Deletion of TSPO Resulted in Change of Metabolomic Profile in Retinal Pigment Epithelial Cells
Abstract
:1. Introduction
2. Results
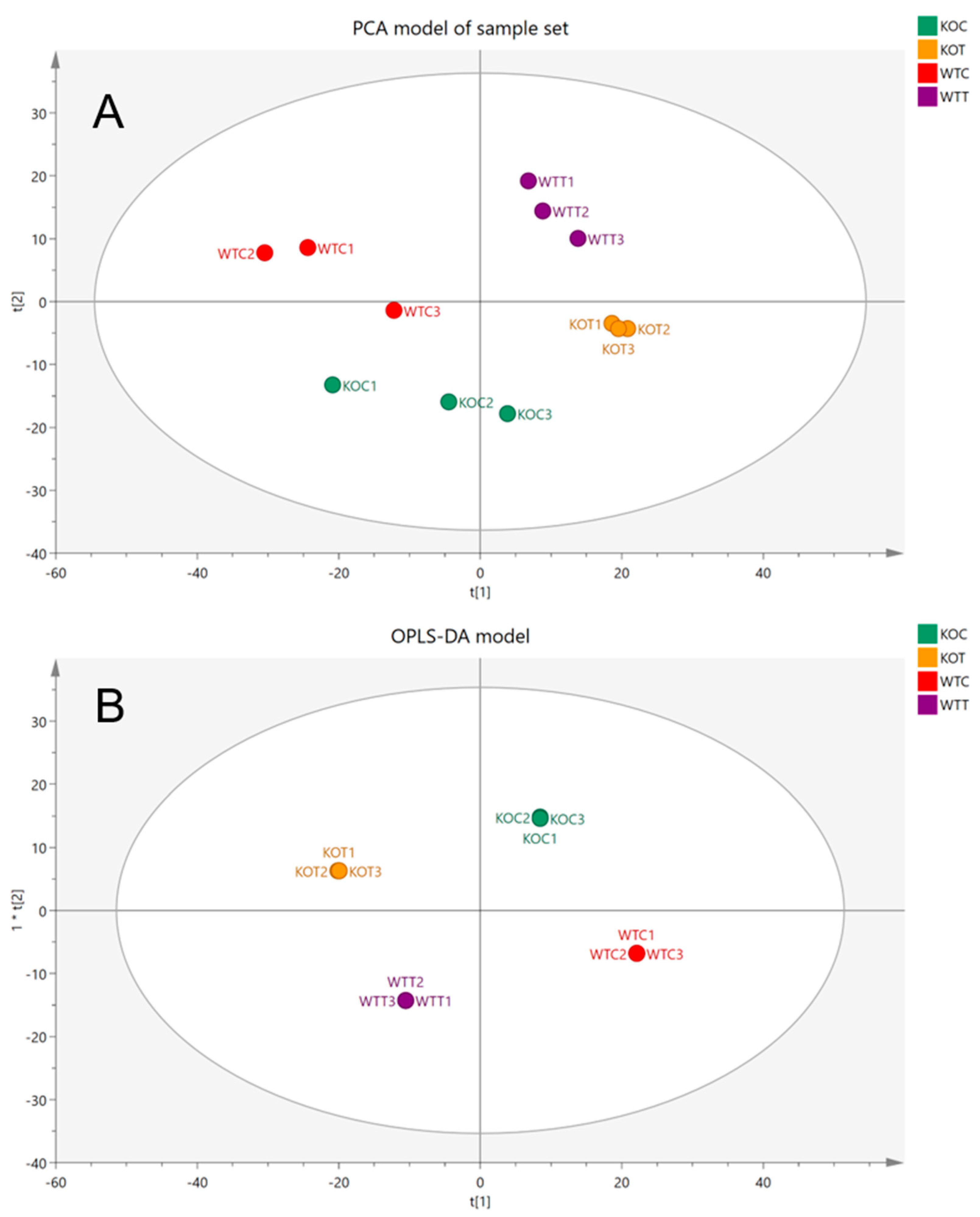
2.1. Principle Component Analysis (PCA) and Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA)
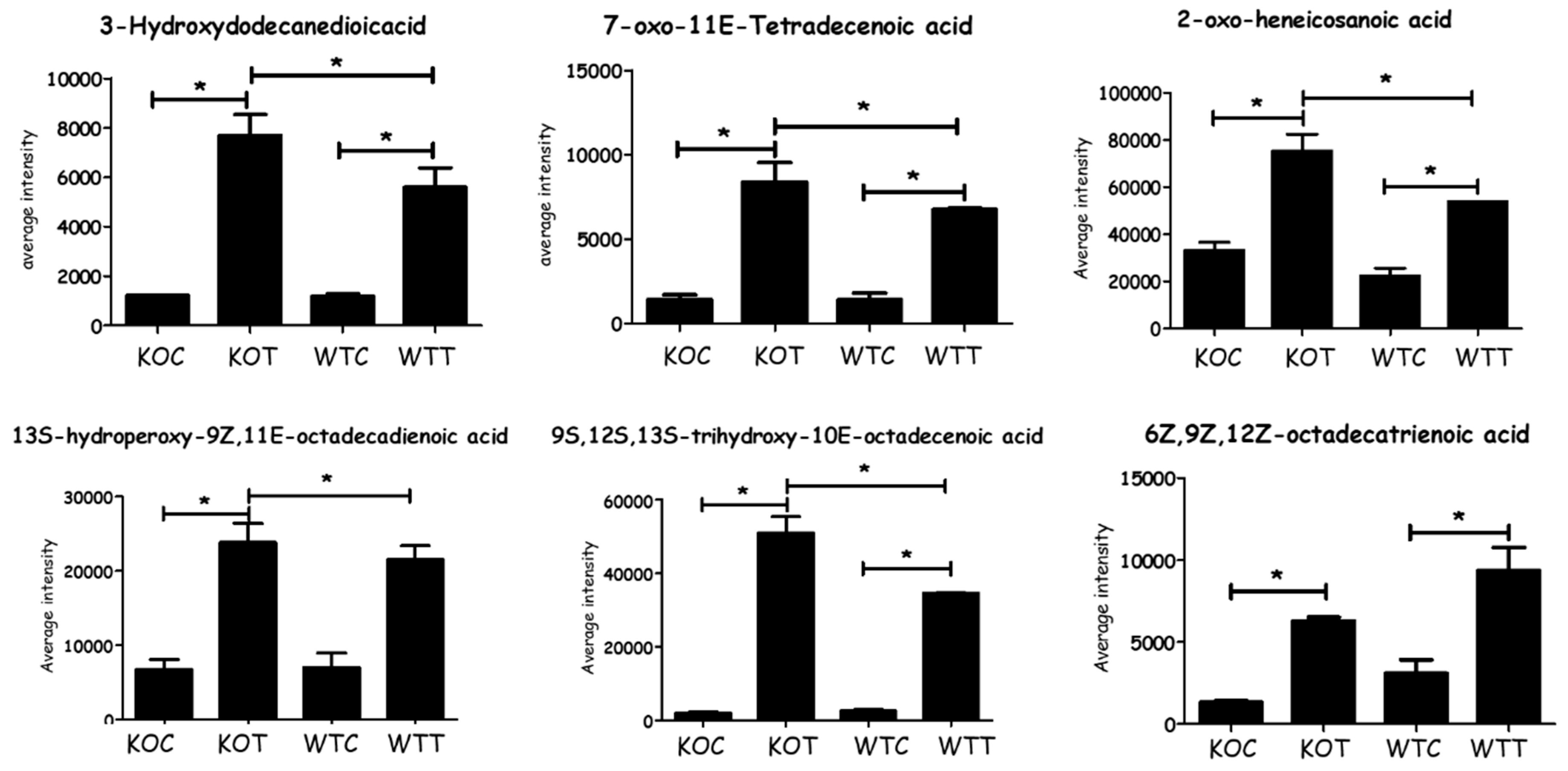
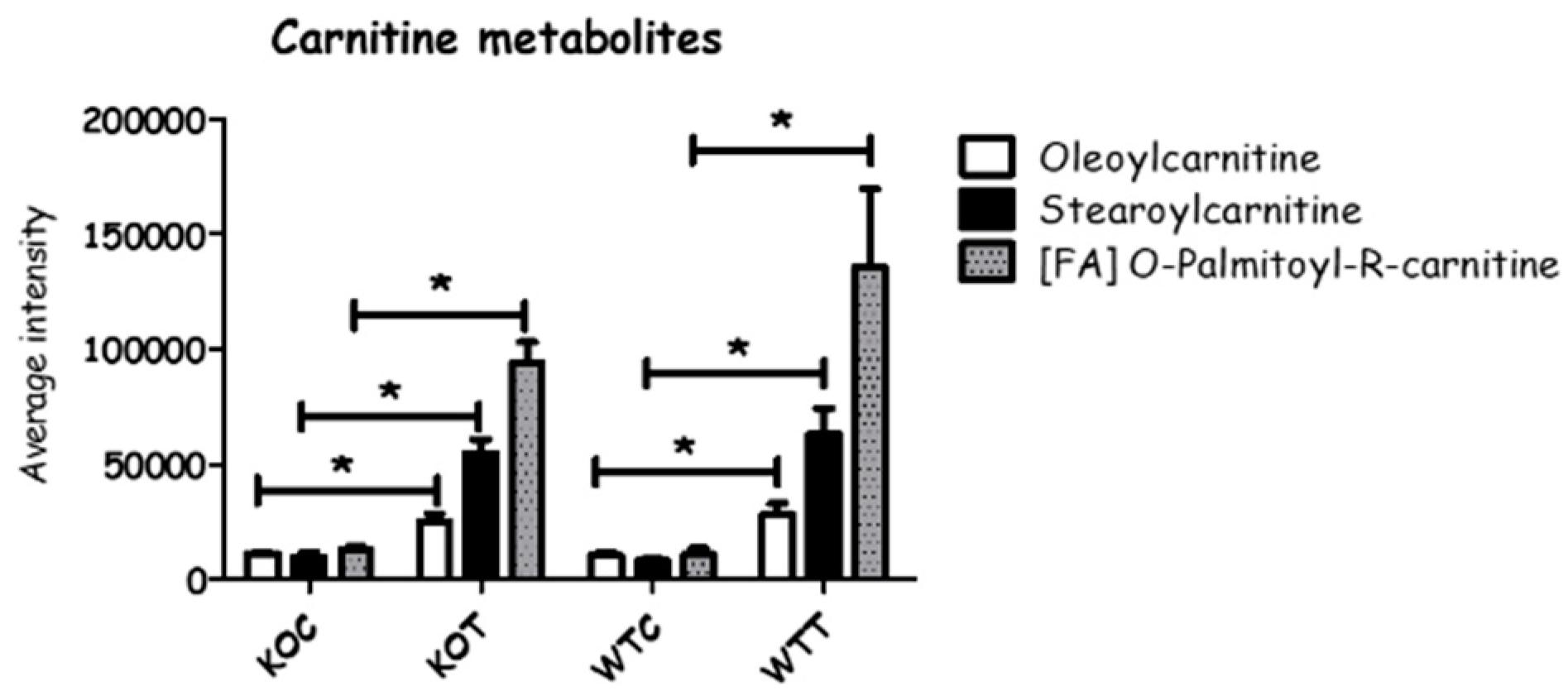
2.2. The Lipid Pathway Was Most Affected
2.3. Metabolic Changes in Glucose Metabolism
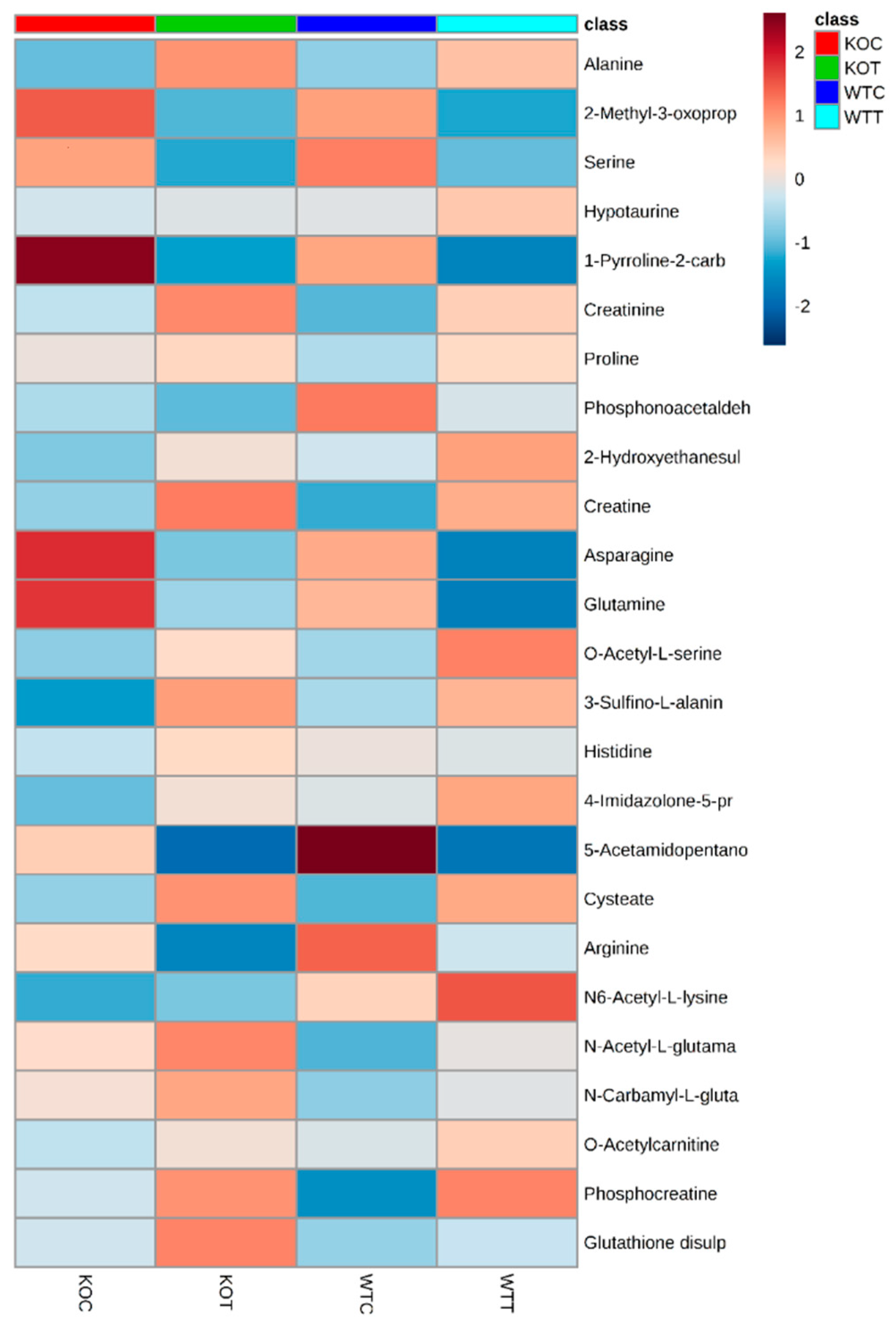
2.4. Metabolic Changes in Amino Acid Metabolism
2.5. Metabolic Changes in Nucleotide Metabolism
2.6. Increased Oxidative Stress in OxLDL-Treated Cells
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Liquid Chromatography Mass Spectrometry (LC/MS) Analysis
4.3. Data Extraction and Processing
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Ferris, F.L., 3rd; Wilkinson, C.P.; Bird, A.; Chakravarthy, U.; Chew, E.; Csaky, K.; Sadda, S.R. Beckman Initiative for Macular Research Classification Committee. Clinical classification of age-related macular degeneration. Ophthaology 2013, 120, 844–851. [Google Scholar] [CrossRef]
- Pikuleva, I.A.; Curcio, C.A. Cholesterol in the retina: The best is yet to come. Prog. Retin. Eye Res. 2014, 41, 64–89. [Google Scholar] [CrossRef] [Green Version]
- Handa, J.T.; Cano, M.; Wang, L.; Datta, S.; Liu, T. Lipids, oxidized lipids, oxidation-specific epitopes, and Age-related Macular Degeneration. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 430–440. [Google Scholar] [CrossRef]
