Branched-chain Amino Acid Biosensing Using Fluorescent Modified Engineered Leucine/Isoleucine/Valine Binding Protein
Abstract
:1. Introduction
2. Results and Discussion
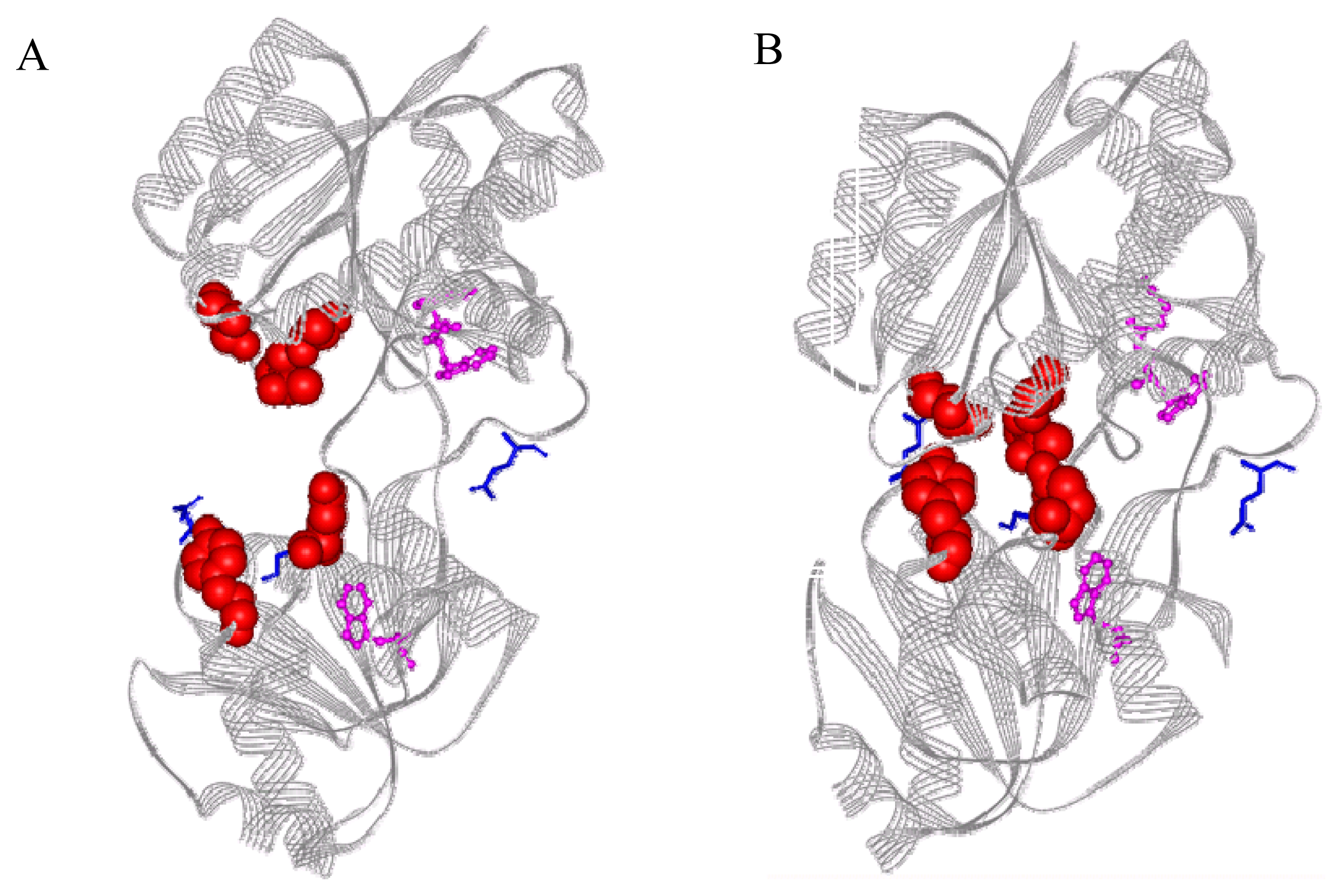
2.1 Fluorophore modification sites
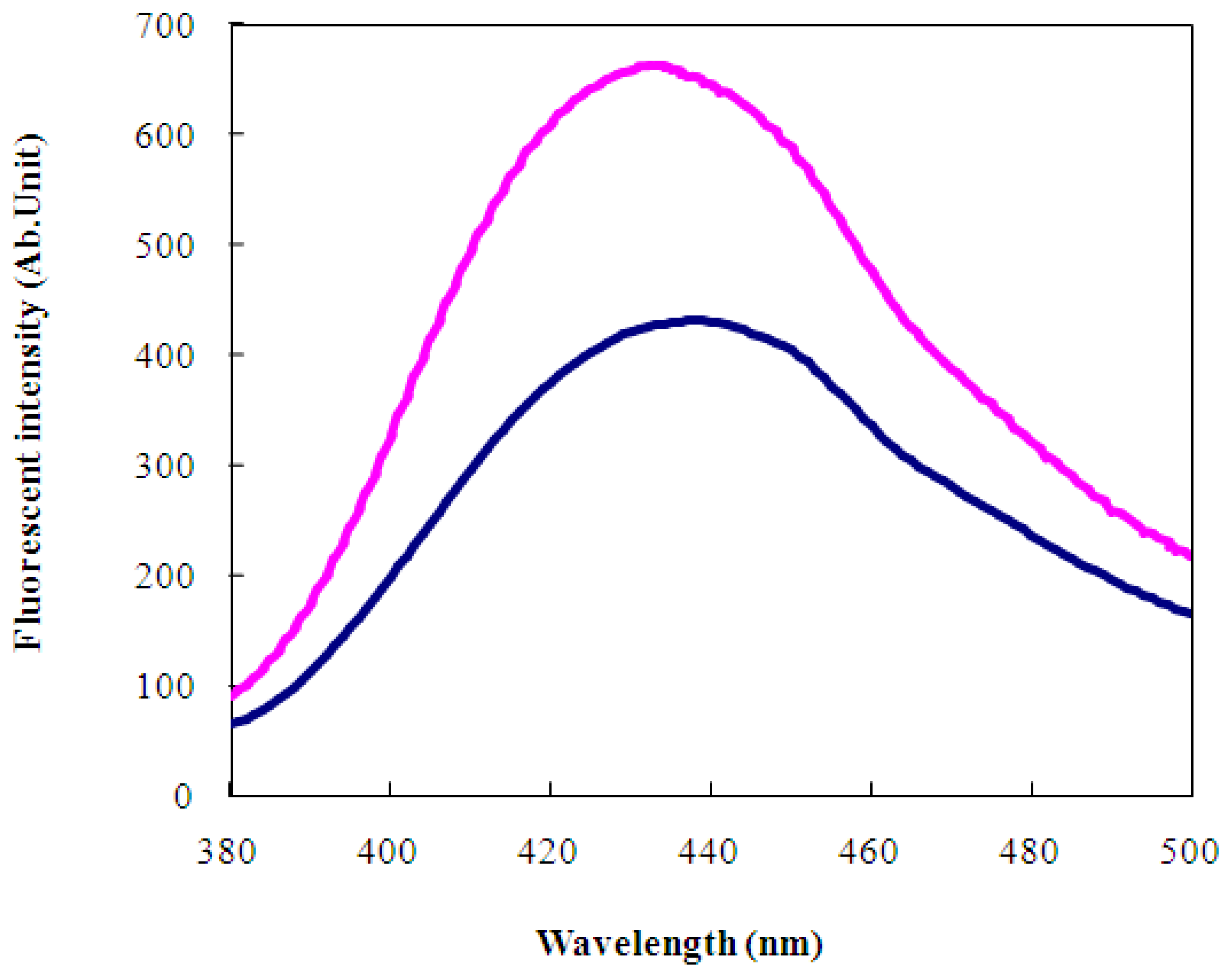
2.2 Characterization of wild-type and engineered LIVBPs by autofluorescence
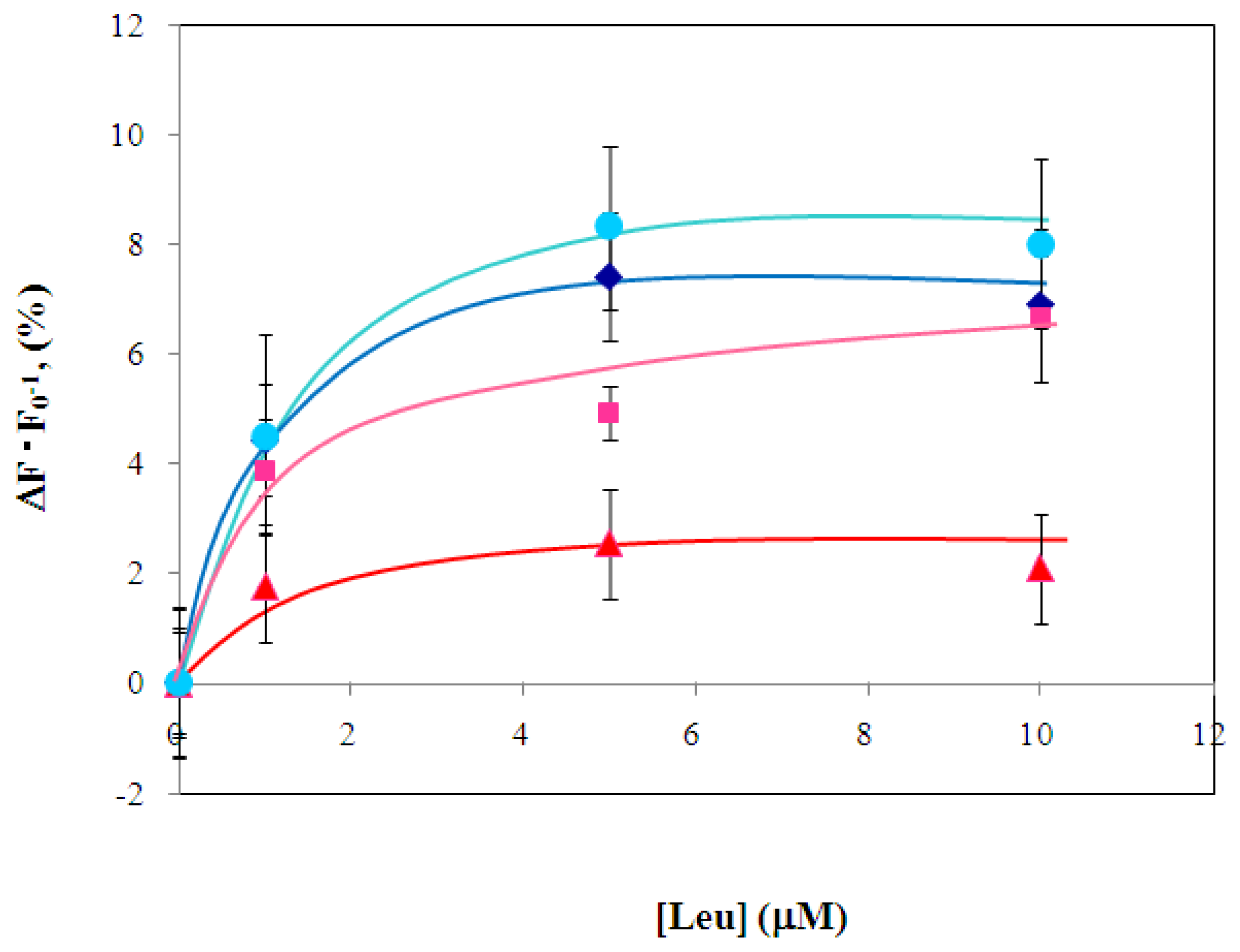
2.3 Characterization of environmentally sensitive fluorescence probe-modified LIVBPs
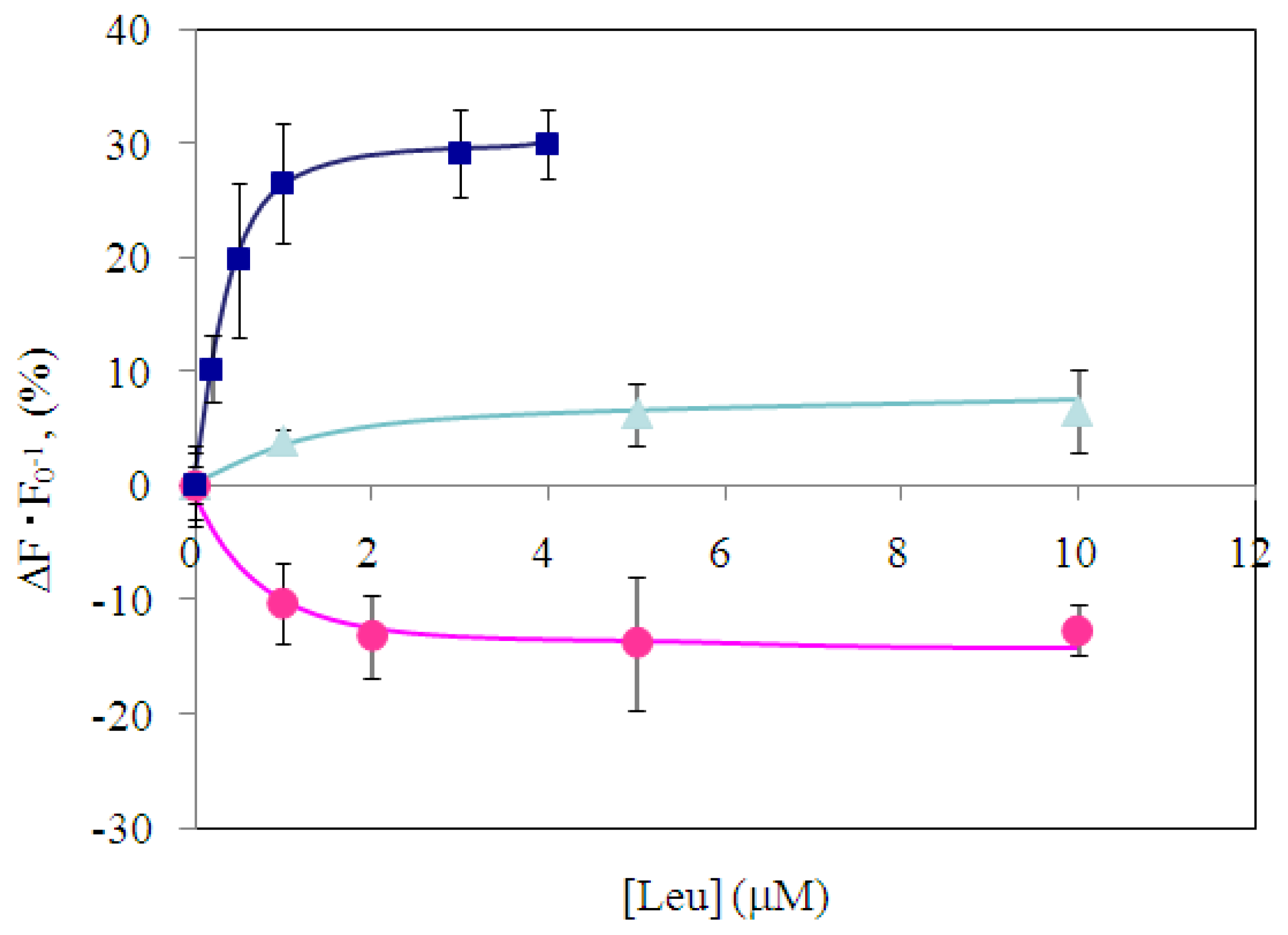
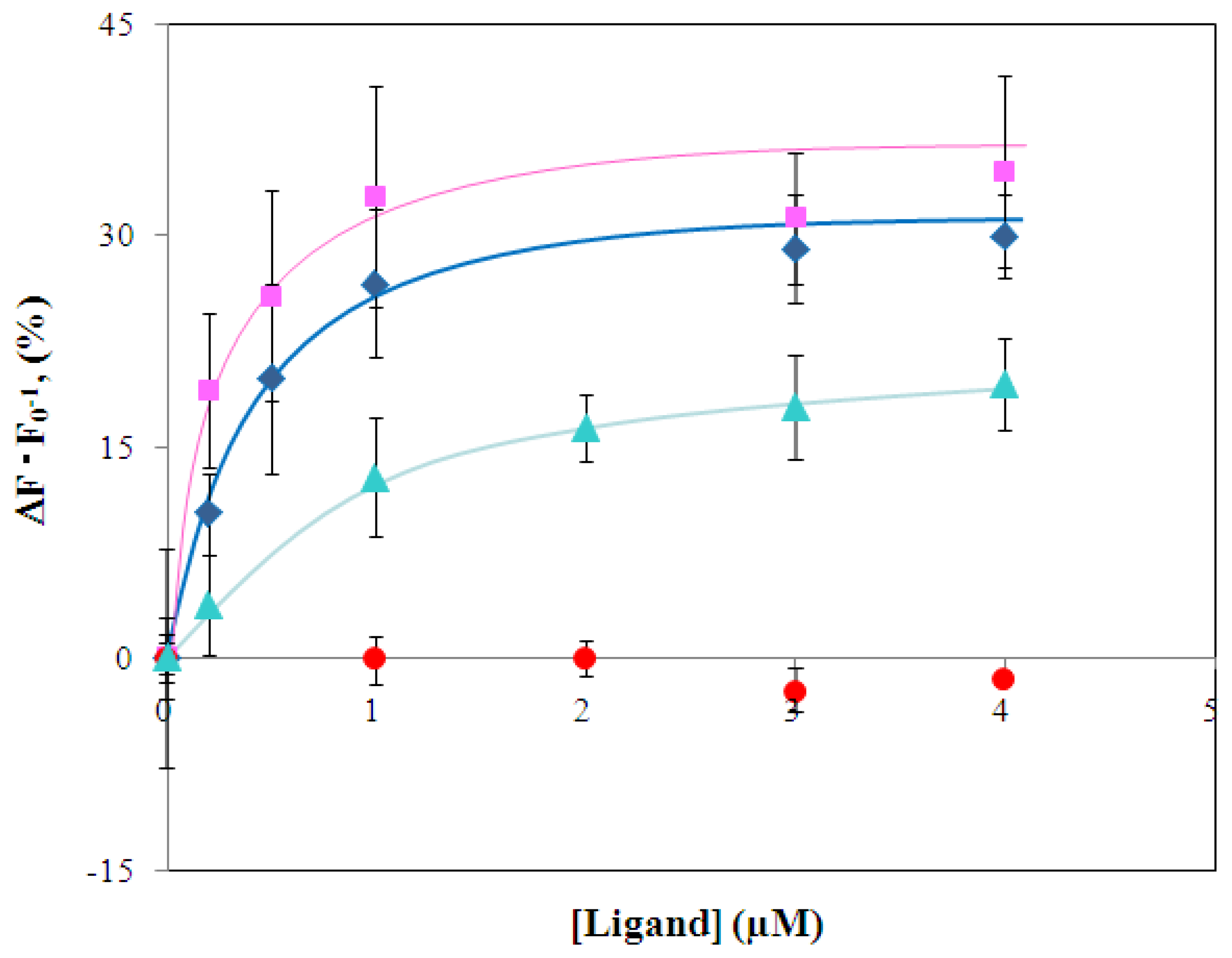
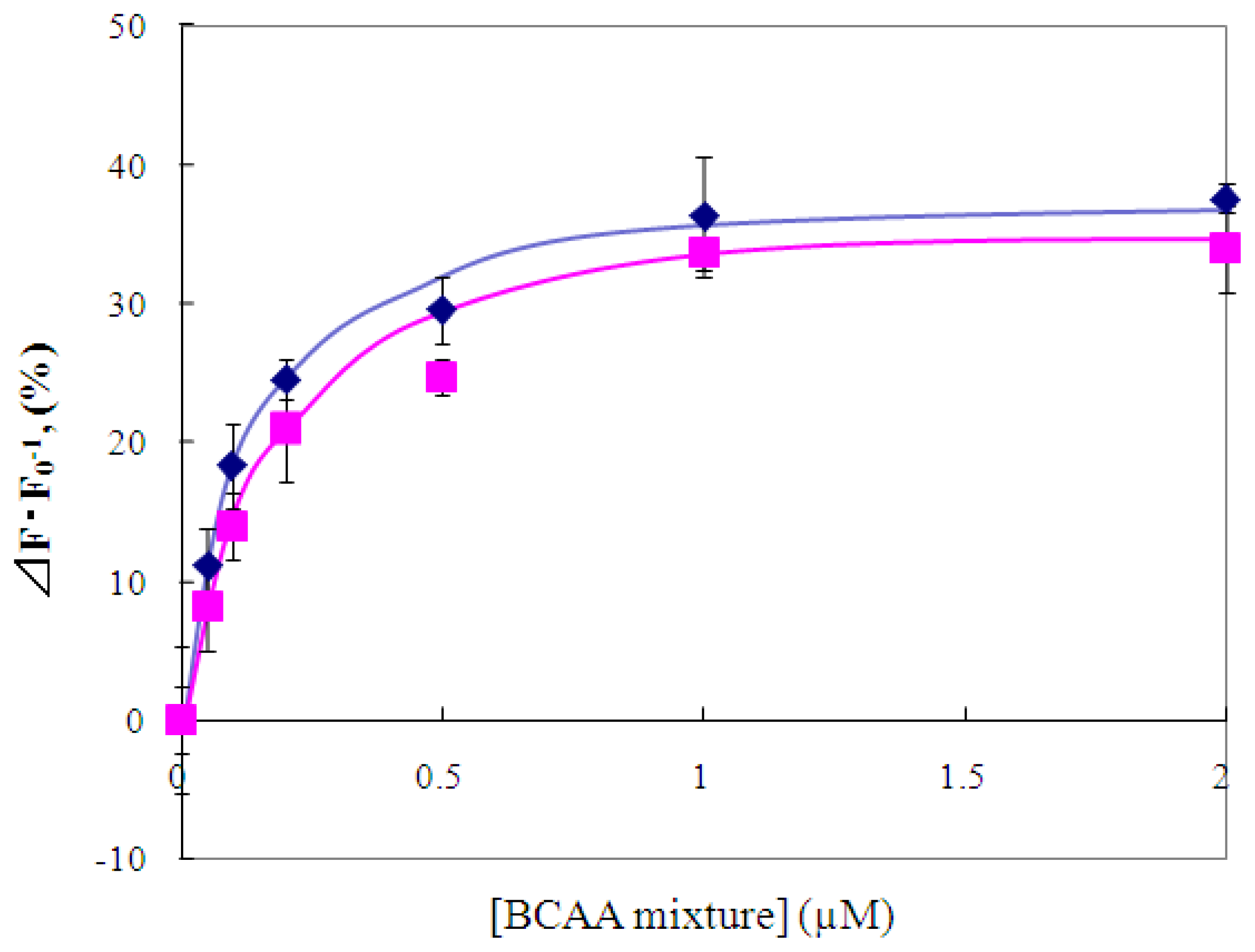
2.4 Fluorescence measurement of branched-chain amino acids using Gln149Cys-MIANS
3. Experimental Section
3.1 Chemicals, enzymes and bacterial strain
3.2 Cloning and recombinant expression of wild-type LIVBP
3.3 Construction of engineered LIVBPs
3.4 Purification of recombinant LIVBPs
3.5 Autofluorescence monitoring
3.6 Fluorescence modification of the engineered LIVBPs
