Early Developmental Zebrafish Embryo Extract to Modulate Senescence in Multisource Human Mesenchymal Stem Cells
Abstract
:1. Introduction
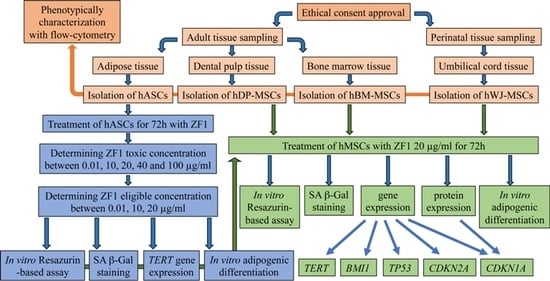
2. Results
2.1. hMSC Isolation and Phenotype Characterization
2.2. Evaluation of Different ZF1 Concentrations Cytotoxicity in hASCs
2.3. ZF1 Does Not Affect Proliferation in hASCs
2.4. ZF1 Decreases SA β-Gal Staining and Increases TERT Gene Expression in hASCs
2.5. ZF1 Promotes Adipogenesis in hASCs
2.6. ZF1 and Modulation of Cell Proliferation in hMSCs Isolated from Four Different Sources
2.7. ZF1 Reduces SA β-Gal Staining in hMSCs Isolated from Four Different Sources
2.8. ZF1 and Transcriptional Modulation of Stem Cell Senescence in Multisource hMSCs
2.9. ZF1 and p21 Expression in Multisource hMSCs
2.10. ZF1 and Adipogenic Commitment in hMSCs
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. hMSCs Harvesting and Culture
4.3. Zebrafish Embryo Extract, Dose Analysis and Treatments
4.4. BCA Protein Assay
4.5. In Vitro Resazurin-Based Toxicology Assay
4.6. Senescence-Associated β-Galactosidase Staining
4.7. Adipogenic Differentiation
4.8. RNA Extraction and RT-PCR
4.9. Real-Time PCR
4.10. Protein Extraction and Western Blot
4.11. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| α-MEM | alfa-Minimal Essential Medium |
| BMI1 | BMI1 proto-oncogene, polycomb ring finger |
| BM | bone marrow |
| CDKN1A (p21) | cyclin dependent kinase inhibitor 1A |
| CDKN2A (p16) | cyclin dependent kinase inhibitor 2A |
| c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
| CTR | control |
| DP | dental pulp |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| FBS | Fetal Bovine Serum |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| h | hour |
| hpf | hours post fertilization |
| hMSCs | human mesenchymal stem cells |
| hASCs | human adipose tissue-derived stem cells |
| HPRT1 | hypoxanthine phosphoribosyl transferase 1 |
| iPS | induced pluripotent stem |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| OCT-4 | POU domain class 5 homeobox 1 (POU5F1), alias Oct-4 |
| qPCR | quantitative relative real-time PCR |
| SA β-gal | Senescence-Associated β-Galactosidase |
| SDS-PAGE | Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis |
| SOLV | solvent |
| TBP | TATA box binding protein |
| TERT | telomerase reverse transcriptase |
| TP53 | tumor protein p53 |
| WJ | Wharton’s Jelly |
| ZF1 | Zebrafish extract |
References
- Bryder, D.; Rossi, D.J.; Weissman, I.L. Hematopoietic stem cells: The paradigmatic tissue-specific stem cell. Am. J. Pathol. 2006, 169, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Asahara, T.; Kawamoto, A. Endothelial progenitor cells for postnatal vasculogenesis. Am. J. Physiol. Cell Physiol. 2004, 287, C572–C579. [Google Scholar] [CrossRef] [Green Version]
- Murry, C.E.; Field, L.J.; Menasche, P. Cell-based cardiac repair: Reflections at the 10-year point. Circulation 2005, 112, 3174–3183. [Google Scholar] [CrossRef]
- Lindvall, O.; Kokaia, Z.; Martinez-Serrano, A. Stem cell therapy for human neurodegenerative disorders-how to make it work. Nat. Med. 2004, 10, S42–S50. [Google Scholar] [CrossRef]
- Bonner-Weir, S.; Weir, G.C. New sources of pancreatic beta-cells. Nat. Biotechnol. 2005, 23, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.; Riminucci, M.; Gronthos, S.; Robey, P.G. Bone marrow stromal stem cells: Nature, biology, and potential applications. Stem Cells 2001, 19, 180–192. [Google Scholar] [CrossRef]