- Yamada, Y.; Tian, J.; Yang, Y.; Cutler, R.G.; Wu, T.; Telljohann, R.S.; Mattson, M.P.; Handa, J.T. Oxidized low density lipoproteins induce a pathologic response by retinal pigmented epithelial cells. J. Neurochem. 2008, 105, 1187–1197. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, K.B.; Fijalkowski, N.; Cano, M.; Handa, J.T. Decreased membrane complement regulators in the retinal pigmented epithelium contributes to age-related macular degeneration. J. Pathol. 2013, 229, 729–742. [Google Scholar] [CrossRef]
- Conley, Y.P.; Thalamuthu, A.; Jakobsdottir, J.; Weeks, D.E.; Mah, T.; Ferrell, R.E.; Gorin, M.B. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum. Mol. Genet. 2005, 14, 1991–2002. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Stambolian, D.; Edwards, A.O.; Branham, K.E.; Othman, M.; Jakobsdottir, J.; Tosakulwong, N.; Pericak-Vance, M.A.; Campochiaro, P.A.; Klein, M.L.; et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2010, 107, 7401–7406. [Google Scholar] [CrossRef]
- Neale, B.M.; Fagerness, J.; Reynolds, R.; Sobrin, L.; Parker, M.; Raychaudhuri, S.; Tan, P.L.; Oh, E.C.; Merriam, J.E.; Souied, E.; et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc. Natl. Acad. Sci. USA 2010, 107, 7395–7400. [Google Scholar] [CrossRef] [Green Version]
- Storti, F.; Raphael, G.; Griesser, V.; Klee, K.; Drawnel, F.; Willburger, C.; Scholz, R.; Langmann, T.; von Eckardstein, A.; Fingerle, J.; et al. Regulated efflux of photoreceptor outer segment-derived cholesterol by human RPE cells. Exp. Eye Res. 2017, 165, 65–77. [Google Scholar] [CrossRef]
- Biswas, L.; Zhou, X.; Dhillon, B.; Graham, A.; Shu, X. Retinal pigment epithelium cholesterol efflux mediated by the 18 kDa translocator protein, TSPO, a potential target for treating age-related macular degeneration. Hum. Mol. Genet. 2017, 26, 4327–4339. [Google Scholar] [CrossRef]