3.7 Fluorescence measurement of BCAAs using the engineered LIVBPs
| Name | Sequence | |
|---|---|---|
| livJ F | 5′-GGATTCTCCATATGAACATAAAGGGTAAAGC-3′ | Cloning |
| livJ R | 5′-GATTAAAAGCTTCTTCGCATCGGTCGCCGTG-3′ | Cloning |
| Q149CF | 5′-GTTCACGACAAACAGTGCTACGGCGAAGGTC TGGCG-3′ | For Gln149Cys mutant |
| Q149CR | 5′-CGCCAGACCTTCGCCGTAGCACTGTTTGTCGT GAAC-3′ | For Gln149Cys mutant |
| G227CF | 5′-GTTTATGGGGCCGGAATGCGTGGCTAACGTTTC G-3′ | For Gly227Cys mutant |
| G227CR | 5′-CGAAACGTTAGCCACGCATTCCGGCCCCATAA AC-3′ | For Gly227Cys mutant |
| Q254CF | 5′-CAAGCCGAAGAACTACGATTGCGTTCCGGCGA ACAAACC-3′ | For Apn254Cys mutant |
| Q254CR | 5′-GGTTTGTTCGCCGGAACGCAATCGTAGTTCTTC GGCTTG-3′ | For Apn254Cys mutant |
References and Notes
- Wandersman, C.; Schwartz, M.; !Ferenci, T. Escherichia coli mutants impaired in maltodextrin transport. J. Bacteriol 1979, 140, 1–13. [Google Scholar]
- Oh, B.H.; Kang, C.H.; De Bondt, H.; Kim, S.H.; Nikaido, K.; Joshi, A.K.; !Ames, G.F. The bacterial periplasmic histidine-binding protein. structure/function analysis of the ligand-binding site and comparison with related proteins. J. Biol. Chem 1994, 269, 4135–4143. [Google Scholar]
