TPS Genes Silencing Alters Constitutive Indirect and Direct Defense in Tomato
Abstract
:1. Introduction
2. Results
2.1. Development of Transgenic Tomato Lines and Confirmation of TPS Genes Silencing
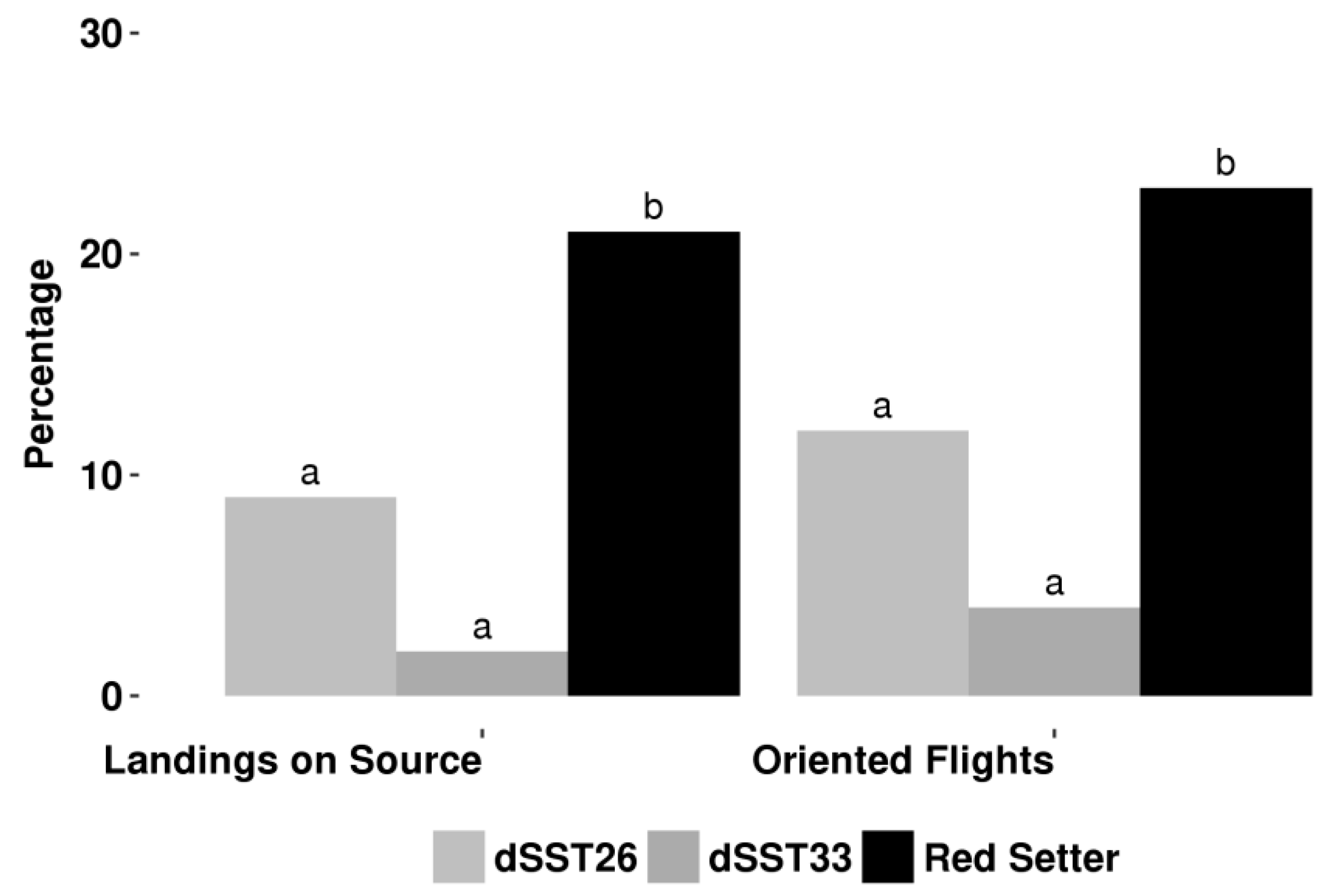
2.2. TPS Genes Silencing Compromises Indirect Defence
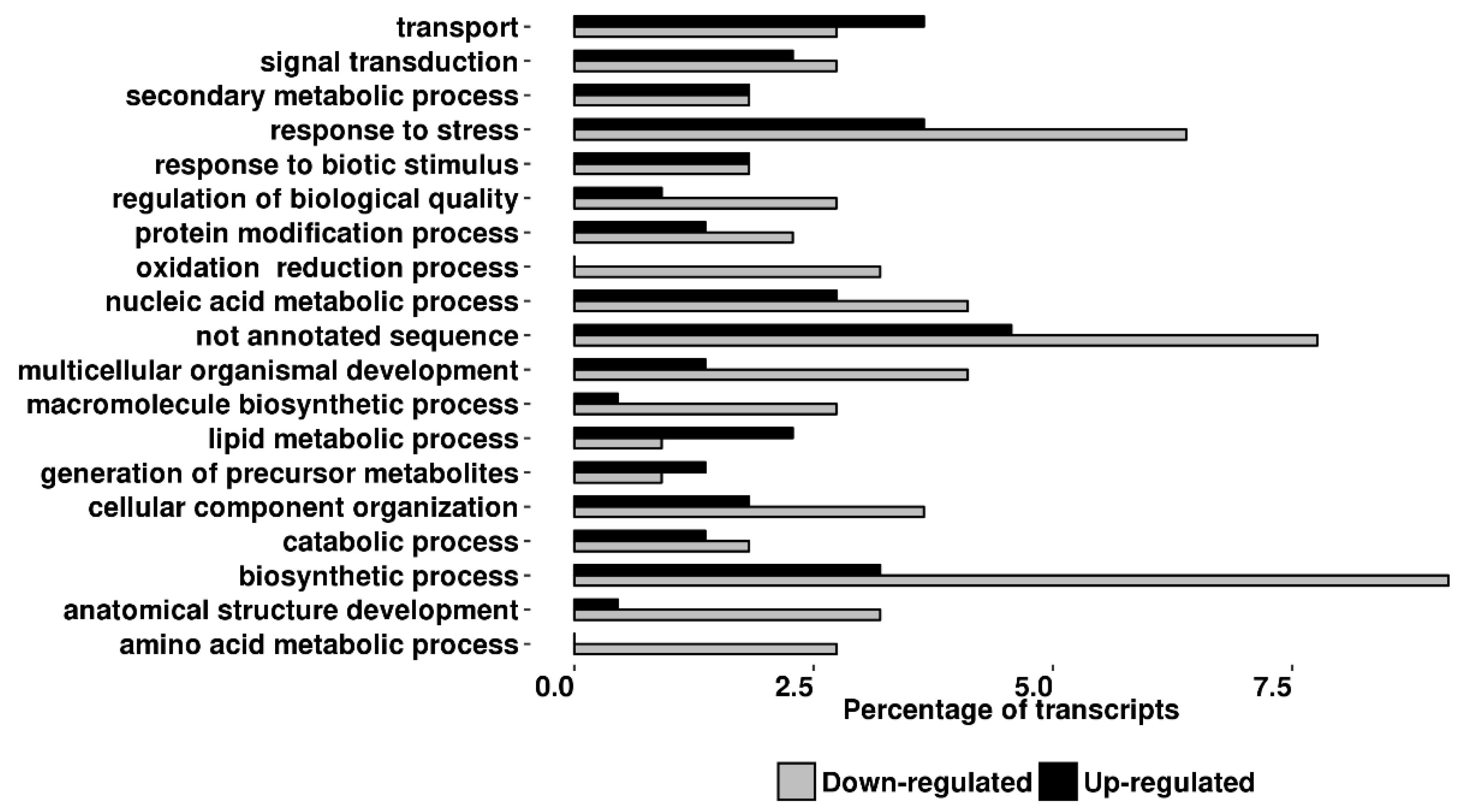
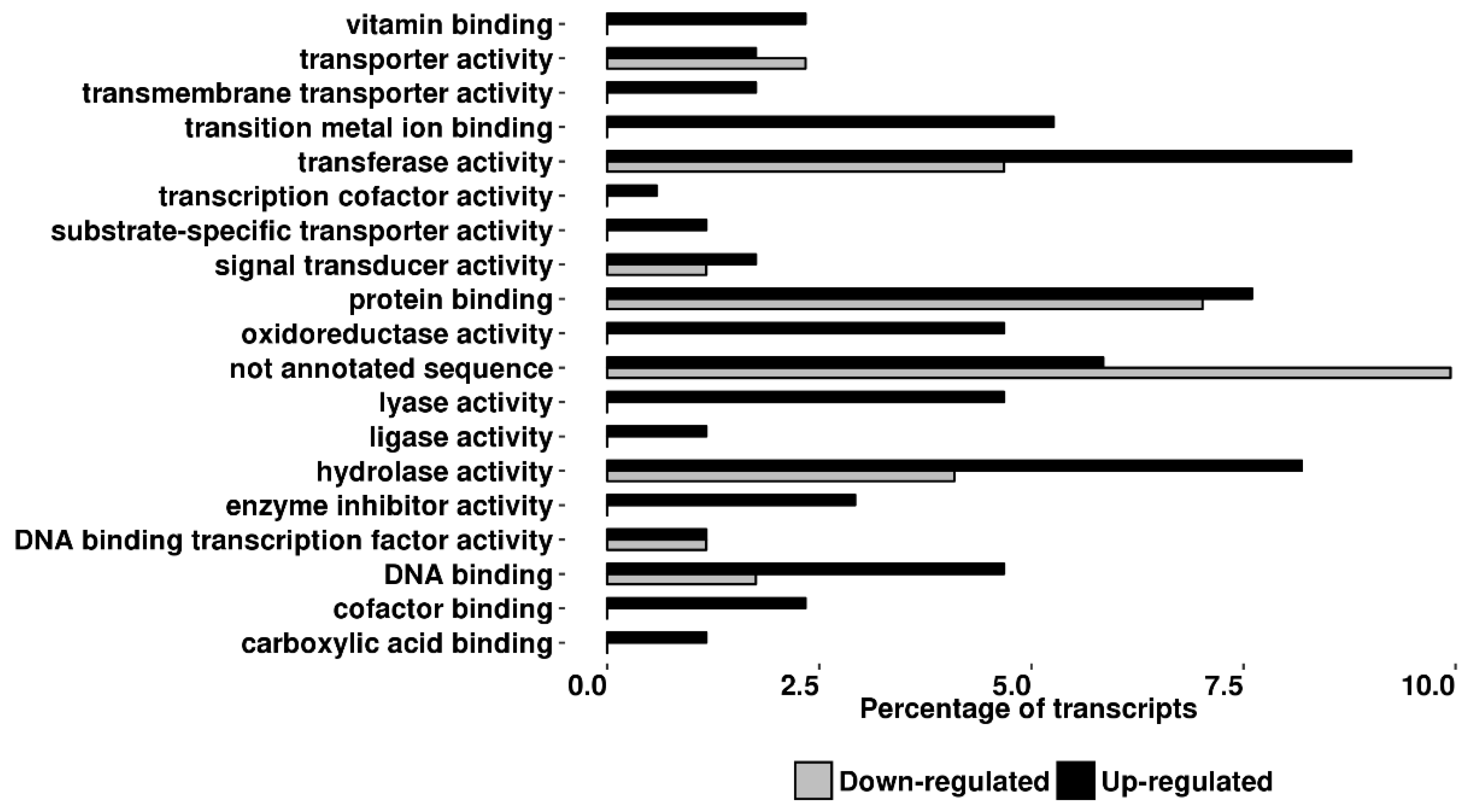
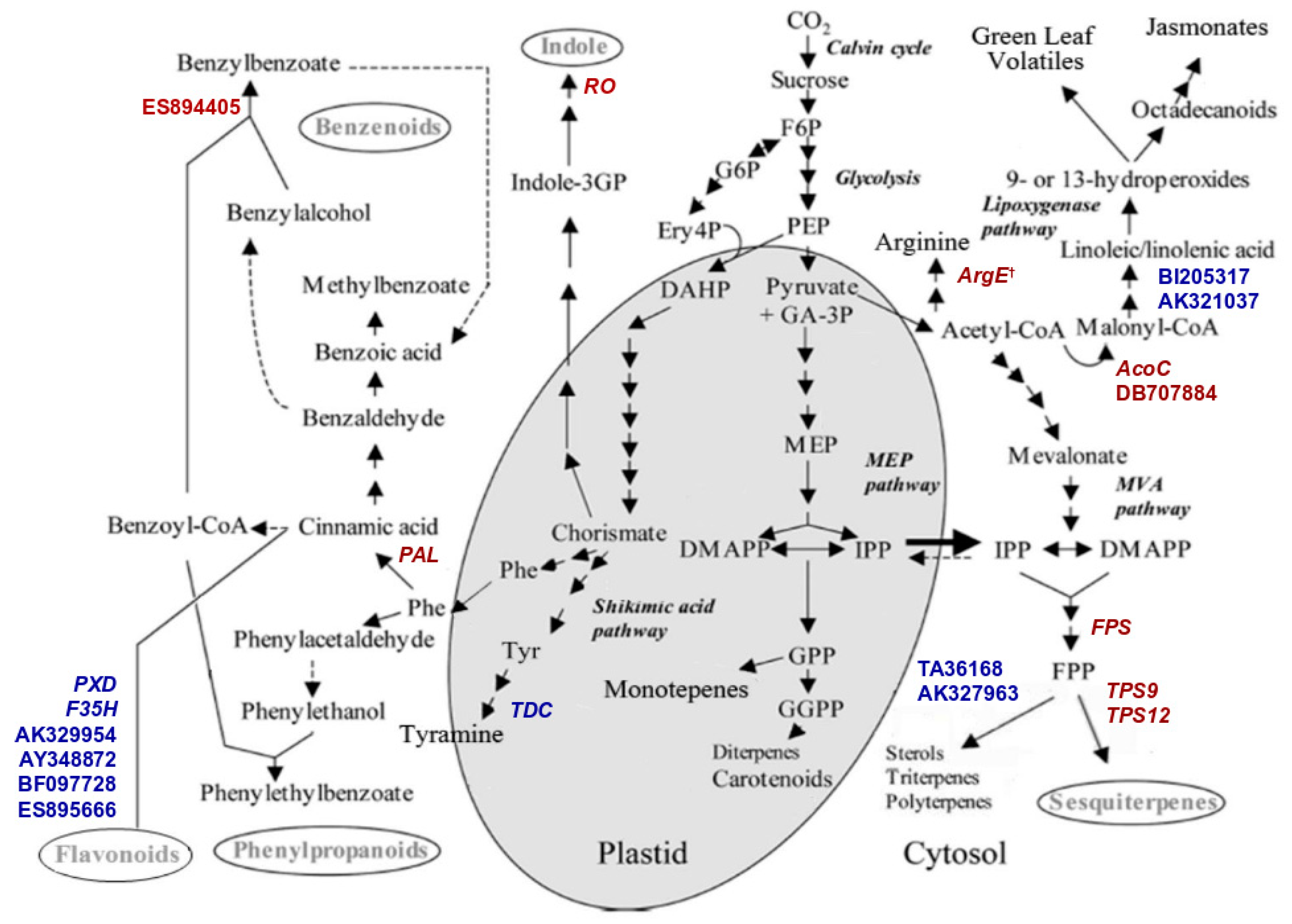
2.3. Transcriptomic Analyses
2.4. TPS Genes Silencing Influences Direct Defence
3. Discussion
4. Material and Methods
4.1. Genetic Construct and Tomato Transformation
4.2. RNA Extraction, cDNA Synthesis and Real-Time RT-PCR Analysis
4.3. Indirect Defence Bioassay
4.4. VOC Collection and Analysis
4.5. Microarray and Functional Analysis
4.6. Direct Defence Biossay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Standards Used for the Identification of Volatiles Collected by Air-Entrainment of Head Space from Tomato Plants
References
- Ponzio, C.; Cascone, P.; Cusumano, A.; Weldegergis, B.T.; Fatouros, N.E.; Guerrieri, E.; Dicke, M.; Gols, R. Volatile-mediated foraging behaviour of three parasitoid species under conditions of dual insect herbivore attack. Anim. Behav. 2016, 111, 197–206. [Google Scholar] [CrossRef]
- Aartsma, Y.; Bianchi, F.J.J.A.; Van Der Werf, W.; Poelman, E.H.; Dicke, M. Herbivore-induced plant volatiles and tritrophic interactions across spatial scales. New Phytol. 2017, 216, 1054–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turlings, T.C.J.; Erb, M. Tritrophic interactions mediated by herbivore-induced plant volatiles: Mechanisms, ecological relevance, and application potential. Annu. Rev. Entomol. 2018, 63, 433–452. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Karban, R. Explaining evolution of plant communication by airborne signals. Trends Ecol. Evol. 2010, 25, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Catola, S.; Centritto, M.; Cascone, P.; Ranieri, A.; Loreto, F.; Calamai, L.; Balestrini, R.; Guerrieri, E. Effects of single or combined water deficit and aphid attack on tomato volatile organic compound (VOC) emission and plant-plant communication. Environ. Exp. Bot. 2018, 153. [Google Scholar] [CrossRef]