- Zekavat, S.M.; Lu, J.; Maugeais, C.; Mazer, N.A. An in silico model of retinal cholesterol dynamics (RCDmodel): Insights into the pathophysiology of dry AMD. J. Lipid Res. 2017, 58, 1325–1337. [Google Scholar] [CrossRef]
- Bonsack, F.; Sukumari-Ramesh, S. TSPO: An Evolutionarily Conserved Protein with Elusive Functions. Int. J. Mol. Sci. 2018, 19, 1694. [Google Scholar] [CrossRef]
- Laíns, I.; Gantner, M.; Murinello, S.; Lasky-Su, J.A.; Miller, J.W.; Friedlander, M.; Husain, D. Metabolomics in the study of retinal health and disease. Prog. Retin. Eye Res. 2018. [Google Scholar] [CrossRef]
- Osborn, M.P.; Park, Y.; Parks, M.B.; Burgess, L.G.; Uppal, K.; Lee, K.; Jones, D.P.; Brantley, M.A., Jr. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS ONE 2013, 8, e72737. [Google Scholar] [CrossRef]
- Luo, D.; Deng, T.; Yuan, W.; Deng, H.; Jin, M. Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol. 2017, 17, 165. [Google Scholar] [CrossRef]
- Mitchell, S.L.; Uppal, K.; Williamson, S.M.; Liu, K.; Burgess, L.G.; Tran, V.; Umfress, A.C.; Jarrell, K.L.; Cooke Bailey, J.N.; Agarwal, A.; et al. The Carnitine Shuttle Pathway is Altered in Patients with Neovascular Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4978–4985. [Google Scholar] [CrossRef]
- Laíns, I.; Duarte, D.; Barros, A.S.; Martins, A.S.; Gil, J.; Miller, J.B.; Marques, M.; Mesquita, T.; Kim, I.K.; Cachulo, M.D.L.; et al. Human plasma metabolomics in age-related macular degeneration (AMD) using nuclear magnetic resonance spectroscopy. PLoS ONE 2017, 12, e0177749. [Google Scholar] [CrossRef]
- Laíns, I.; Kelly, R.S.; Miller, J.B.; Silva, R.; Vavvas, D.G.; Kim, I.K.; Murta, J.N.; Lasky-Su, J.; Miller, J.W.; Husain, D. Human Plasma Metabolomics Study across All Stages of Age-Related Macular Degeneration Identifies Potential Lipid Biomarkers. Ophthalmology 2018, 125, 245–254. [Google Scholar] [CrossRef]
