1. Introduction
Although viral vectors are able to remain relatively efficient gene delivery
in vivo, and generally yield higher gene expression, there are much concerns over the immunogenic, cytotoxic, and recombinogenic potentials of viral gene transfer [
1,
2]. Due to the improved safety profile and ease of preparation and manipulation, synthetic alternatives for gene delivery have been extensively explored [
3,
4]. In particular, much progress has been made with cationic polymers such as poly(lysine) [
5], polyethylenimine [
6] and cationic liposomes [
7]. These systems, however, have significant cytotoxicity issues, in part due to the poor biocompatibility of nondegradable polymers [
8,
9]. Consequently, recent attention has focused on the development of biodegradable cationic polymers [
10–
17], but gene delivery using these biodegradable polycations has only achieved limited success. One of the most successful and widely studied gene delivery polymers reported to date is polyethylenimine (PEI). It is a cationic polymer which has proven to be an effective transfection agent both in vitro [
18,
19] and in vivo [
20,
21]. Due to its relatively high gene delivery efficiency [
19,
22,] and ready availability, branched, 25-kDa PEI has become a benchmark to which other polymers [
23], especially newly designed and synthesized materials, are often compared. Recent reports indicate that a degradable analogue of 25-kDa branched PEI was produced by conjugating amino groups on 800 Da PEI to diacrylates and this molecule showed high efficient gene delivery capability together with low toxicity [
17]. Inspired by this molecule, we designed the molecules to be endowed with both the favorable low toxicity properties of the low molecular weight PEI and the higher transfection efficiency of high molecular weight PEI by coupling low molecular weight 2000 Da PEIs together to form conjugates of high molecular weight PEI using short diacrylate linkages. The molecules were provided with biodegradable ability because of containing ester bonds, it was proved that the ester bonds can be hydrolyzed under physiological conditions. This will facilitate gene transfection of the cross-linked PEIs