- Powlowski, J.; !Sahlman, L. Reactivity of the two essential cysteine residues of the periplasmic mercuric ion-binding protein, MerP. J. Biol. Chem 1999, 274, 33320–33326. [Google Scholar]
- Jacobson, B.L.; He, J.J.; Vermersch, P.S.; Lemon, D.D.; !Quiocho, F.A. Engineered interdomain disulfide in the periplasmic receptor for sulfate transport reduces flexibility. Site-directed mutagenesis and ligand-binding studies. J. Biol. Chem 1991, 266, 5220–5225. [Google Scholar]
- Davies, T.G.; Hubbard, R.E.; !Tame, J.R. Relating structure to thermodynamics: the crystal structures and binding affinity of eight OppA-peptide complexes. Protein. Sci 1999, 8, 1432–1444. [Google Scholar]
- Dwyer, M.A.; Hellinga, H.W. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr. Opin. Struct. Biol 2004, 14, 495–504. [Google Scholar]
- de Lorimier, R.M.; Smith, J.J.; Dwyer, M.A.; Looger, L.L.; Sali, K.M.; Paavola, C.D.; Rizk, S.S.; Sadigov, S.; Conrad, D.W.; Loew, L.; Hellinga, H.W. Construction of a fluorescent biosensor family. Protein. Sci 2002, 11, 2655–2675. [Google Scholar]
- Oxender, D.L.; Anderson, J.J.; Daniels, C.J.; Landick, R.; Gunsalus, R.P.; Zurawski, G.; Selker, E.; Yanofsky, C. Structural and functional analysis of cloned DNA containing genes responsible for branched-chain amino acid transport in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1980, 77, 1412–1416. [Google Scholar]
- Vorotyntseva, T.I.; Surin, A.M.; Trakhanov, S.D.; Nabiev, I.R.; Antonov, V.K. Spectral properties of the leucine-isoleucine-valine binding protein and its complexes with substrates. Bioorg. Khim 1981, 7, 45–57. [Google Scholar]
- Fischer, J.E.; Rosen, H.M.; Ebeid, A.M.; James, J.H.; Keane, J.M.; Soeters, P.B. The effect of normalization of plasma amino acids on hepatic encephalopathy in man. Surgery 1976, 80, 77–91. [Google Scholar]