- Li, M.; Zhang, X.; Liao, N.; Ye, B.; Peng, Y.; Ji, Y.; Wen, F. Analysis of the serum lipid profile in polypoidal choroidal vasculopathy. Sci. Rep. 2016, 6, 38342. [Google Scholar] [CrossRef]
- Tanito, M.; Anderson, R. Dual roles of polyunsaturated fatty acids in retinal physiology and pathophysiology associated with retinal degeneration. Clin. Lipidol. 2009, 4, 821–827. [Google Scholar] [CrossRef]
- Miyamura, N.; Ogawa, T.; Boylan, S.; Morse, L.S.; Handa, J.T.; Hjelmeland, L.M. Topographic and age-dependent expression of heme oxygenase-1 and catalase in the human retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1562–1565. [Google Scholar] [CrossRef]
- Liles, M.R.; Newsome, D.A.; Oliver, P.D. Antioxidant enzymes in the aging human retinal pigment epithelium. Arch. Ophthalmol. 1991, 109, 1285–1288. [Google Scholar] [CrossRef]
- Tu, L.N.; Zhao, A.H.; Hussein, M.; Stocco, D.M.; Selvaraj, V. Translocator protein (TSPO) affects mitochondrial fatty acid oxidation in steroidogenic cells. Endocrinology 2016, 157, 1110–1121. [Google Scholar] [CrossRef]
- Wyss, M.; Kaddurah-Daouk, R. Creatine and creatinine metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef]
- Zhao, C.; Yasumura, D.; Li, X.; Matthes, M.; Lloyd, M.; Nielsen, G.; Ahern, K.; Snyder, M.; Bok, D.; Dunaief, J.L.; et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Investig. 2011, 121, 369–383. [Google Scholar] [CrossRef]
- Winkler, B.S.; Arnold, M.J.; Brassell, M.A.; Puro, D.G. Energy metabolism in human retinal Müller cells. Investig. Ophthalmol. Vis. Sci. 2000, 41, 3183–3190. [Google Scholar]