- Crisan, M.; Yap, S.; Casteilla, L.; Chen, C.W.; Corselli, M.; Park, T.S.; Andriolo, G.; Sun, B.; Zheng, B.; Zhang, L.; et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008, 3, 301–313. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yang, X.; Han, Z.P.; Qu, F.F.; Shao, L.; Shi, Y.F. Mesenchymal stem cells: A new trend for cell therapy. Acta Pharmacol. Sin. 2013, 34, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Mahla, R.S. Stem cells applications in regenerative medicine and disease therapeutics. Int. J. Cell Biol. 2016, 2016, 6940283. [Google Scholar] [CrossRef]
- Bai, L.; Lennon, D.P.; Caplan, A.I.; DeChant, A.; Hecker, J.; Kranso, J.; Zaremba, A.; Miller, R.H. Hepatocyte growth factor mediates mesenchymal stem cell-induced recovery in multiple sclerosis models. Nat. Neurosci. 2012, 15, 862–870. [Google Scholar] [CrossRef]
- Chang, C.; Wang, X.; Niu, D.; Zhang, Z.; Zhao, H.; Gong, F. Mesenchymal stem cells adopt beta-cell fate upon diabetic pancreatic microenvironment. Pancreas 2009, 38, 275–281. [Google Scholar] [CrossRef]
- Kuo, Y.R.; Goto, S.; Shih, H.S.; Wang, F.S.; Lin, C.C.; Wang, C.T.; Huang, E.Y.; Chen, C.L.; Wei, F.C.; Zheng, X.X.; et al. Mesenchymal stem cells prolong composite tissue allotransplant survival in a swine model. Transplantation 2009, 87, 1769–1777. [Google Scholar] [CrossRef]
- Souza, B.S.; Nogueira, R.C.; de Oliveira, S.A.; de Freitas, L.A.; Lyra, L.G.; Ribeiro dos Santos, R.; Lyra, A.C.; Soares, M.B. Current status of stem cell therapy for liver diseases. Cell Transplant. 2009, 18, 1261–1279. [Google Scholar] [CrossRef]
- Quevedo, H.C.; Hatzistergos, K.E.; Oskouei, B.N.; Feigenbaum, G.S.; Rodriguez, J.E.; Valdes, D.; Pattany, P.M.; Zambrano, J.P.; Hu, Q.; McNiece, I.; et al. Allogeneic mesenchymal stem cells restore cardiac function in chronic ischemic cardiomyopathy via trilineage differentiating capacity. Proc. Natl. Acad. Sci. USA 2009, 106, 14022–14027. [Google Scholar] [CrossRef] [Green Version]
- Facchin, F.; Bianconi, E.; Canaider, S.; Basoli, V.; Biava, P.M.; Ventura, C. Tissue Regeneration without Stem Cell Transplantation: Self-Healing Potential from Ancestral Chemistry and Physical Energies. Stem Cells Int. 2018, 2018, 7412035. [Google Scholar] [CrossRef]
- Castorina, A.; Szychlinska, M.A.; Marzagalli, R.; Musumeci, G. Mesenchymal stem cells-based therapy as a potential treatment in neurodegenerative disorders: Is the escape from senescence an answer? Neural Regen. Res. 2015, 10, 850–858. [Google Scholar] [CrossRef]
- Szychlinska, M.A.; Stoddart, M.J.; D’Amora, U.; Ambrosio, L.; Alini, M.; Musumeci, G. Mesenchymal Stem Cell-Based Cartilage Regeneration Approach and Cell Senescence: Can We Manipulate Cell Aging and Function? Tissue Eng. Part. B Rev. 2017, 23, 529–539. [Google Scholar] [CrossRef]
- Tal, T.L.; Franzosa, J.A.; Tanguay, R.L. Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish—A mini-review. Gerontology 2010, 56, 231–240. [Google Scholar] [CrossRef]
- Shi, W.; Fang, Z.; Li, L.; Luo, L. Using zebrafish as the model organism to understand organ regeneration. Sci. China Life Sci. 2015, 58, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Goessling, W.; North, T.E. Repairing quite swimmingly: Advances in regenerative medicine using zebrafish. Dis. Model. Mech. 2014, 7, 769–776. [Google Scholar] [CrossRef]
- Brockerhoff, S.E.; Fadool, J.M. Genetics of photoreceptor degeneration and regeneration in zebrafish. Cell Mol. Life Sci. 2011, 68, 651–659. [Google Scholar] [CrossRef]
- Becker, T.; Becker, C.G. Axonal regeneration in zebrafish. Curr. Opin. Neurobiol. 2014, 27, 186–191. [Google Scholar] [CrossRef]