- De Moraes, C.M.; Mescher, M.C.; Tumlinson, J.H. Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 2001, 410, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Digilio, M.C.; Cascone, P.; Iodice, L.; Guerrieri, E. Interactions between tomato volatile organic compounds and aphid behaviour. J. Plant Interact. 2012, 7, 322–325. [Google Scholar] [CrossRef] [Green Version]
- Dudareva, N.; Negre, F.; Nagegowda, D.A.; Orlova, I. Plant volatiles: Recent advances and future perspectives. CRC Crit. Rev. Plant Sci. 2006, 25, 417–440. [Google Scholar] [CrossRef]
- Sasso, R.; Iodice, L.; Digilio, M.C.; Carretta, A.; Ariati, L.; Guerrieri, E. Host-locating response by the aphid parasitoid Aphidius ervi to tomato plant volatiles. J. Plant Interact. 2007, 2, 175–183. [Google Scholar] [CrossRef]
- Raghava, T.; Ravikumar, P.; Hegde, R.; Kush, A. Spatial and temporal volatile organic compound response of select tomato cultivars to herbivory and mechanical injury. Plant Sci. 2010, 179, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Turlings, T.C.J.; Tumlinson, J.H.; Lewis, W.J. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 1990, 250, 1251–1253. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.K.; Cox, M.M.; Bede, J.C.; Inoue, K.; Alborn, H.T.; Tumlinson, J.H.; Korth, K.L. Lepidopteran herbivory and oral factors induce transcripts encoding novel terpene synthases in Medicago truncatula. Arch. Insect Biochem. Physiol. 2005, 58, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Price, P.W.; Bouton, C.E.; Gross, P.; McPheron, B.A.; Thompson, J.N.; Weis, A.E. Interactions among three trophic levels: Influence of plants on interactions between insect herbivores and natural enemies. Annu. Rev. Ecol. Syst. 1980, 11, 41–65. [Google Scholar] [CrossRef]
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science 2001, 291, 2141–2144. [Google Scholar] [CrossRef] [PubMed]