- Kurihara, T.; Westenskow, P.D.; Gantner, M.L.; Usui, Y.; Schultz, A.; Bravo, S.; Aguilar, E.; Wittgrove, C.; Friedlander, M.S.; Paris, L.P.; et al. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. Elife 2016, 5, e14319. [Google Scholar] [CrossRef]
- Rohrer, B.; Bandyopadhyay, M.; Beeson, C. Reduced metabolic capacity in aged primary retinal pigment epithelium (rpe) is correlated with increased susceptibility to oxidative stress. In Retinal Degenerative Diseases; Springer: Berlin, Germany, 2016; pp. 793–798. [Google Scholar]
- Farooqui, A.A.; Ong, W.-Y.; Horrocks, L.A. Plasmalogens, docosahexaenoic acid and neurological disorders. In Peroxisomal Disorders and Regulation of Genes; Springer: Berlin, Germany, 2003; pp. 335–354. [Google Scholar]
- Hoffman, D.R.; Uauy, R.; Birch, D.G. Metabolism of omega-3 fatty acids in patients with autosomal dominant retinitis pigmentosa. Exp. Eye Res. 1995, 60, 279–289. [Google Scholar] [CrossRef]
- Krohne, T.U.; Stratmann, N.K.; Kopitz, J.; Holz, F.G. Effects of lipid peroxidation products on lipofuscinogenesis and autophagy in human retinal pigment epithelial cells. Exp. Eye Res. 2010, 90, 465–471. [Google Scholar] [CrossRef]
- Farooqui, A.A.; Horrocks, L.A.; Farooqui, T. Interactions between neural membrane glycerophospholipid and sphingolipid mediators: A recipe for neural cell survival or suicide. J. Neurosci. Res. 2007, 85, 1834–1850. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Bretillon, L. The ins and outs of cholesterol in the vertebrate retina. J. Lipid Res. 2010, 51, 3399–3413. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, I.R.; Larrayoz, I.M. Cholesterol oxidation in the retina: Implications of 7KCh formation in chronic inflammation and age-related macular degeneration. J. Lipid Res. 2010, 51, 2847–2862. [Google Scholar] [CrossRef]