in vivo. Among the degradable PEI derivatives we synthesized, three degradable polyethylenimines were selected and validated with low toxicity and highly efficient gene delivery for B16F10, 293T and 3T3 cell lines.
In this study, we also optimize the conditions for gene delivery to B16F10 cell and other cells using branched, 25-kDa PEI and the degradable analogues of 25-kDa branched PEI produced by addition of amino groups on 2000-Da PEI to diacrylates. The feature of the transfection protocol was carefully optimized including the volume of the PEI/DNA complexation solution, the forming time of PEI/DNA complexation solution and ratio of PEI/DNA. We also studied the effective condensation of DNA by gel retardation assay and dynamic light scattering. The gene transfection introduced by 25 kDa branched PEI and the cross-linked PEIs in mouse leg muscles was observed and assessed by fluorescence microscopy.
3. Discussion
The gene delivery efficiency and cytotoxicity of PEI vary with its molecular weight. For example, the branched 2000 Da PEI is several hundred fold less efficient than the branched 25-kDa PEI at the same concentration [
28], but the former is not cytotoxic whereas the latter is. We are interested in how to combine the advantages of good transfection capability of high molecule weight PEI and nontoxic property of low molecule weight PEI. We hypothesized that high molecular weight conjugates composed of 2000 Da PEI would yield transfection efficiencies comparable to those obtained with high molecular weight PEI. Furthermore, we expected that when the transfection complexes were exposed to the physiological conditions, the ester bonds introduced by the cross-linking reagents would be hydrolyzed, and the hydrolysis of the ester bonds has been validated under physiological conditions by Forrest et al 2003 [
17]. This would promote reversion of the high molecular weight complexes back to their low molecular weight counterparts, presumably leading to lower toxicity and potentially easier access by the transcription machinery. We hoped that optimizing the combination of PEIs and the cross-linking reagents would achieve the enhancement in gene delivery efficiency with minimal cytotoxicity.
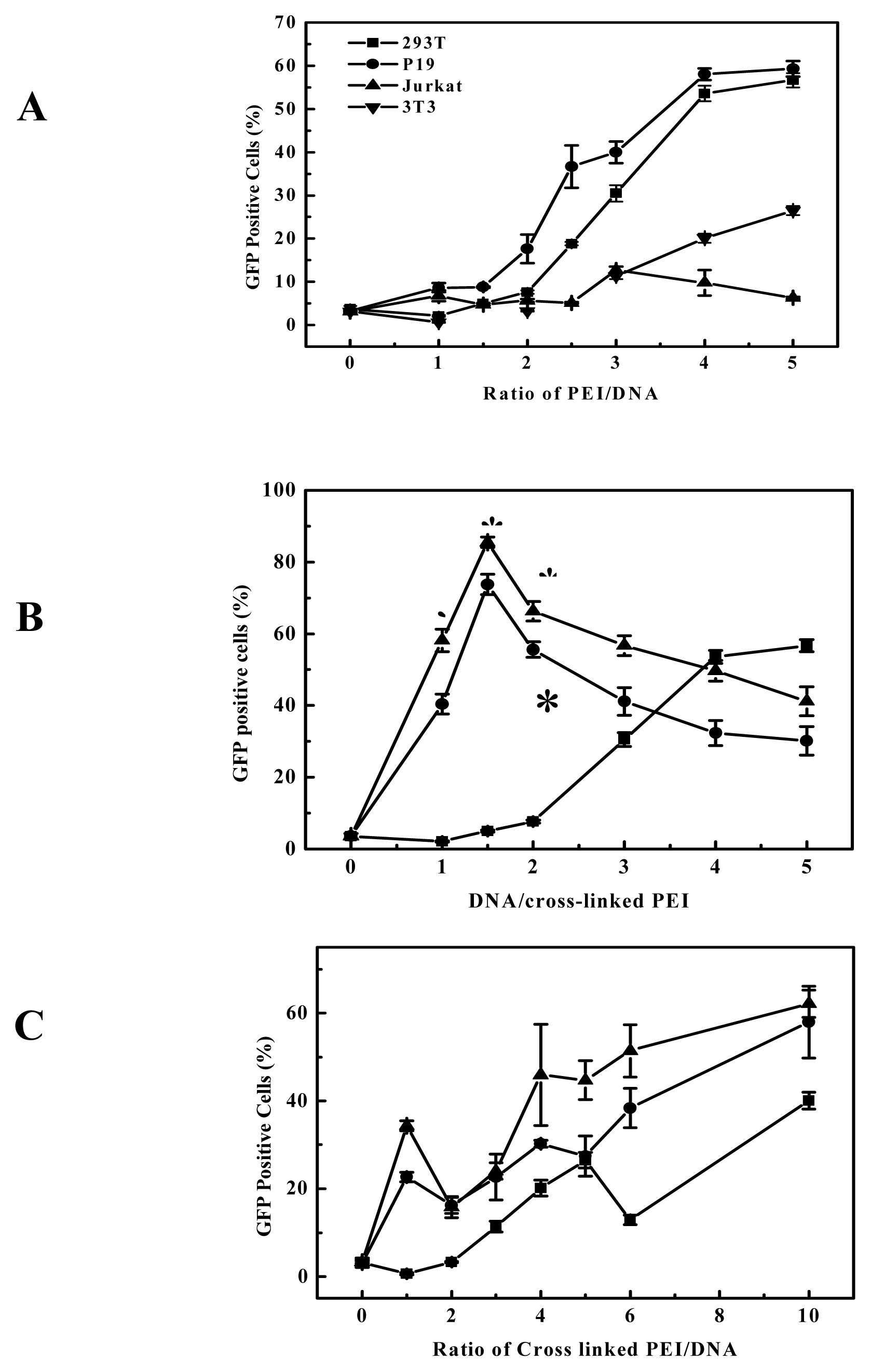
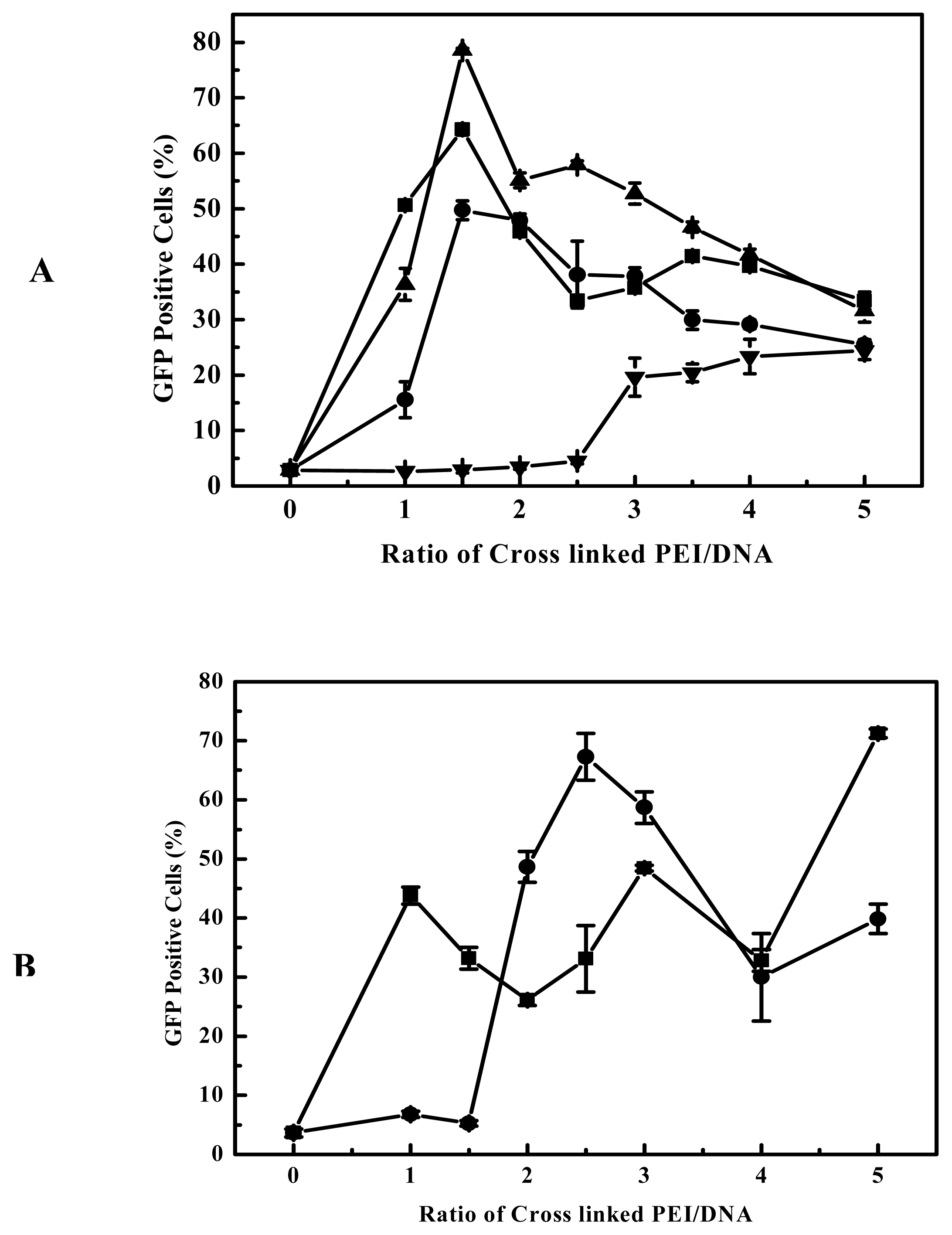