- Arimura, G.I.; Kost, C.; Boland, W. Herbivore-induced, indirect plant defences. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2005, 1734, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Held, M.; Fantaye, C.A.; Turlings, T.C.; Degenhardt, J.; Gershenzon, J. Attractiveness of constitutive and herbivore-induced sesquiterpene blends of maize to the parasitic wasp Cotesia marginiventris (Cresson). J. Chem. Ecol. 2011, 37, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Digilio, M.C.; Corrado, G.; Sasso, R.; Coppola, V.; Iodice, L.; Pasquariello, M.; Bossi, S.; Maffei, M.E.; Coppola, M.; Pennacchio, F.; et al. Molecular and chemical mechanisms involved in aphid resistance in cultivated tomato. New Phytol. 2010, 187, 1089–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farag, M.A.; Paré, P.W. C6-green leaf volatiles trigger local and systemic VOC emissions in tomato. Phytochemistry 2002, 61, 545–554. [Google Scholar] [CrossRef]
- Ament, K. Jasmonic Acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol. 2004, 135, 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hernández, C.; López, M.G.; Délano-Frier, J.P. Reduced levels of volatile emissions in jasmonate-deficient spr2 tomato mutants favour oviposition by insect herbivores. Plant Cell Environ. 2006, 29, 546–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farag, M.A. Jasmonate-deficient plants have reduced direct and indirect defenses against herbivores. Ecol. Lett. 2002, 5, 764–774. [Google Scholar]
- Falara, V.; Akhtar, T.A.; Nguyen, T.T.H.; Spyropoulou, E.A.; Bleeker, P.M.; Schauvinhold, I.; Matsuba, Y.; Bonini, M.E.; Schilmiller, A.L.; Last, R.L.; et al. The tomato terpene synthase gene family. Plant Physiol. 2011, 157, 770–789. [Google Scholar] [CrossRef] [PubMed]
- Colby, S.M.; Crock, J.; Dowdle-Rizzo, B.; Lemaux, P.G.; Croteau, R. Germacrene C synthase from Lycopersicon esculentum cv. VFNT cherry tomato: CDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc. Natl. Acad. Sci. USA 1998, 95, 2216–2221. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, P.M.; Mirabella, R.; Diergaarde, P.J.; VanDoorn, A.; Tissier, A.; Kant, M.R.; Prins, M.; de Vos, M.; Haring, M.A.; Schuurink, R.C. Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proc. Natl. Acad. Sci. USA 2012, 109, 20124–20129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, A.X.; Xiang, C.Y.