- Kopitz, J.; Holz, F.G.; Kaemmerer, E.; Schutt, F. Lipids and lipid peroxidation products in the pathogenesis of age-related macular degeneration. Biochimie 2004, 86, 825–831. [Google Scholar] [CrossRef]
- Kaemmerer, E.; Schutt, F.; Krohne, T.U.; Holz, F.G.; Kopitz, J. Effects of Lipid Peroxidation-Related Protein Modifications on RPE Lysosomal Functions and POS Phagocytosis. Investig. Opthalmol. Vis. Sci. 2007, 48, 1342–1347. [Google Scholar] [CrossRef] [Green Version]
- Dunaief, J.L.; Dentchev, T.; Ying, G.-S.; Milam, A.H. The role of apoptosis in age-related macular degeneration. Arch. Ophthalmol. 2002, 120, 1435–1442. [Google Scholar] [CrossRef]
- Krohne, T.U.; Holz, F.G.; Kopitz, J. Apical-to-Basolateral Transcytosis of Photoreceptor Outer Segments Induced by Lipid Peroxidation Products in Human Retinal Pigment Epithelial Cells. Investig. Opthalmol. Vis. Sci. 2010, 51, 553–560. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Kamei, M.; Itabe, H.; Yoneda, K.; Bando, H.; Kume, N.; Tano, Y. Oxidized phospholipids in the macula increase with age and in eyes with age-related macular degeneration. Mol. Vis. 2007, 13, 772–778. [Google Scholar]
- Suzuki, M.; Tsujikawa, M.; Itabe, H.; Du, Z.J.; Xie, P.; Matsumura, N.; Fu, X.; Zhang, R.; Sonoda, K.H.; Egashira, K.; et al. Chronic photo-oxidative stress and subsequent MCP-1 activation as causative factors for age-related macular degeneration. J. Cell Sci. 2012, 125, 2407–2415. [Google Scholar] [CrossRef] [Green Version]