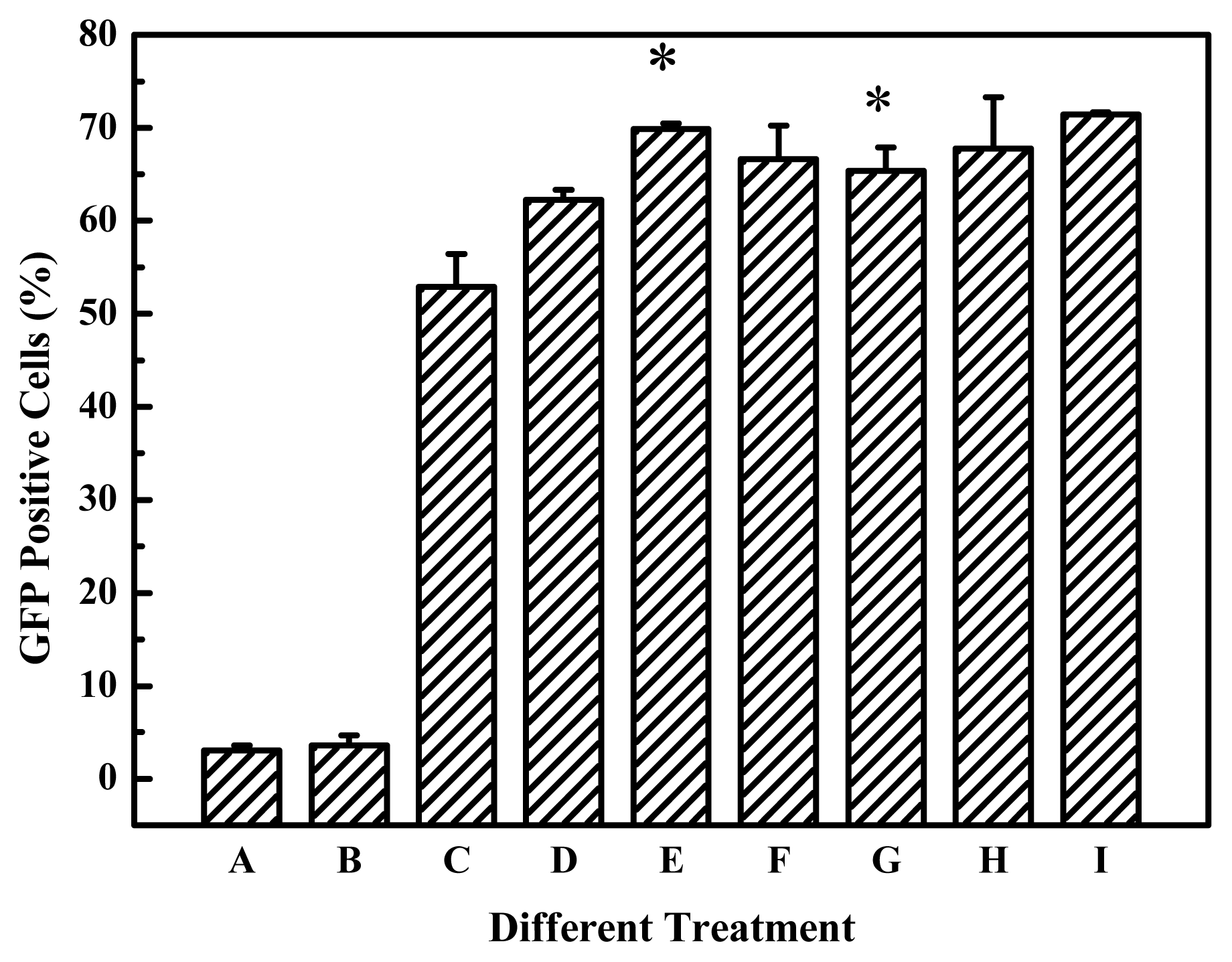
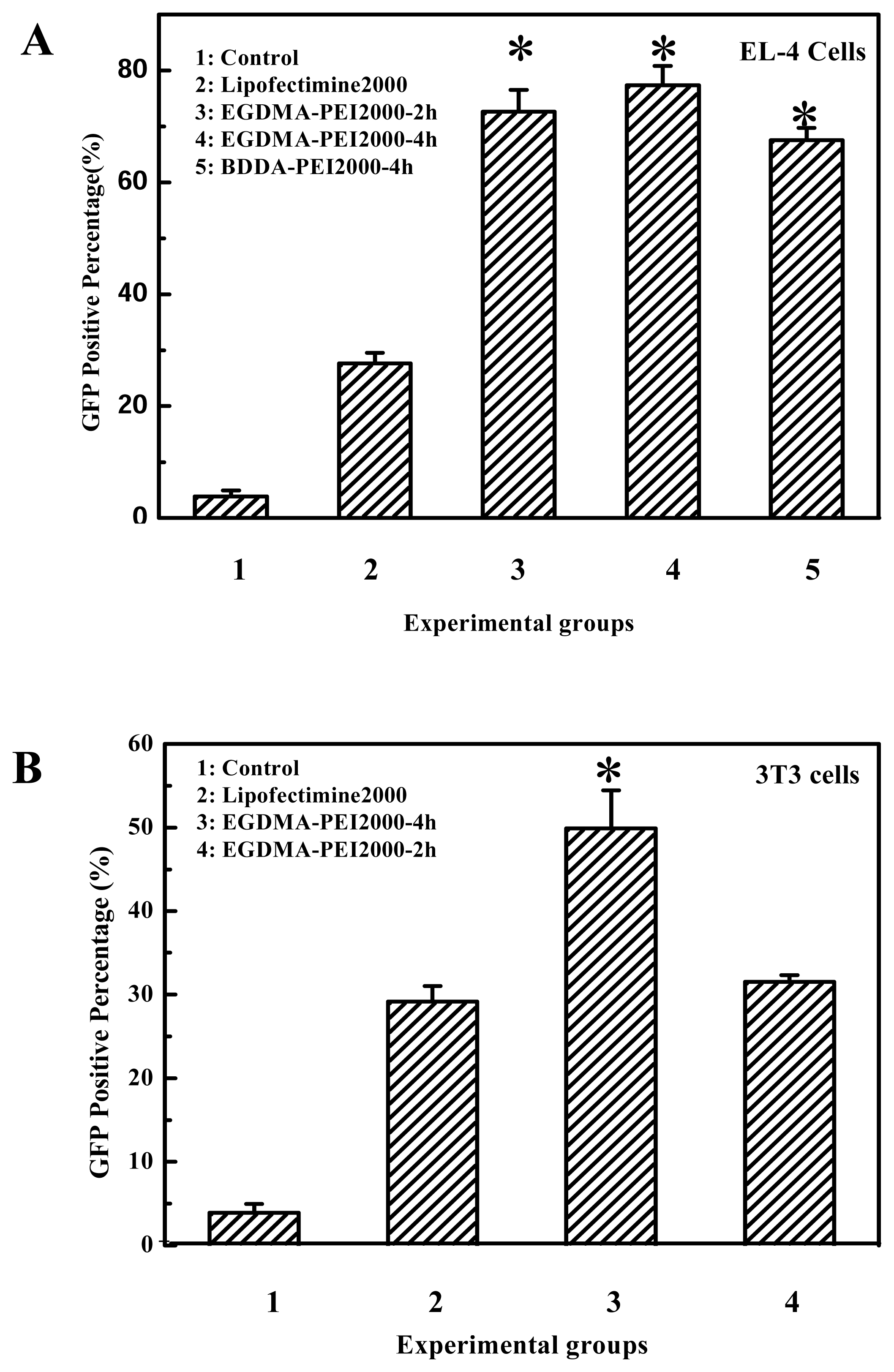
The degradable PEIs reported here appears to retain the beneficial function of 25-kDa PEI, but upon degradation we believe the polymers allow for more efficient transcription and interferes less with normal cell functions. As far as the gene delivery into B16F10 cells is concerned, among all the prepared degradable PEIs, the cross-linked product of branched 2000 Da PEI and EGDMA was more highly efficient, this indicated that branched 2000 Da PEI and EGDMA compose better combination of monomer and cross-linking agent. It was reported that 800 Da PEI and 1,3-butanediol diacrylate/1,6-hexanediol diacrylate was also better combination of monomer and cross-linking agent [
17], but the transfection efficiency is not easy for us to make comparison each other, because the report was confined to the gene expression of luciferase and did not report the percentage of transfected cells. The cross-linked products of 2000 Da PEI and EGDMA was easier to obtain high transfection efficiency in our experiments (
Figure 4). These showed that the high efficiency is dependent on the nature of the linkages and the PEIs used. Although the viscosity, an indicator of molecular weight, increased with the elongation of polymerization time, the polymerization time showed less influential on the transfection efficiency of the products for B16F10 cells (
Figure 4A). For example, the cross-linked products polymerized for 2 to 4 hours all had better transfection efficiency for B16F10 cell, and it also can apply to other cell lines, for example, EGDMA-PEI 2000-4h exhibited the highest transfection efficiency for 3T3 (
Figure 3C), while branched 25-kDa PEI and other cross-linked polymers were less efficient for 3T3. In fact when using the EGFP reporter expression analysis, the complexes of EGDMA-PEI 2000-4h and plasmid in PBS (pH7.4) achieved stable transfection with the average efficiency up to 60% for all the cell lines we studied, including B16F10, 293T, CHO, EL-4 and Hela (data not shown). For 293T and B16F10 cell lines the transfection efficiency was frequently 80–90%. Therefore EGDMA-PEI 2000-4h displays broad application potential in wide range of cell lines.