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Lou, Y.G.; Chen, X.Y. The rice (E)-β-caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry 2007, 68, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Kappers, I.F.; Aharoni, A.; Van Herpen, T.W.J. M.; Luckerhoff, L.L.P.; Dicke, M.; Bouwmeester, H.J. Plant science: Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 2005, 309, 2070–2072. [Google Scholar] [CrossRef] [PubMed]
- Schnee, C.; Kollner, T.G.; Held, M.; Turlings, T.C.J.; Gershenzon, J.; Degenhardt, J. The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. Acad. Sci. USA 2006, 103, 1129–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollner, T.G.; Held, M.; Lenk, C.; Hiltpold, I.; Turlings, T.C.J.; Gershenzon, J.; Degenhardt, J. A maize (E)-β-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most american maize varieties. Plant Cell Online 2008, 20, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Varsha Wesley, S.; Helliwell, C.A.; Smith, N.A.; Wang, M.; Rouse, D.T.; Liu, Q.; Gooding, P.S.; Singh, S.P.; Abbott, D.; Stoutjesdijk, P.A.; et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001, 27, 581–590. [Google Scholar] [CrossRef] [Green Version]
- Olsen, K.M.; Hehn, A.; Jugdé, H.; Slimestad, R.; Larbat, R.; Bourgaud, F.; Lillo, C. Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Mathews, H. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell Online 2003, 15, 1689–1703. [Google Scholar] [CrossRef]
- Liu, J.J.; Sturrock, R.; Ekramoddoullah, A.K.M. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010, 29, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Negre, F. Regulation of Methylbenzoate Emission after pollination in snapdragon and petunia flowers. Plant Cell Online 2003, 15, 2992–3006. [Google Scholar] [CrossRef] [PubMed]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Colquhoun, T.A.; Verdonk, J.C.; Schimmel, B.C.J.; Tieman, D.M.; Underwood, B.A.; Clark, D.G. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry 2010, 71, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.M.; Tantasawat, P.; Steffens, J.C. Tomato polyphenol oxidase B is spatially and temporally regulated during development and in response to ethylene. Molecules 2011, 16, 493–517. