, 51, 501–511. [Google Scholar] [CrossRef]
- Martins-Lopes, P.; Lima-Brito, J.; Gomes, S.; Meirinhos, J.; Santos, L.; Guedes-Pinto, H. RAPD and ISSR molecular markers in Olea europaea L.: Genetic variability and molecular cultivar identification. Genet. Resour. Crop. Evol. 2007, 54, 117–128. [Google Scholar] [CrossRef]
- Belaj, A.; Satovic, Z.; Rallo, L.; Trujillo, I. Genetic diversity and relationships in olive (Olea europaea L.) germplasm collections as determined by randomly amplified polymorphic DNA. Theor. Appl. Genet. 2002, 105, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Terzopoulos, P.J.; Kolano, B.; Bebeli, P.J.; Kaltsikes, P.J. Identification of Olea europaea L. cultivars using inter simple sequence repeat markers. Sci. Hortic. 2005, 105, 45–51. [Google Scholar] [CrossRef]
- Belaj, A.; Muñoz-Diez, C.; Baldoni, L.; Satovic, Z.; Barranco, D. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Sci. Hortic. 2010, 124, 323–330. [Google Scholar] [CrossRef]
- Bakkali, A.; Haouane, H.; Hadiddou, A.; Oukabli, A.; Santoni, S.; Udupa, S.M.; Van Damme, P.; Khadari, B. Genetic diversity of on-farm selected olive trees in Moroccan traditional olive orchards. Plant Genet. Resour. 2012, 11, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Díez, C.M.; Trujillo, I.; Barrio, E.; Belaj, A.; Barranco, D.; Rallo, L. Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 2011, 108, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Ouazzani, N.; Lumaret, R.; Villemur, P. Genetic variation in the olive tree (Olea europaea L.) cultivated in Morocco. Euphytica 1996, 91, 9–20. [Google Scholar] [CrossRef]