- Fischer, J.E.; Yoshimura, N.; Aguirre, A.; James, J.H.; Cummings, M.G.; Abel, R.M.; Deindoerfer, F. Plasma amino acids in patients with hepatic encephalopathy. Effects of amino acid infusions. Am. J. Surg 1974, 127, 40–47. [Google Scholar]
- Soeters, P.B.; Fischer, J.E. Insulin, glucagon, amino acid imbalance, and hepatic encephalopathy. Lancet 1976, 2, 880–882. [Google Scholar]
- Klassen, P.; Furst, P.; Schulz, C.; Mazariegos, M.; Solomons, N.W. Plasma free amino acid concentrations in healthy Guatemalan adults and in patients with classic dengue. Am. J. Clin. Nutr 2001, 73, 647–52. [Google Scholar]
- Nishitani, S.; Ijichi, C.; Takehana, K.; Fujitani, S.; Sonaka, I. Pharmacological activities of branched-chain amino acids: specificity of tissue and signal transduction. Biochem Biophys Res Commun 2004, 313, 387–9. [Google Scholar]
- Nieuwoudt, M.; Kunnike, R.; Smuts, M.; Becker, J.; Stegmann, GF.; Van der Walt, C.; Neser, J.; Van der Merwe, S. Standardization criteria for an ischemic surgical model of acute hepatic failure in pigs. Biomaterials 2006, 27, 3836–45. [Google Scholar]