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Kapteyn, J.; Gang, D.R. A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J. 2008, 54, 349–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichersky, E.; Noel, J.P.; Dudareva, N. Biosynthesis of plant volatiles: Nature’s diversity and ingenuity. Science 2006, 311, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.S.J. Flavonoid-insect interactions: Recent advances in our knowledge. Phytochemistry 2003, 64, 21–30. [Google Scholar] [CrossRef]
- Simmonds, M.S.J. Importance of flavonoids in insect- plant interactions: Feeding and oviposition. Phytochemistry 2001, 56, 245–252. [Google Scholar] [CrossRef]
- Hatano, E.; Kunert, G.; Michaud, J.P.; Weisser, W.W. Chemical cues mediating aphid location by natural enemies. Eur. J. Entomol. 2008, 105, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Elzen, G.W.; Williams, H.J.; Vinson, S.B. Wind tunnel flight responses by hymenopterous parasitoid Campoletis sonorensis to cotton cultivars and lines. Entomol. Exp. Appl. 1986, 42, 285–289. [Google Scholar] [CrossRef]
- Elzen, G.W.; Williams, H.J.; Vinson, S.B.; Powell, J.E. Comparative flight behavior of parasitoids Campoletis sonorensis and Microplitis croceipes. Entomol. Exp. Appl. 1987, 45, 175–180. [Google Scholar] [CrossRef]
- Kariyat, R.R.; Mauck, K.E.; De Moraes, C.M.; Stephenson, A.G.; Mescher, M.C. Inbreeding alters volatile signalling phenotypes and influences tri-trophic interactions in horsenettle (Solanum carolinense L.). Ecol. Lett. 2012, 15, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, D.C.; Refi-Hind, S.; Stratmann, J.W.; Lincoln, D.E. Systemin and jasmonic acid regulate constitutive and herbivore-induced systemic volatile emissions in tomato, Solanum lycopersicum. Phytochemistry 2010, 71, 2024–2037. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, L.; Zhao, J.; Li, C.; Ge, F.; Kang, L. Ecological trade-offs between jasmonic acid-dependent direct and indirect plant defences in tritrophic interactions. New Phytol. 