- Khadari, B.; Charafi, J.; Moukhli, A.; Ater, M. Substantial genetic diversity in cultivated Moroccan olive despite a single major cultivar: A paradoxical situation evidenced by the use of SSR loci. Tree Genet. Genomes 2008, 4, 213–221. [Google Scholar] [CrossRef]
- Corrado, G.; La Mura, M.; Ambrosino, O.; Pugliano, G.; Varrichio, P.; Rao, R. Relationships of Campanian olive cultivars: Comparative analysis of molecular and phenotypic data. Genome 2009, 52, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Bellon, M.R.; Gotor, E.; Caracciolo, F. Conserving landraces and improving livelihoods: How to assess the success of on-farm conservation projects? Int. J. Agric. Sustain. 2015, 13, 167–182. [Google Scholar] [CrossRef]
- Zhang, D.; Gardini, E.A.; Motilal, L.A.; Baligar, V.; Bailey, B.; Zuñiga-Cernades, L.; Arevalo-Arevalo, C.E.; Meinhardt, L. Dissecting genetic structure in farmer selections of Theobroma cacao in the Peruvian Amazon: Implications for on farm conservation and rehabilitation. Trop. Plant Biol. 2011, 4, 106–116. [Google Scholar] [CrossRef]
- Baldoni, L.; Tosti, N.; Ricciolini, N.; Belaj, A.; Arcioni, S.; Pannelli, G.; Germana, M.A.; Mulas, M.; Porceddu, A. Genetic structure of wild and cultivated olives in the Central Mediterranean Basin. Ann. Bot. 2006, 98, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Achtak, H.; Ater, M.; Oukabli, A.; Santoni, S.; Kjellberg, F.; Khadari, B. Traditional agroecosystems as