- Sanvicens, N.; Cotter, T.G. Ceramide is the key mediator of oxidative stress-induced apoptosis in retinal photoreceptor cells. J. Neurochem. 2006, 98, 1432–1444. [Google Scholar] [CrossRef] [Green Version]
- Howe, C.C.; Alshehri, A.; Muggeridge, D.; Mullen, A.B.; Boyd, M.; Spendiff, O.; Moir, H.J.; Watson, D.G. Untargeted metabolomics profiling of an 80.5 km simulated treadmill ultramarathon. Metabolites 2018, 8, 14. [Google Scholar] [CrossRef]
- Scheltema, R.A.; Jankevics, A.; Jansen, R.C.; Swertz, M.A.; Breitling, R. Peakml/mzmatch: A file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 2011, 83, 2786–2793. [Google Scholar] [CrossRef]
- Creek, D.J.; Jankevics, A.; Breitling, R.; Watson, D.G.; Barrett, M.P.; Burgess, K.E. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography–mass spectrometry: Improved metabolite identification by retention time prediction. Anal. Chem. 2011, 83, 8703–8710. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach tomultiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamri, A.; Biswas, L.; Watson, D.G.; Shu, X. Deletion of TSPO Resulted in Change of Metabolomic Profile in Retinal Pigment Epithelial Cells. Int. J. Mol. Sci. 2019, 20, 1387. https://doi.org/10.3390/ijms20061387
Alamri A, Biswas L, Watson DG, Shu X. Deletion of TSPO Resulted in Change of Metabolomic Profile in Retinal Pigment Epithelial Cells. International Journal of Molecular Sciences. 2019; 20(6):1387. https://doi.org/10.3390/ijms20061387
Chicago/Turabian StyleAlamri, Abdulwahab, Lincoln Biswas, David G. Watson, and Xinhua Shu. 2019. "Deletion of TSPO Resulted in Change of Metabolomic Profile in Retinal Pigment Epithelial Cells" International Journal of Molecular Sciences 20, no. 6: 1387. https://doi.org/10.3390/ijms20061387