- Pozzoli, O.; Vella, P.; Iaffaldano, G.; Parente, V.; Devanna, P.; Lacovich, M.; Lamia, C.L.; Fascio, U.; Longoni, D.; Cotelli, F.; et al. Endothelial fate and angiogenic properties of human CD34+ progenitor cells in zebrafish. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1589–1597. [Google Scholar] [CrossRef]
- Canaider, S.; Maioli, M.; Facchin, F.; Bianconi, E.; Santaniello, S.; Pigliaru, G.; Ljungberg, L.; Burigana, F.; Bianchi, F.; Olivi, E.; et al. Human stem cell exposure to developmental stage zebrafish extracts: A novel strategy for tuning stemness and senescence patterning. CellR4 2014, 2, e1226. [Google Scholar]
- Facchin, F.; Canaider, S.; Bianconi, E.; Maioli, M.; Santoro, U.; Santaniello, S.; Basoli, V.; Biava, P.M.; Ventura, C. Zebrafish embryo extract counteracts human stem cell senescence. Front. Biosci. (Schol Ed.) 2019, 11, 89–104. [Google Scholar] [CrossRef]
- Mathavan, S.; Lee, S.G.; Mak, A.; Miller, L.D.; Murthy, K.R.; Govindarajan, K.R.; Tong, Y.; Wu, Y.L.; Lam, S.H.; Yang, H.; Ruan, Y.; et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005, 1, 260–276. [Google Scholar] [CrossRef]
- Yao, Y.; Ma, L.; Jia, Q.; Deng, W.; Liu, Z.; Zhang, Y.; Ren, J.; Xue, Y.; Jia, H.; Yang, Q. Systematic characterization of small RNAome during zebrafish early developmental stages. BMC Genom. 2014, 15, 117. [Google Scholar] [CrossRef]
- Palmblad, M.; Henkel, C.V.; Dirks, R.P.; Meijer, A.H.; Deelder, A.M.; Spaink, H.P. Parallel deep transcriptome and proteome analysis of zebrafish larvae. BMC Res. Notes 2013, 6, 428. [Google Scholar] [CrossRef]
- Haggard, D.E.; Noyes, P.D.; Waters, K.M.; Tanguay, R.L. Transcriptomic and phenotypic profiling in developing zebrafish exposed to thyroid hormone receptor agonists. Reprod. Toxicol. 2018, 77, 80–93. [Google Scholar] [CrossRef]
- Huang, X.; Agrawal, I.; Li, Z.; Zheng, W.; Lin, Q.; Gong, Z. Transcriptomic Analyses in Zebrafish Cancer Models for Global Gene Expression and Pathway Discovery. Adv. Exp. Med. Biol. 2016, 916, 147–168. [Google Scholar] [CrossRef]
- Li, C.; Tan, X.F.; Lim, T.K.; Lin, Q.; Gong, Z. Comprehensive and quantitative proteomic analyses of zebrafish plasma reveals conserved protein profiles between genders and between zebrafish and human. Sci. Rep. 2016, 6, 24329. [Google Scholar] [CrossRef] [Green Version]
- Tay, T.L.; Lin, Q.; Seow, T.K.; Tan, K.H.; Hew, C.L.; Gong, Z. Proteomic analysis of protein profiles during early development of the zebrafish, Danio rerio. Proteomics 2006, 6, 3176–3188. [Google Scholar] [CrossRef]
- Link, V.; Shevchenko, A.; Heisenberg, C.P. Proteomics of early zebrafish embryos. BMC Dev. Biol. 2006, 6, 1–9. [Google Scholar] [CrossRef]
- Lucitt, M.B.; Price, T.S.; Pizarro, A.; Wu, W.; Yocum, A.K.; Seiler, C.; Pack, M.A.; Blair, I.A.; Fitzgerald, G.A.; Grosser, T. Analysis of the zebrafish proteome during embryonic development. Mol. Cell Proteom. 2008, 7, 981–994. [Google Scholar] [CrossRef]
- Vesterlund, L.; Jiao, H.; Unneberg, P.; Hovatta, O.; Kere, J. The zebrafish transcriptome during early development. BMC Dev. Biol. 2011, 11, 30. [Google Scholar] [CrossRef]
- Forné, I.; Abián, J.; Cerdà, J. Fish proteome analysis: Model organisms and non-sequenced species. Proteomics 2010, 10, 858–872. [Google Scholar] [CrossRef]
- Saxena, S.; Singh, S.K.; Lakshmi, M.G.; Meghah, V.; Sundaram, C.S.; Swamy, C.V.; Idris, M.M. Proteome profile of zebrafish kidney. J. Proteom. 2011, 74, 2937–2947. [Google Scholar] [CrossRef]
- Smidak, R.; Aradska, J.; Kirchberger, S.; Distel, M.; Sialana, F.J.; Wackerlig, J.; Mechtcheriakova, D.; Lubec, G. A detailed proteomic profiling of plasma membrane from zebrafish brain. Proteom. Clin. Appl. 2016, 10, 1264–1268. [Google Scholar] [CrossRef]