As discussed above, the transfection activity of the cross linked PEI is critically dependent on their structure, although molecular weight of polymers is important [
29,
22]. Thomas M.
et al reported that moderate enhancement in hydrophobicity increased the transfection efficiency of 25-kDa PEI [
28], and Daniel G.
et al reported that the most effective cross linked polymers for gene delivery was composed of monomer containing alcohol groups and cross-linking agent of hydrophobic acrylates [
30]. But the water-solubility of cross-linked products of PEI 2000 and EGDMA/BDDA was less than 25-kDa PEI, and was also likely less than poly(β-amino esters) with hydroxy groups. Therefore the cross-linking agent of hydrophilic acrylates seemed to be benifical for gene delivery in our experiments. Zhong Zhiyuan reported that the hydrophobic segments in the polymers likely facilitate cellular uptake on one hand, and lead to cell membrane damage on the other hand via hydrophobic interactions with cell membranes [
31]. Thus for the transfection efficiency and cytotoxicity, the hydrophilic-hydrophobic balance of the polycation is also important. Although branched 25-kDa PEI had high gene delivery efficiency to B16F10 and 293T cell at the optimal polymer/DNA ratio between 3 and 4 (
Figure 5 and
Figure 3A), in general it showed evident cytotoxicity when branched 25-kDa PEI was used in gene delivery to some cell lines with polymer/DNA ratio over 3. Compared to branched 25-kDa PEI, cross-linked PEIs not only had high gene delivery efficiency to B16F10 and 293 cells, but also can delivery gene at lower optimal polymer/DNA ratios (
Figure 5 and
Figure 3B), so they had low cytotoxicity. Generally, lower optimal polymer/DNA ratios are indicative of the greater DNA condensation ability of the polycations and thus the cross-linked PEIs are suitable candidates for