conservatories and incubators of cultivated plant varietal diversity: The case of fig (Ficus carica L.) in Morocco. BMC Plant Biol. 2010, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Almandanim, M.C.; Baleiras-Couto, M.M.; Pereira, H.S.; Carneiro, L.C.; Fevereiro, P.; Eiras-Dias, J.E.; Morais-Cecílio, L.; Viegas, W.; Veloso, M.M. Genetic diversity of the grapevine (Vitis vinifera L.) cultivars most utilized for wine production in Portugal. Vitis 2007, 46, 116–119. [Google Scholar]
- Queiroz, A.; Assunção, A.; Ramadas, I.; Viegas, W.; Veloso, M.M. Molecular characterization of Portuguese pear landraces (Pyrus communis L.) using SSR markers. Sci. Hortic. 2015, 183, 72–76. [Google Scholar] [CrossRef]
- Monteiro, F.; Vidigal, P.; Barros, A.B.; Monteiro, A.; Oliveira, H.R.; Viegas, W. Genetic distinctiveness of rye in situ accessions from Portugal unveils a hotspot of unexplored genetic resources. Front. Plant Sci. 2016, 7, 1334. [Google Scholar] [CrossRef] [PubMed]
- Leitão, S.T.; Dinis, M.; Veloso, M.M.; Satovic, Z.; Vaz-Patto, M.C. Establishing the bases for introducing the unexplored portuguese common bean germplasm into the breeding world. Front. Plant Sci. 2017, 8, 1296. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, A.; Santos, M.L.; Morais, N.; Miranda, A. As variedades de oliveira, Portugal oleícola. In O Grande Livro da Oliveira e do Azeite, 1st ed.; Bohm, J., Godinho, C., Coelho, F., Eds.; Dinalivro: Lisboa, Portugal, 2013; pp. 188–220. ISBN 9789725766200. [Google Scholar]