- Dickinson, J.C.; Rosenblum, H.; Hamilton, P.B. Ion Exchange Chromatography of the Free Amino Acids in the Plasma of the Newborn Infant. Pediatrics 1965, 36, 2–13. [Google Scholar]
- Hughes, G.J.; Winterhalter, K.H.; Boller, W.; Wilson, K.J. Amino acid analysis using standard high-performance liquid chromatography equipment. J. Chromatogr 1982, 235, 417–426. [Google Scholar]
- Azuma, Y.; Maekawa, M.; Kuwabara, Y.; Nakajima, T.; Taniguchi, K.; Kanno, T. Determination of branched-chain amino acids and tyrosine in serum of patients with various hepatic diseases, and its clinical usefulness. Clin. Chem 1989, 35, 1399–1403. [Google Scholar]
- Nakamura, T.; Mori, M.; Yoshida, T.; Murakami, N.; Kato, T.; Sugihara, J.; Saito, K.; Moriwaki, H.; Tomita, E.; Muto, Y.; et al. Enzymatic determination of a molar ratio of free branched-chain amino acids to tyrosine (BTR) and its clinical significance in plasma of patients with various liver diseases. Rinsho Byori 1989, 37, 911–917. [Google Scholar]
- Dattelbaum, J.D.; Lakowicz, J.R. Optical determination of glutamine using a genetically Engineered protein. Anal. Biochem 2001, 291, 89–95. [Google Scholar]
- Salins, L.L.; Goldsmith, E.S.; Ensor, C.M.; Daunert, S. A fluorescence-based sensing system for the environmental monitoring of nickel using the nickel binding protein from Escherichia coli. Anal. Bioanal. Chem 2002, 372, 174–80. [Google Scholar]
- Morgan, M.Y.; Marshall, A.W.; Milsom, J.P.; Sherlock, S. Plasma amino-acid patterns in liver disease. Gut 1982, 23, 362–370. [Google Scholar]
- Abe, A.; Okuno, M.; Noma, A.; Imamine, T.; Nakamura, T.; Muto, Y. Enzymatic Assay of tyrosine and branched-chain Amino Acids in Plasma and Its Application to Liver Diseases. Jpn. J. Clin. Chem 1990, 19, 124–130. [Google Scholar]
- Landick, R.; Oxender, D.L. The complete nucleotide sequences of the Escherichia coli LIV-BP and LS-BP genes. Implications for the mechanism of high-affinity branched-chain amino acid transport. J. Biol. Chem 1985, 260, 8257–8261. [Google Scholar]
© 2007 by MDPI Reproduction is permitted for noncommercial purposes.
Share and Cite
Chino, S.; Sakaguchi, A.; Yamoto, R.; Ferri, S.; Sode, K. Branched-chain Amino Acid Biosensing Using Fluorescent Modified Engineered Leucine/Isoleucine/Valine Binding Protein. Int. J. Mol. Sci. 2007, 8, 513-525. https://doi.org/10.3390/i8060513
Chino S, Sakaguchi A, Yamoto R, Ferri S, Sode K. Branched-chain Amino Acid Biosensing Using Fluorescent Modified Engineered Leucine/Isoleucine/Valine Binding Protein. International Journal of Molecular Sciences. 2007; 8(6):513-525. https://doi.org/10.3390/i8060513
Chicago/Turabian StyleChino, Sakura, Akane Sakaguchi, Rie Yamoto, Stefano Ferri, and Koji Sode. 2007. "Branched-chain Amino Acid Biosensing Using Fluorescent Modified Engineered Leucine/Isoleucine/Valine Binding Protein" International Journal of Molecular Sciences 8, no. 6: 513-525. https://doi.org/10.3390/i8060513