2011, 189, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Han, B.Y.; Chen, Z.M. Composition of the volatiles from intact and mechanically pierced tea aphid-tea shoot complexes and their attraction to natural enemies of the tea aphid. J. Agric. Food Chem. 2002, 50, 2571–2575. [Google Scholar] [CrossRef] [PubMed]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. USA 2010, 107, 3600–3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolosova, N. Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant Cell Online 2001, 13, 2333–2347. [Google Scholar] [CrossRef]
- Verdonk, J.C.; De Vos, C.H.R.; Verhoeven, H.A.; Haring, M.A.; Van Tunen, A.J.; Schuurink, R.C. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 2003, 62, 997–1008. [Google Scholar] [CrossRef]
- Spitzer, B.; Zvi, M.M.B.; Ovadis, M.; Marhevka, E.; Barkai, O.; Edelbaum, O.; Marton, I.; Masci, T.; Alon, M.; Morin, S.; et al. Reverse genetics of floral scent: Application of tobacco rattle virus-based gene silencing in Petunia. Plant Physiol. 2007, 145, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Henkes, S.; Sonnewald, U.; Badur, R.; Flachmann, R.; Stitt, M. A Small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 2001, 13, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual (No. Ed. 2); Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; ISBN 0879693096. [Google Scholar]
- Bevan, M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984, 12, 8711–8721. [Google Scholar] [CrossRef] [PubMed]
- Van Roekel, J.S.C.; Damm, B.; Melchers, L.S.; Hoekema, A. Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep. 1993, 12, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Coppola, M.; Cascone, P.; Madonna, V.; Di Lelio, I.; Esposito, F.; Avitabile, C.; Romanelli, A.; Guerrieri, E.; Vitiello, A.; Pennacchio, F.; et al. Plant-To-plant communication triggered by systemin primes anti-herbivore resistance in tomato. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pokalsky, A.R.; Hiatt, W.R.; Ridge, N.; Rasmussen, R.; Houck, C.M.; Shewmaker, C.K. Structure and expression of elongation factor 1α in tomato. Nucleic Acids Res. 1989, 17, 4661–4673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrado, G.; Sasso, R.; Pasquariello, M.; Iodice, L.; Carretta, A.; Cascone, P.; Ariati, L.; Digilio, M.C.; Guerrieri, E.; Rao, R. Systemin