- Cordeiro, A.; Inês, C.; Morais, N. Principais cultivares de oliveira existentes em Portugal. In Boas Práticas No Olival e No Lagar, 1st ed.; InstitutoNacional de Investigação Agrária e Veterinária: Oeiras, Portugal, 2014; pp. 44–54. ISBN 978-972-579-041-0. [Google Scholar]
 50 ha of olive tree with 60 trees/ha;
50 ha of olive tree with 60 trees/ha;  50 ha of olive tree with 61 to 100 trees/ha;
50 ha of olive tree with 61 to 100 trees/ha;  50 ha of olive tree with 101 to 300 trees/ha;
50 ha of olive tree with 101 to 300 trees/ha;  50 ha of olive tree with more than 300 trees/ha.
50 ha of olive tree with more than 300 trees/ha.
 50 ha of olive tree with 60 trees/ha;
50 ha of olive tree with 60 trees/ha;  50 ha of olive tree with 61 to 100 trees/ha;
50 ha of olive tree with 61 to 100 trees/ha;  50 ha of olive tree with 101 to 300 trees/ha;
50 ha of olive tree with 101 to 300 trees/ha;  50 ha of olive tree with more than 300 trees/ha.
50 ha of olive tree with more than 300 trees/ha.
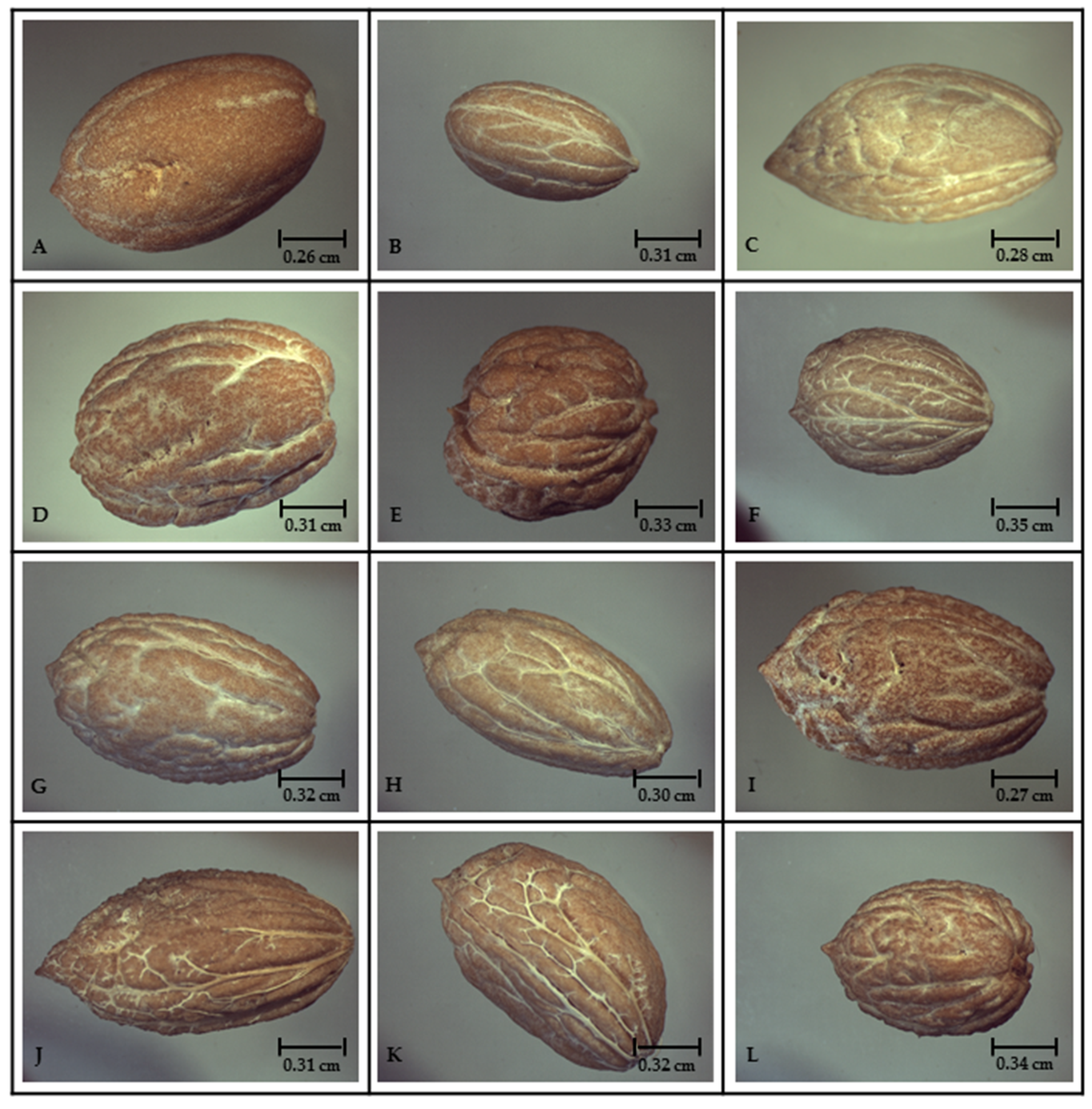

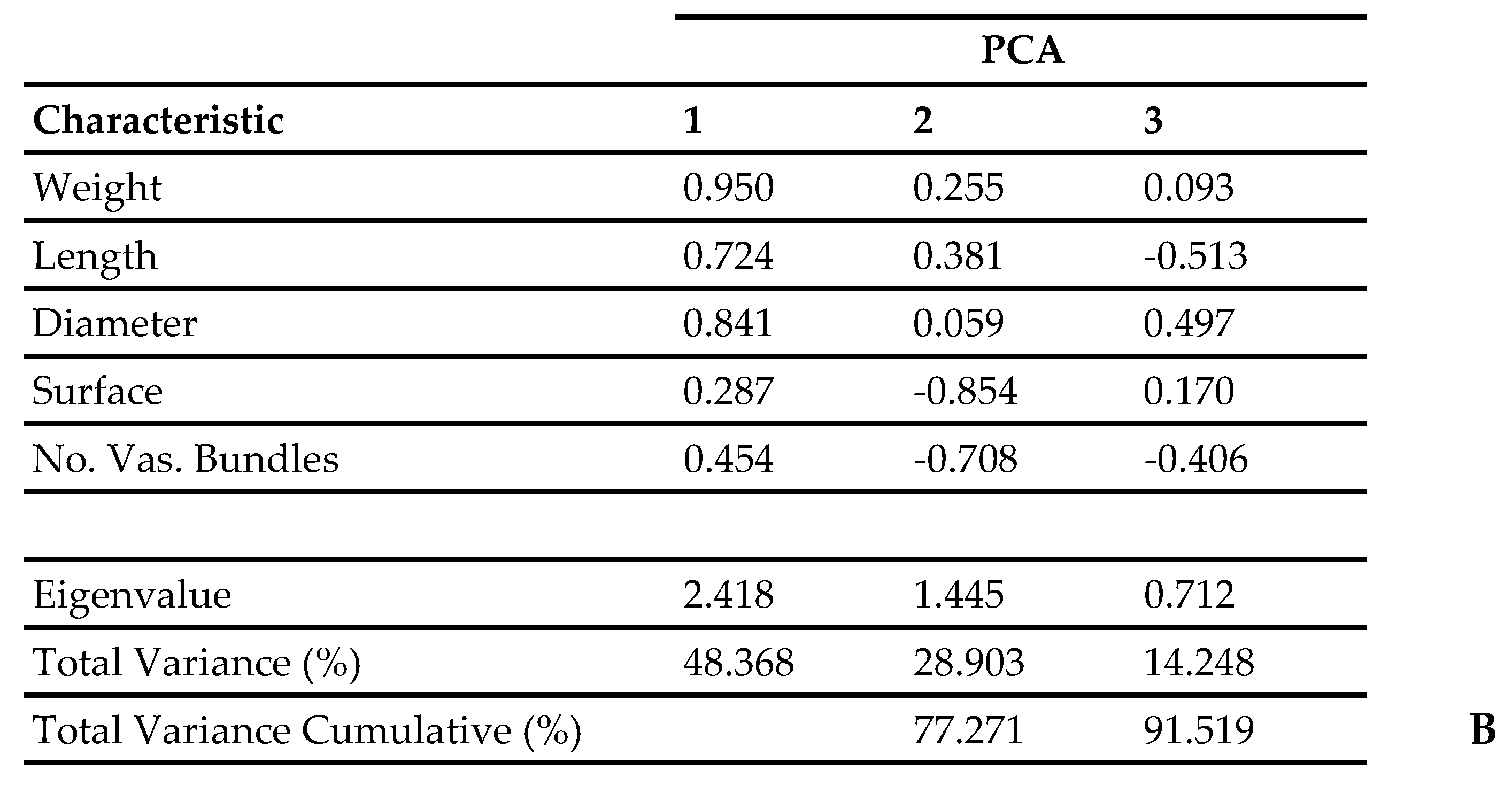
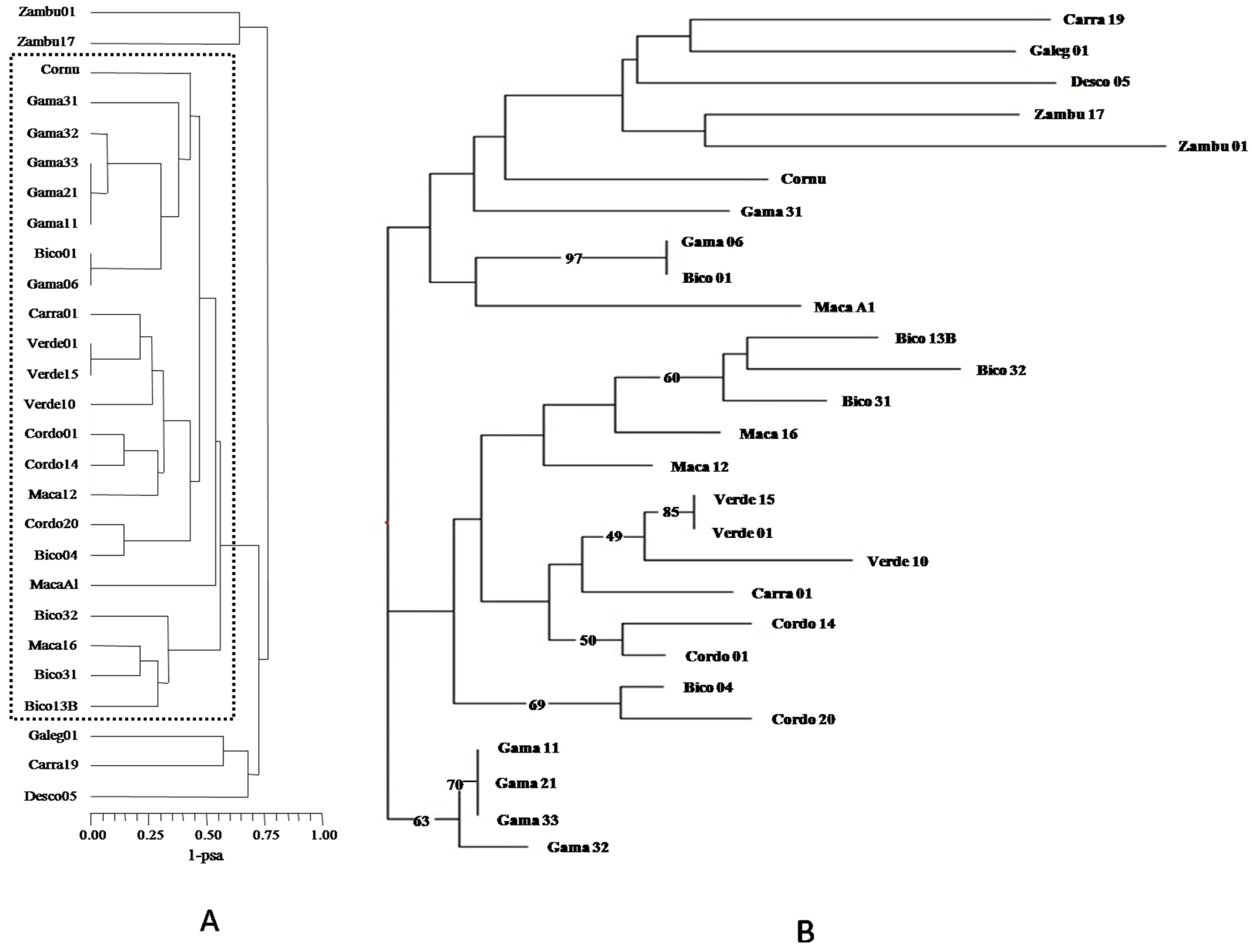

| Olive Tree | Designations | Area of Cultivation |
|---|---|---|
| Local Cultivars | Bico de Corvo | Traditional orchards, Ficalho |
| Carrasquenha | ||
| Cordovil de Serpa | ||
| Galega | ||
| Gama | ||
| Maçanilha | ||
| Verdeal Alentejana | ||
| From other Regions | Cornezuelo | Germplasm collection, Herdade da Abóboda * |
| Maçanilha Algarvia | ||
| Unnamed Tree | Desconhecida | Traditional orchards, Ficalho |