in vivo gene delivery. The cross-linked PEI (EGDMA-PEI 2000-2h) for
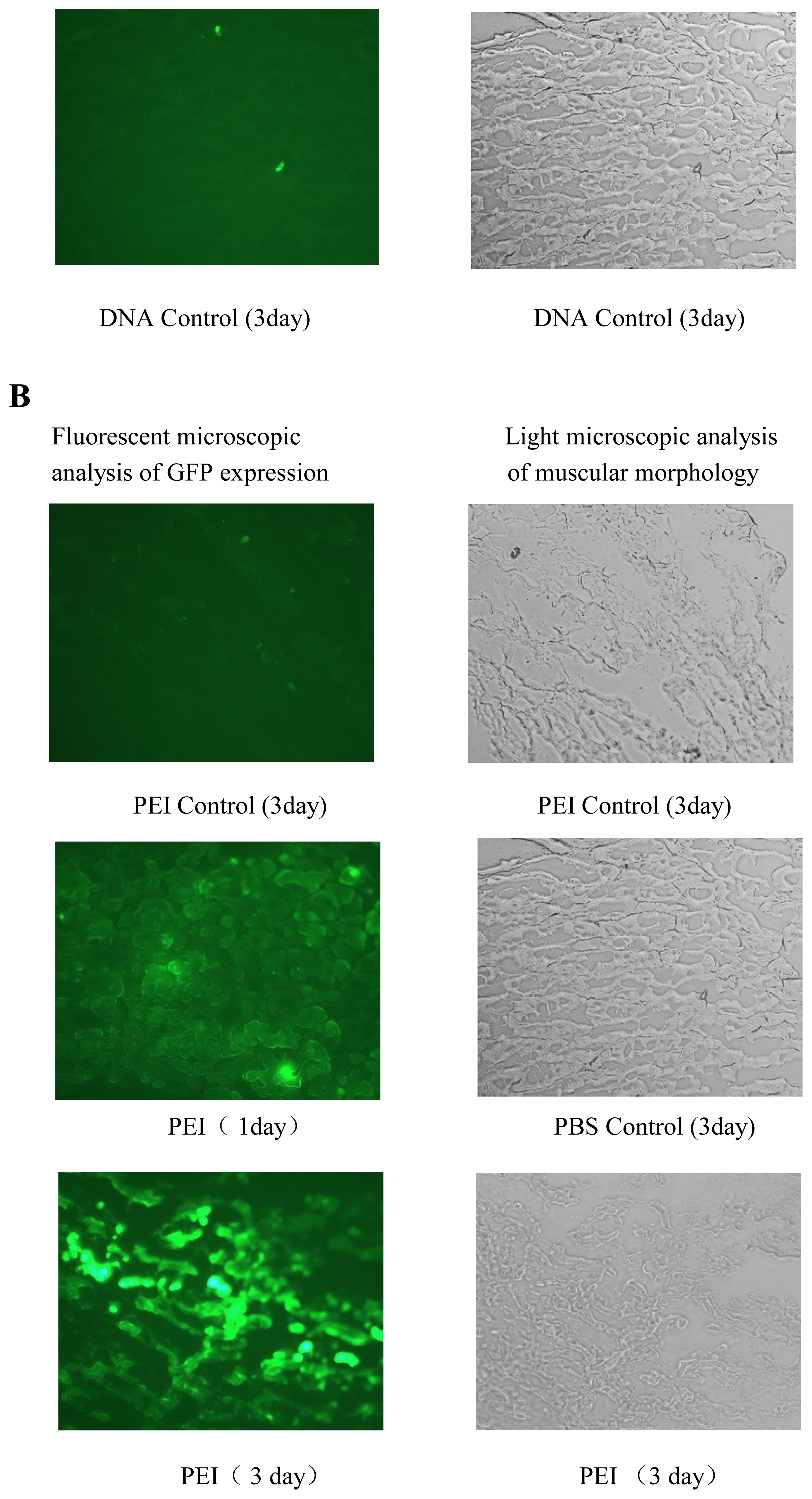
in vivo gene delivery was tested for its efficiency in gene delivery into mouse muscle (
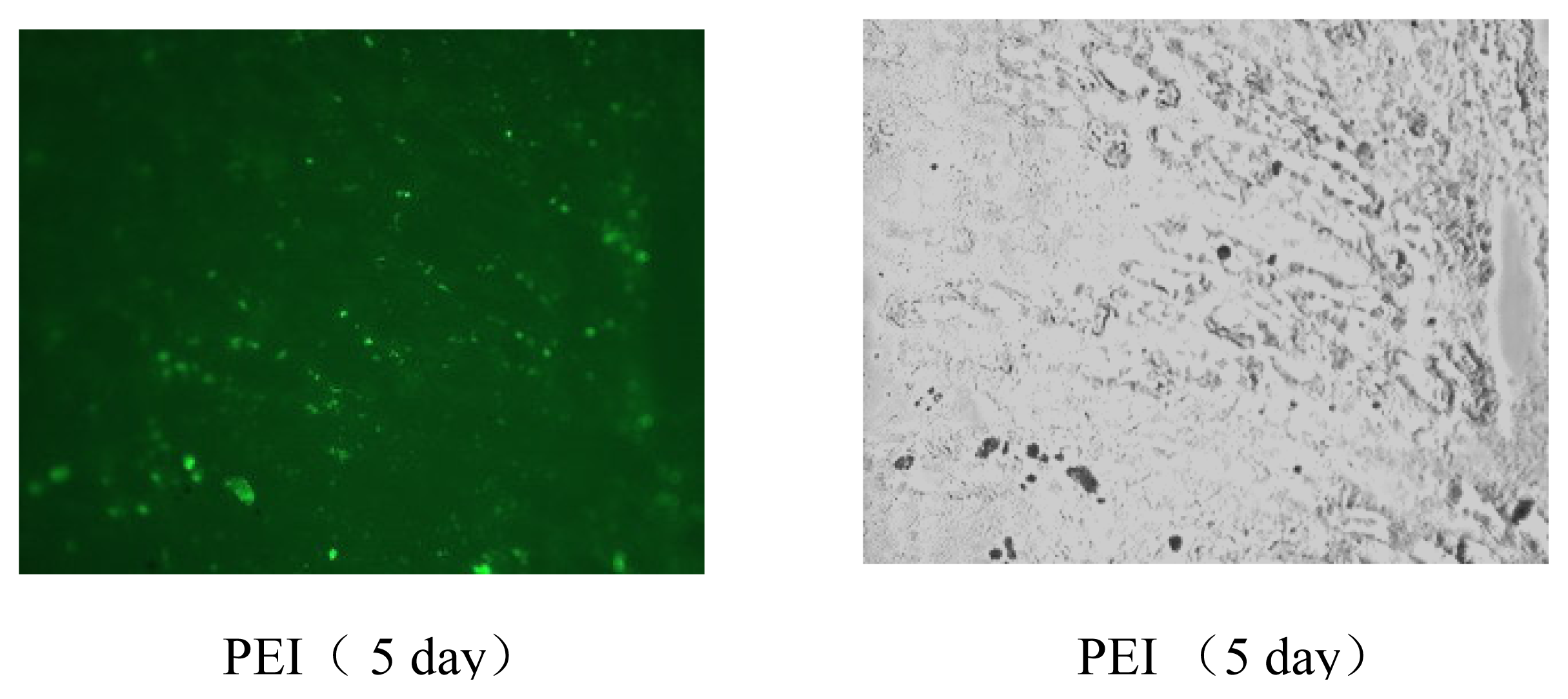
Figure 7) and tumor models (data not shown). The results of gene delivery in mouse muscle were showed in
Figure 7, the transfection efficiency for muscle is similar to that of Massimo Don’s electro-transfection of genes in adult skeletal muscle [
32], but our procedure is easier and equipment-free. The gene delivery of mouse tumor model also was studied in our lab and the cross-linked PEI mediated obvious tumor inhibition of RNA interfering plamids [
33].
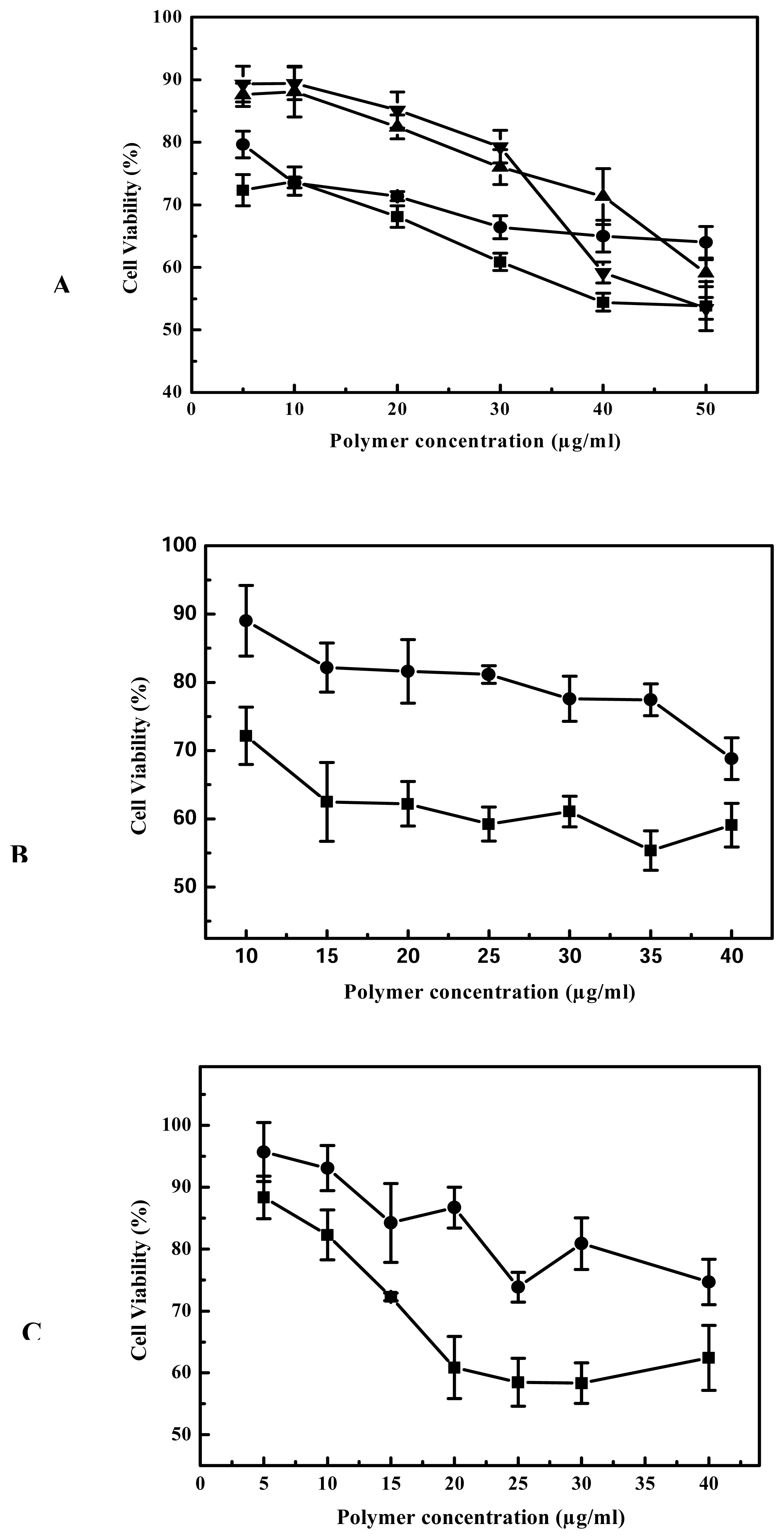
25-kDa PEI has been widely studied as DNA condensing agent and transfection vector [
3] and is the standard to which new polymeric vectors is often compared [
23]. Unfortunately, as outlined above, it is also associated with obvious cytotoxicity and high level of gene expression is usually realized at a substantial cost to cell viability. To determine the toxicity profile of cross-linked PEIs, we conducted MTT assay for B16F10, 293T and 3T3 cell lines, and the results are compared with 25-kDa PEI’s. The results of MTT assay showed that the cell viability of cross-linked PEIs was more than that of 25-kDa PEI with the increase of 21%, 22%, 33% for B16F10, 3T3 and 293T respectively, the cytotoxicity of cross-linked PEI was evidently less than 25 kDa PEI in the process of gene transfection. On the other hand biodegradability is very beneficial when these polymers used
in vovo, this can be proved in the gene transfection experiment of mouse’s muscle (
Figure 7). Detailed investigations of the molecular weight weight and the degradation rate of the synthesized polymers with different nature of the linkage groups are in progress.