- Carneiro, M.; Gutiérrez-Praena, D.; Osório, H.; Vasconcelos, V.; Carvalho, A.P.; Campos, A. Proteomic analysis of anatoxina acute toxicity in zebrafish reveals gender specific responses and additional mechanisms of cell stress. Ecotoxicol. Environ. Saf. 2015, 120, 93–101. [Google Scholar] [CrossRef]
- Biava, P.M.; Canaider, S.; Facchin, F.; Bianconi, E.; Ljungberg, L.; Rotilio, D.; Burigana, F.; Ventura, C. Stem cell differentiation stage factors from zebrafish embryo: A novel strategy to modulate the fate of normal and pathological human (stem) cells. Curr. Pharm. Biotechnol. 2015, 16, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Paradisi, M.; Alviano, F.; Pirondi, S.; Lanzoni, G.; Fernandez, M.; Lizzo, G.; Giardino, L.; Giuliani, A.; Costa, R.; Marchionni, C.; Bonsi, L.; Calza, L. Human mesenchymal stem cells produce bioactive neurotrophic factors: Source, individual variability and differentiation issues. Int. J. Immunopathol. Pharmacol. 2014, 27, 391–402. [Google Scholar] [CrossRef]
- Bianchi, F.; Maioli, M.; Leonardi, E.; Olivi, E.; Pasquinelli, G.; Valente, S.; Mendez, A.J.; Ricordi, C.; Raffaini, M.; Tremolada, C.; Ventura, C. A new nonenzymatic method and device to obtain a fat tissue derivative highly enriched in pericyte-like elements by mild mechanical forces from human lipoaspirates. Cell Transplant. 2013, 22, 2063–2077. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, Y.; Liu, D.; Zhao, J.; Xu, J.; Ren, J.; Hu, Y.; Wang, Z.; Hou, Y.; Zhao, G. MiR-495 promotes senescence of mesenchymal stem cells by targeting Bmi-1. Cell Physiol. Biochem. 2017, 42, 780–796. [Google Scholar] [CrossRef] [PubMed]
- Molofsky, A.V.; Pardal, R.; Iwashita, T.; Park, I.K.; Clarke, M.F.; Morrison, S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 2003, 425, 962–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rando, T.A. Stem cells, ageing and the quest for immortality. Nature 2006, 441, 1080–1086. [Google Scholar] [CrossRef]
- Da Silva, M.L.; Chagastelles, P.C.; Nardi, N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 2006, 119, 2204–2213. [Google Scholar] [CrossRef] [Green Version]
- Neirinckx, V.; Coste, C.; Rogister, B.; Wislet-Gendebien, S. Concise review: Adult mesenchymal stem cells, adult neural crest stem cells, and therapy of neurological pathologies: A state of play. Stem Cells Transl. Med. 2013, 2, 284–296. [Google Scholar] [CrossRef]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed]
- Parolini, O.; Alviano, F.; Bagnara, G.P.; Bilic, G.; Bühring, H.J.; Evangelista, M.; Hennerbichler, S.; Liu, B.; Magatti, M.; Mao, N.; et al. Concise review: Isolation and characterization of cells from human term placenta: Outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells 2008, 26, 300–311. [Google Scholar] [CrossRef]
- La Rocca, G.; Lo Iacono, M.; Corsello, T.; Corrao, S.; Farina, F.; Anzalone, R. Human Wharton’s jelly mesenchymal stem cells maintain the expression of key immunomodulatory molecules when subjected to osteogenic, adipogenic and chondrogenic differentiation in vitro: New perspectives for cellular therapy. Curr. Stem Cell Res. Ther. 2013, 8, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Laurent, L.C.; Ulitsky, I.; Slavin, I.; Tran, H.; Schork, A.; Morey, R.; Lynch, C.; Harness, J.V.; Lee, S.; Barrero, M.J.; et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 2011, 8, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, A.; Rossi, M.; Alviano, F.; Poggi, P.; Zannini, C.; Marchionni, C.; Ricci, F.; Tazzari, P.L.; Taglioli, V.; Calder, P.C.; Bonsi, L. Restored in vivo-like membrane lipidomics positively influence in vitro features of