| Wild | Zambujeiro | Traditional orchards, Ficalho |
| Locus | N | Na | Ne | Npa | Ho | He | PIC |
|---|---|---|---|---|---|---|---|
| OeUA-DCA04 | 27 | 6.000 | 2.666 | 1 | 0.222 | 0.578 | 0.538 |
| OeUA-DCA05 | 27 | 5.000 | 1.211 | 3 | 0.185 | 0.177 | 0.170 |
| OeUA-DCA14 | 27 | 7.000 | 2.285 | 1 | 0.667 | 0.671 | 0.618 |
| OeUA-DCA16 | 27 | 14.000 | 5.080 | 6 | 0.667 | 0.819 | 0.782 |
| OeUA-DCA18 | 27 | 9.000 | 3.636 | 4 | 0.630 | 0.739 | 0.684 |
| GAPU71-B | 27 | 5.000 | 4.599 | - | 1.000 | 0.797 | 0.747 |
| GAPU103-A | 27 | 6.000 | 3.984 | 1 | 0.704 | 0.753 | 0.693 |
| Axes | 1 | 2 | 3 |
|---|---|---|---|
| % | 39.78 | 19.63 | 12.74 |
| Cum % | 39.78 | 59.41 | 72.15 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veloso, M.M.; Simões-Costa, M.C.; Carneiro, L.C.; Guimarães, J.B.; Mateus, C.; Fevereiro, P.; Pinto-Ricardo, C. Olive Tree (Olea europaea L.) Diversity in Traditional Small Farms of Ficalho, Portugal. Diversity 2018, 10, 5. https://doi.org/10.3390/d10010005
Veloso MM, Simões-Costa MC, Carneiro LC, Guimarães JB, Mateus C, Fevereiro P, Pinto-Ricardo C. Olive Tree (Olea europaea L.) Diversity in Traditional Small Farms of Ficalho, Portugal. Diversity. 2018; 10(1):5. https://doi.org/10.3390/d10010005
Chicago/Turabian StyleVeloso, Maria Manuela, Maria Cristina Simões-Costa, Luís C. Carneiro, Joana B. Guimarães, Célia Mateus, Pedro Fevereiro, and Cândido Pinto-Ricardo. 2018. "Olive Tree (Olea europaea L.) Diversity in Traditional Small Farms of Ficalho, Portugal" Diversity 10, no. 1: 5. https://doi.org/10.3390/d10010005