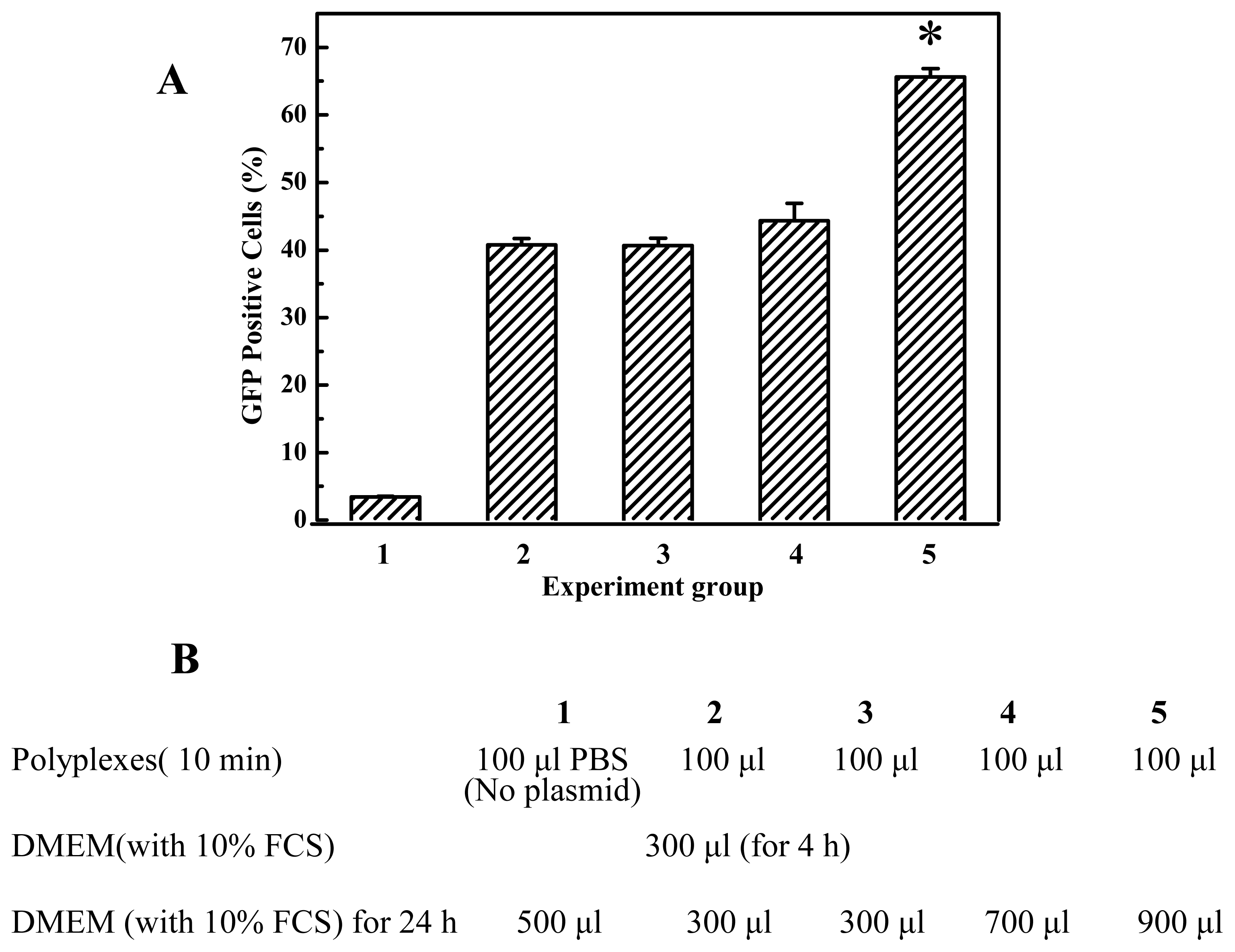
In general, serum was usually present in the transfection medium. The transfection activity of cross-linked PEIs prepared here was preserved or increased in the presence of serum proteins (
Figure 8), this is convenient for application
in vitro, and this characteristic is also important, even necessary, for its application
in vivo. We therefore investigated the effect of different DMEM (with/without 10% FCS) on polyplex particle size and the influence of polyplex particle size on transfection efficiency. As shown in
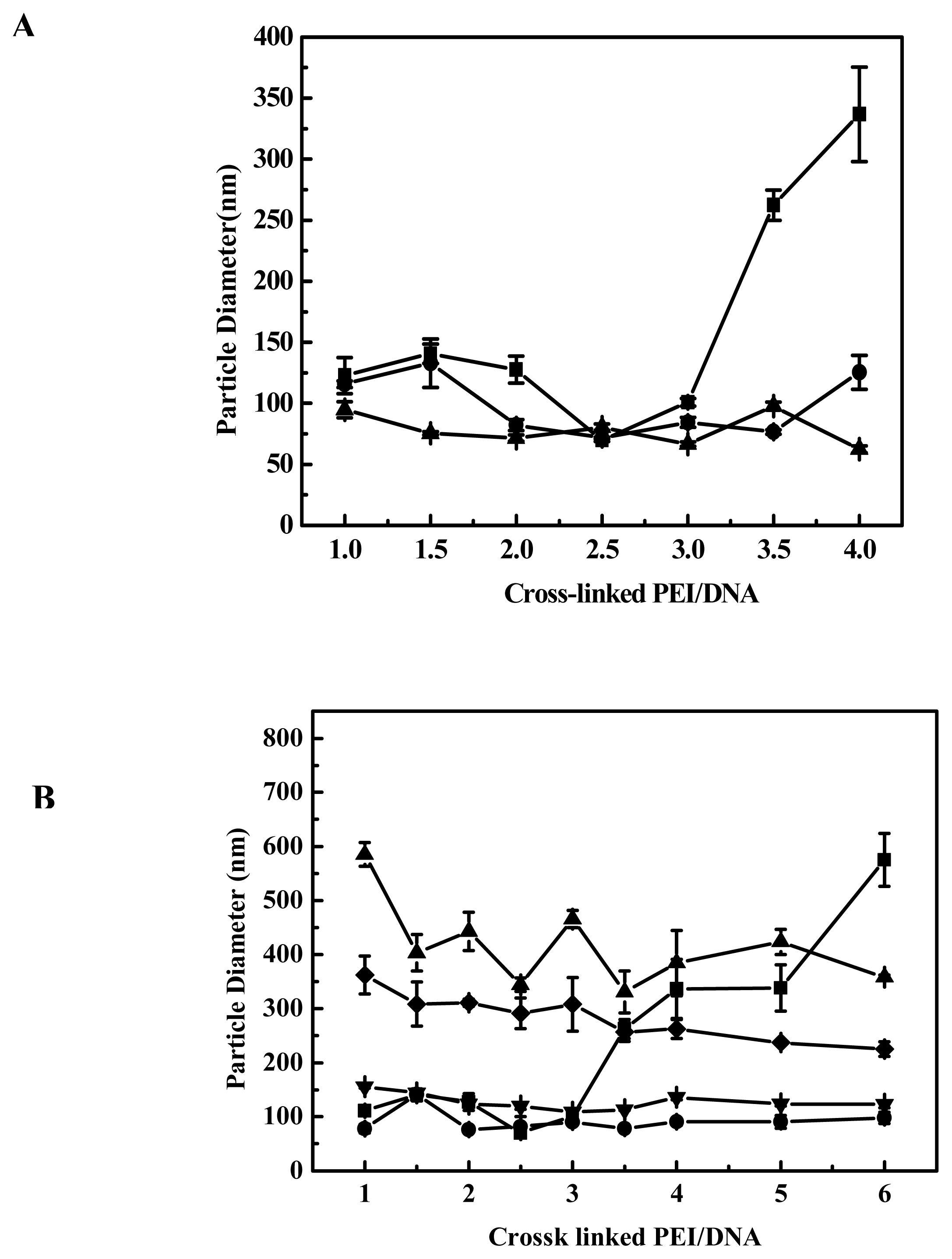
Figure 2B, when the polyplexes were prepared in PBS with EGDMA-PEI 2000-4h and DNA, after 10–15 minutes they were added into DMEM with 10% fetal calf serum and the particle size of the polyplexes was as small as that of polyplexes prepared in ddH
2O. The particle size did not increase during a 4-h period after mixing DNA and EGDMA-PEI 2000-4h, but after 10 h we found that aggregation has occurred and all polyplexes sank into the bottom of cuvette. The particle size was larger (>300 nm) and a gradual increase in particle size was observed when polyplexes were added to serum-free DMEM, and aggregated over a period of hours to yield larger complexes with diameters in the range of 1–2 μm. But the transfection efficiency was high, this showed that small particle size is not an absolute criterion for transfection [
27,
34]. When DMEM supplemented with 10% FCS was added, no particle size growth occurred, suggesting that serum protein could stabilize the particle against aggregation, thus preventing size increase. Similarly Sharma V. K. reported that dilution itself reduced the rate of aggregation of PEI/DNA complexes [
35]. Thus we investigated the effect of the amount of DMEM with 10% FCS on gene transfection efficiency and found that it is beneficial that PEI/DNA complexation solution was diluted by a great deal of growth media when the PEI/DNA complexes were formed after 10–15 min. Prolongation of transfection duration will not improve transfection efficiency, for example, the transfection efficiency of cells transfected for 24 hours did not show much difference from that transfected for 4 hours (
Figure 8). It implies that transfection process occurred essentially within 4 hours, and within 4 hours, did not severely aggregate when transfected in free serum medium or with serum medium (
Figure 2B). In fact the transfection efficiency of EGDMA-PEI2000- 4h, EGDMA-PEI2000-2h and BDDA-PEI2000-4h was serum independent, for B16F10 cells and 293T cells, the three polymers was identical with Lipofectamine™ 2000 (data not shown) when the transfection was carried out in growth media with 10% FCS, but the polymers could mediate more efficient expression of reporter gene than the Lipofectamine™ 2000 at their optimal transfection condition for EL-4 and 3T3 cell lines (