regulates both systemic and volatile signaling in tomato plants. J. Chem. Ecol. 2007, 33, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shewmaker, C.K.; Ridge, N.P.; Pokalsky, A.R.; Rose, R.E.; Hiatt, W.R.; Street, F. Nucleotide tomato sequence of an EF-1α genomic clone from tomato. Nucleic Acids Res. 1990, 18, 4276. [Google Scholar] [CrossRef] [PubMed]
- Alfano, G.; Vitiello, C.; Caccioppoli, C.; Caramico, T.; Carola, A.; Szego, M.J.; McInnes, R.R.; Auricchio, A.; Banfi, S. Natural antisense transcripts associated with genes involved in eye development. Hum. Mol. Genet. 2005, 14, 913–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrieri, E.; Poppy, G.M.; Powell, W.; Rao, R.; Pennacchio, F. Plant-to-plant communication mediating in-flight orientation of Aphidius ervi. J. Chem. Ecol. 2002, 28, 1703–1715. [Google Scholar] [CrossRef] [PubMed]
- Petterson, J. An aphid sex attractant 1. Insect Syst. Evol. 1970, 1, 63–73. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. Biometry: The Principles and Practice of Statistics in Biological Research; W.H. Freeman and Company: San Francisco, SF, USA, 1995; ISBN 0-71 67-241 1-1. [Google Scholar]
- Cascone, P.; Iodice, L.; Maffei, M.E.; Bossi, S.; Arimura, G.I.; Guerrieri, E. Tobacco overexpressing β-ocimene induces direct and indirect responses against aphids in receiver tomato plants. J. Plant Physiol. 2015, 173, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and opls statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Coppola, M.; Corrado, G.; Coppola, V.; Cascone, P.; Martinelli, R.; Digilio, M.C.; Pennacchio, F.; Rao, R. Prosystemin overexpression in tomato enhances resistance to different biotic stresses by activating genes of multiple signaling pathways. Plant Mol. Biol. Rep. 2015, 33. [Google Scholar] [CrossRef] [PubMed]




| Mean Value of Volatiles Organic Compounds VOC (ng mg−1 fr wt) ± SE | ||||
|---|---|---|---|---|
| Compound Groups | Compound | dSST26 | dSST33 | Red Setter |
| Norisoprenes | 2,4 dimethyl 1 heptene | 8.21 ± 1.32 | 8.32 ± 1.35 | 8.27 ± 1.55 |
| 6 methyl 5 hepten 2 one | 0.04 ± 0.03 | 0.13 ± 0.11 | 0.15 ± 0.08 | |