cultured mesenchymal stromal/stem cells derived from human placenta. Stem Cell Res. Ther. 2017, 8, 31. [Google Scholar] [CrossRef]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Comm Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Samsonraj, R.M.; Raghunath, M.; Nurcombe, V.; Hui, J.H.; van Wijnen, A.J.; Cool, S.M. Concise Review: Multifaceted Characterization of Human Mesenchymal Stem Cells for Use in Regenerative Medicine. Stem Cells Transl. Med. 2017, 6, 2173–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Liu, Y.; Sun, Y.; Wang, B.; Xiong, Y.; Lin, W.; Wei, Q.; Wang, H.; He, W.; Wang, B.; Li, G. Tissue source determines the differentiation potentials of mesenchymal stem cells: A comparative study of human mesenchymal stem cells from bone marrow and adipose tissue. Stem Cell Res. Ther. 2017, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Shay, J.W. Actions of human telomerase beyond telomeres. Cell Res. 2008, 18, 725–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, R.; Mustafi, S.B.; Street, M.; Dey, A.; Dwivedi, S.K. Bmi-1: At the crossroads of physiological and pathological biology. Genes Dis. 2015, 2, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Lee, Y.D.; Wagers, A.J. Stem cell aging: Mechanisms, regulators and therapeutic opportunities. Nat. Med. 2014, 20, 870–880. [Google Scholar] [CrossRef]
- Stampfer, M.R.; Garbe, J.; Nijjar, T.; Wigington, D.; Swisshelm, K.; Yaswen, P. Loss of p53 function accelerates acquisition of telomerase activity in indefinite lifespan human mammary epithelial cell lines. Oncogene 2003, 22, 5238–5251. [Google Scholar] [CrossRef] [Green Version]
- Artandi, S.E.; Attardi, L.D. Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem. Biophys. Res. Commun. 2005, 331, 881–890. [Google Scholar] [CrossRef]
- Shats, I.; Milyavsky, M.; Tang, X.; Stambolsky, P.; Erez, N.; Brosh, R.; Kogan, I.; Braunstein, I.; Tzukerman, M.; Ginsberg, D.; Rotter, V. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 2004, 279, 50976–50985. [Google Scholar] [CrossRef]
- Liu, T.M.; Ng, W.M.; Tan, H.S.; Vinitha, D.; Yang, Z.; Fan, J.B.; Zou, Y.; Hui, J.H.; Lee, E.H.; Lim, B. Molecular basis of immortalization of human mesenchymal stem cells by combination of p53 knockdown and human telomerase reverse transcriptase overexpression. Stem Cells Dev. 2013, 22, 268–278. [Google Scholar] [CrossRef]
- Sarin, K.Y.; Cheung, P.; Gilison, D.; Lee, E.; Tennen, R.I.; Wang, E.; Artandi, M.K.; Oro, A.E.; Artandi, S.E. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 2005, 436, 1048–1052. [Google Scholar] [CrossRef]
- Choi, J.; Southworth, L.K.; Sarin, K.Y.; Venteicher, A.S.; Ma, W.; Chang, W.; Cheung, P.; Jun, S.; Artandi, M.K.; Shah, N.; Kim, S.K.; Artandi, S.E. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008, 4, e10. [Google Scholar] [CrossRef]
- Bieback, K.; Brinkmann, I. Mesenchymal stromal cells from human perinatal tissues: From biology to cell therapy. World J. Stem Cells. 2010, 2, 81–92. [Google Scholar] [CrossRef]
- Marchionni, C.; Bonsi, L.; Alviano, F.; Lanzoni, G.; Di Tullio, A.; Costa, R.; Montanari, M.; Tazzari, P.L.; Ricci, F.; Pasquinelli, G.; et al. Angiogenic potential of human dental pulp stromal (stem) cells. Int. J. Immunopathol. Pharmacol. 2009, 22, 699–706. [Google Scholar] [CrossRef]
- Pierdomenico, L.; Bonsi, L.; Calvitti, M.; Rondelli, D.; Arpinati, M.; Chirumbolo, G.; Becchetti, E.; Marchionni, C.; Alviano, F.; Fossati, V.; et al. Multipotent mesenchymal stem cells with immunosuppressive activity can be easily isolated from dental pulp. Transplantation 2005, 80, 836–842. [Google Scholar] [CrossRef]