Figure 9).
In conclusion, the three cross-linked polymers investigated here are desirable for practical gene transfer application. By employing a properly selected combination of PEIs and potentially biodegradable cross-linking agents, both the in vitro and in vivo gene delivery by PEIs are evidently enhanced, and it is likely to obtain some degradable polyethylenimine derivatives with low toxicity for highly efficient gene delivery when the methodology described herein is adapted to high-throughput synthesis. We are currently further elaborating on the structure-activity-cytotoxicity relationships for this class of polymers.
4. Experimental Section (materials and methods)
Plasmid DNA
Transfections were performed using 2 μg per well (in 24-well plates) of plasmids containing the enhanced green fluorescent protein gene (pEGFP-C1) (Clontech, USA). The plasmids were amplified and purified according to standard molecular biology techniques.
Cell culture
Murine B16F10 melanoma cell lines were obtained from ATCC. NIH 3T3 murine fibroblasts cell lines and 293T (human embryonal kidney) cell lines were purchased from the Institute of Shanghai Biochemistry and Cell Biology, Chinese Academy of Science. NIH-3T3, 293T and B16F10 were cultured in DMEM with 10% fetal calf serum (FCS). All cell culture media were supplemented with 2 mM L-glutamine (Gibco BRL, USA), 100 units/ml penicillin (Gibco BRL, USA), and 100 μg/ml streptomycin (Gibco BRL, USA). Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere. When 80% confluent, cells were detached with trypsin/EDTA (Gibco BRL, USA), then dispersed and cultured in normal growth media.
Polymer synthesis
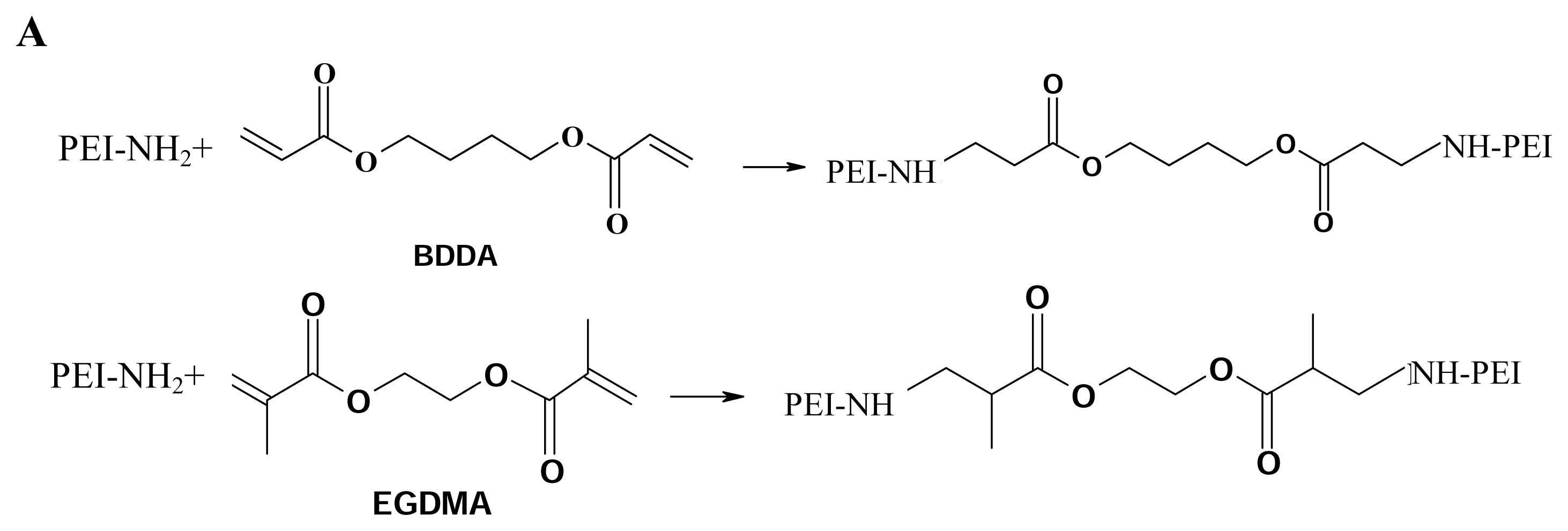
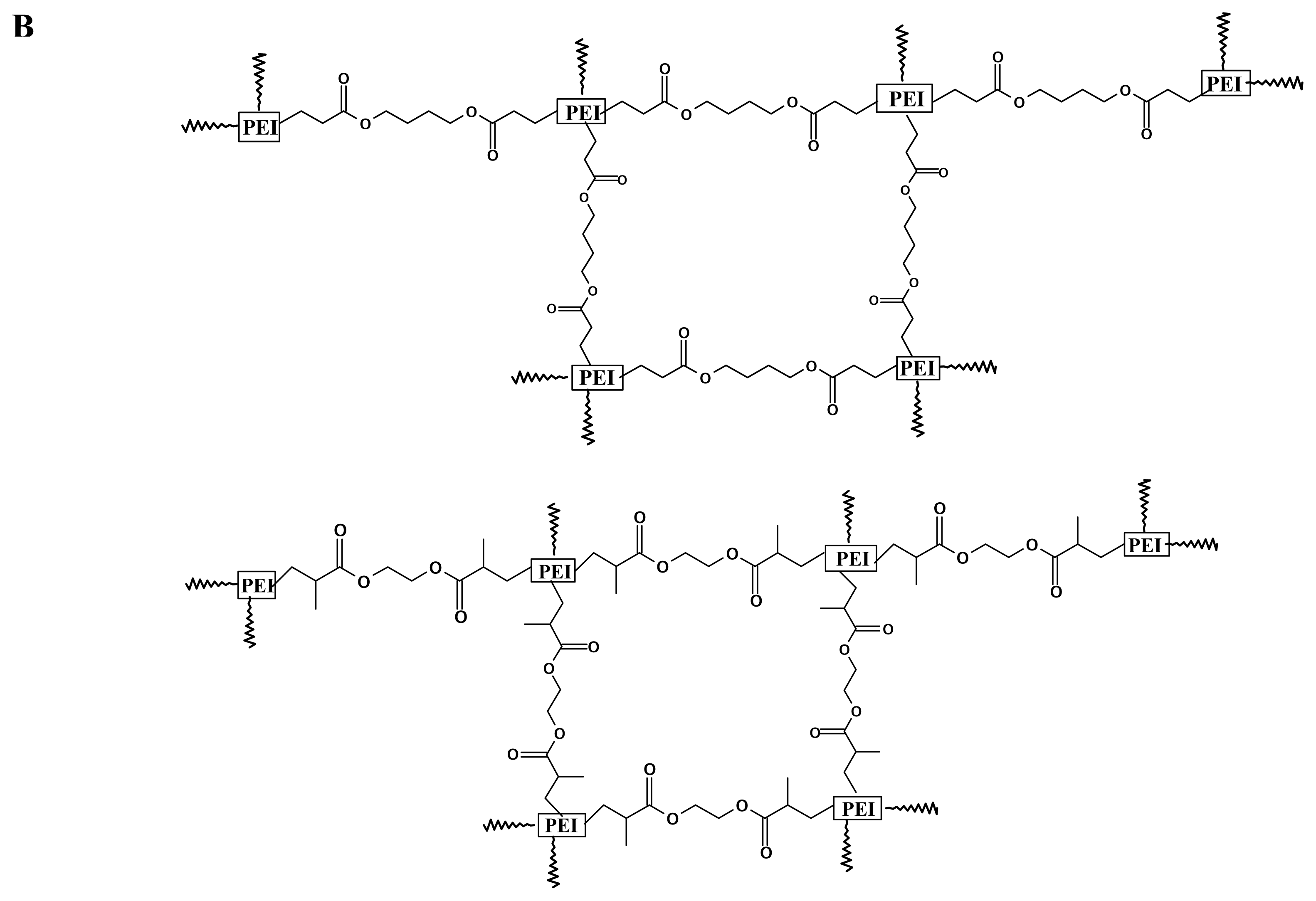
Branched 2000 Da PEI, branched 25-kDa PEI, ethyleneglycol dimethacrylate (EGDMA) and 1, 4- butanediol diacrylate (BDDA) were purchased from Sigma-Aldrich USA, and used without further purification unless noted otherwise. Cross-linking was performed following the procedure of Lynn et al. [
36,
37]. One gram of PEI (branched, 2000 Da) was transferred to a 10ml vial and dissolved in 3 ml of freshly distilled methylene chloride. An equimolar amount of diacrylate linker (ethyleneglycol dimethacrylate or 1, 4-butanediol diacrylate) was added, and the flask was sealed with a solvent-resistant cap. The reaction was carried out at 45°C, with shaking, for several hours. The polymer was then precipitated with ether, lyophilized (Labconco 18L Freeze-Dry System, Kansas City, MO, USA), and stored at -70 °C.
Cytotoxicity assay
The cytotoxicity of the polymers was measured by MTT assay. B16F10 cells or other cells were seeded at a density of 1×104 cells/well in 96-well plates and incubated for 24 h. Prepared polymer complexes were added into cells in the absence of 10% fetal calf serum. After 4 hours, the polymer-containing medium was removed, 100 μl conventional growth media and 20 μl of 5 mg/ml MTT in PBS buffer were added. Plates were incubated for additional 4 h at 37°C. MTT-containing medium was removed and 100 μl of DMSO was added to dissolve the formazan crystal formed by live cells. Absorbance was measured at 570 nm using a microplate reader (Molecular Devices Co., Menlo Park, CA, USA) and recorded as a percentage relative to untreated control cells.
Particle size measurements
Polyplex sizes were measured by dynamic light scattering using a Brookhaven 90PLUS particle size analyzer (Brookhaven Instruments Corporation, Holtsville, NY, USA) at 25°C. Polyplex sizes are expressed as effective diameters (based on intensity weighted analysis). Complexes were prepared as in transfection experiments, except that the amounts of PEI and DNA were scaled up to a total volume of 1.5 ml of polyplexes from 100 μl. Specifically, appropriate amounts of PEI (for various polymer/DNA ratios) in 750 μl of ddH2O/PBS were added to 30 μg of DNA in 750 μl of ddH2O/PBS. This procedure was followed by pipette mixing and incubation at room temperature for 10–15 min, and the polyplexes were then subjected to light scattering experiments. The results were expressed as a mean ± SD, n = 3.
Gene delivery in mice
pEGFP-C1 DNAs (Clontech, USA) in ddH2O, mixed with EGDMA-PEI 2000-2h solution (1 mg/ml) in PBS at indicated polymer/DNA ratios, were incubated at room temperature for 10–15 min. Mouse legs were exposed by removing hair and fifty microliters of the mixture containing 10 μg of DNA were administered into the leg muscles of 8-week-old male C57BL/6J mice (Shanghai Laboratory Animal Center, China) using a Hamilton syringe in a proximal to distal direction. Mice were sacrificed at different time points (1, 3, 5, and 7 days) and muscles (n=6 for each point) were removed, and stored at -70 °C, then frozen sections were immediately made to evaluate the gene transfection efficiency of 25-kDa branched PEI and the cross-linked PEI through observing the EGFP expression by fluorescence microscopy.
Statistics and Presentation of Data
All experiments were repeated at least three times. All numerical data were expressed as mean± S.D. Data were analyzed using the two-tailed t test.