- La Rocca, G.; Anzalone, R.; Corrao, S.; Magno, F.; Loria, T.; Lo Iacono, M.; Di Stefano, A.; Giannuzzi, P.; Marasà, L.; Cappello, F.; et al. Isolation and characterization of Oct-4+/HLA-G+ mesenchymal stem cells from human umbilical cord matrix: Differentiation potential and detection of new markers. Histochem. Cell Biol. 2009, 131, 267–282. [Google Scholar] [CrossRef]
- Livraghi, T.; Meloni, F.; Frosi, A.; Lazzaroni, S.; Bizzarri, T.M.; Frati, L.; Biava, P.M. Treatment with stem cell differentiation stage factors of intermediate-advanced hepatocellular carcinoma: An open randomized clinical trial. Oncol. Res. 2005, 15, 399–408. [Google Scholar] [CrossRef]
- Rinaldi, S.; Maioli, M.; Pigliaru, G.; Castagna, A.; Santaniello, S.; Basoli, V.; Fontani, V.; Ventura, C. Stem cell senescence. Effects of REAC technology on telomerase-independent and telomerase-dependent pathways. Sci. Rep. 2014, 4, 6373. [Google Scholar] [CrossRef] [Green Version]
- Facchin, F.; Bianconi, E.; Romano, M.; Impellizzeri, A.; Alviano, F.; Maioli, M.; Canaider, S.; Ventura, C. Comparison of Oxidative Stress Effects on Senescence Patterning of Human Adult and Perinatal Tissue-Derived Stem Cells in Short and Long-term Cultures. Int. J. Med. Sci. 2018, 15, 1486–1501. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Alviano, F.; Ricci, F.; Vignoli, F.; Marchionni, C.; Valente, S.; Zannini, C.; Tazzari, P.L.; Vignoli, M.; Bartoletti, E.; Bonsi, L. In vitro multilineage potential and immunomodulatory properties of adipose derived stromal/stem cells obtained from nanofat lipoaspirates. CellR4 2016, 4, e2212. [Google Scholar]
- Casadei, R.; Piovesan, A.; Vitale, L.; Facchin, F.; Pelleri, M.C.; Canaider, S.; Bianconi, E.; Frabetti, F.; Strippoli, P. Genome-scale analysis of human mRNA 5’ coding sequences based on expressed sequence tag (EST) database. Genomics 2012, 100, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Facchin, F.; Vitale, L.; Bianconi, E.; Piva, F.; Frabetti, F.; Strippoli, P.; Casadei, R.; Pelleri, M.C.; Piovesan, A.; Canaider, S. Complexity of bidirectional transcription and alternative splicing at human RCAN3 locus. PLoS ONE 2011, 6, e24508. [Google Scholar] [CrossRef]
- Beraudi, A.; Bianconi, E.; Catalani, S.; Canaider, S.; De Pasquale, D.; Apostoli, P.; Bordini, B.; Stea, S.; Toni, A.; Facchin, F. In vivo response of heme-oxygenase-1 to metal ions released from metal-on-metal hip prostheses. Mol. Med. Rep. 2016, 14, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; Wittwer, C.T. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]












© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Facchin, F.; Alviano, F.; Canaider, S.; Bianconi, E.; Rossi, M.; Bonsi, L.; Casadei, R.; Biava, P.M.; Ventura, C. Early Developmental Zebrafish Embryo Extract to Modulate Senescence in Multisource Human Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 2646. https://doi.org/10.3390/ijms20112646
Facchin F, Alviano F, Canaider S, Bianconi E, Rossi M, Bonsi L, Casadei R, Biava PM, Ventura C. Early Developmental Zebrafish Embryo Extract to Modulate Senescence in Multisource Human Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2019; 20(11):2646. https://doi.org/10.3390/ijms20112646
Chicago/Turabian StyleFacchin, Federica, Francesco Alviano, Silvia Canaider, Eva Bianconi, Martina Rossi, Laura Bonsi, Raffaella Casadei, Pier Mario Biava, and Carlo Ventura. 2019. "Early Developmental Zebrafish Embryo Extract to Modulate Senescence in Multisource Human Mesenchymal Stem Cells" International Journal of Molecular Sciences 20, no. 11: 2646. https://doi.org/10.3390/ijms20112646