| 8.25 ± 1.35 | 8.45 ± 1.46 | 8.42 ± 1.63 | ||
| Aldehydes | nonanal | 0.33 ± 0.08 | 0.39 ± 0.05 | 0.47 ± 0.07 |
| Monoterpenes | linalool | 0.27 ± 0.02 | 0.22 ± 0.03 | 0.3 ± 0.04 |
| α pinene | 0.19 ± 0.01 | 0.17 ± 0.02 | 0.21 ± 0.03 | |
| limonene | 0.09 ± 0.01 | 0.07 ± 0.01 | 0.08 ± 0.01 | |
| β phellandrene | 0.12 ± 0.03 | 0.08 ± 0.02 | 0.13 ± 0.02 | |
| p cymene | 0.1 ± 0.04 | 0.07 ± 0.04 | 0.01 ± 0.01 | |
| camphor | 0.06 ± 0.03 | 0.07 ± 0.03 | 0.05 ± 0.03 | |
| 0.83 ± 0.14 | 0.68 ± 0.15 | 0.78 ± 0.14 | ||
| Hydrocarbons | dodecene | 1.02 ± 0.41 | 1.31 ± 0.37 | 2.32 ± 0.37 |
| 4 methyl nonane | 0.4 ± 0.14 | 0.62 ± 0.17 | 0.51 ± 0.17 | |
| 1.42 ± 0.55 | 1.93 ± 0.54 | 2.83 ± 0.54 | ||
| Alcohols | 2 ethyl 1 hexanol | 0.84 ± 0.4 | 1.05 ± 0.36 | 1.04 ± 0.28 |
| benzil alcol | 0.05 ± 0.05 | 0.31 ± 0.13 | 0.07 ± 0.07 | |
| 0.89 ± 0.45 | 1.36 ± 0.49 | 1.11 ± 0.35 | ||
| Benzenoids | ethylbenzene | 0.41 ± 0.06 | 0.61 ± 0.14 | 0.52 ± 0.1 |
| benzaldehyde | 0.12 ± 0.08 | 0.26 ± 0.11 | 0.61 ± 0.22 | |
| 4 methyl benzaldehyde | 3.6 ± 0.81 | 3.36 ± 0.81 | 2.01 ± 0.46 | |
| 2,4 dimethyl benzaldehyde | 0 ± 0 | 0.01 ± 0.01 | 0.05 ± 0.03 | |
| 2,5-dimethyl benzaldehyde | 0 ± 0 | 0.06 ± 0.04 | 0 ± 0 | |
| trimethyl benzene | 0.09 ± 0.06 | 0.13 ± 0.06 | 0.26 ± 0.1 | |
| 1,4 dichlorobenzene | 0.29 ± 0.25 | 0.62 ± 0.38 | 0.16 ± 0.09 | |
| benzothiazole | 0.09 ± 0.04 | 0.12 ± 0.06 | 0.13 ± 0.05 | |
| methyl benzoate | 0.22 ± 0.1 | 0.38 ± 0.08 | 0.35 ± 0.07 | |
| acetophenone | 0.49 ± 0.12 | 0.47 ± 0.11 | 0.41 ± 0.06 | |
| naphthalene | 2.52 ± 1.4 | 3.06 ± 1.3 | 1.5 ± 0.31 | |
| 4 vinylphenol | 0 ± 0 | 0 ± 0 | 0.01 ± 0.01 | |
| 2 phenoxyethanol | 0.24 ± 0.17 | 0.04 ± 0.03 | 0.19 ± 0.11 | |
| pXylene | 1.66 ± 0.26 | 1.6 ± 0.31 | 1.98 ± 0.28 | |
| 9.73 ± 3.35 | 10.72 ± 3.44 | 8.18 ± 1.89 | ||
| Total VOC | 21.45 ± 5.92 | 23.53 ± 6.13 | 21.79 ± 4.62 | |
| Plant Weight (g) | 2.43 ± 0.24 | 2.35 ± 0.29 | 2.50 ± 0.12 | |
| Primers | Sequence 5′ to 3′ | Ta (°C) | Lenght (base pairs) | Gene or Expressed Sequence Tag | National Center for Biotechnology Information |
|---|---|---|---|---|---|
| GCSFw GCSRw | TTGGTGAAGCCTTAACTCAGCC GCAAATGGTGGTGTGCATCAT | 60 | 101 | GCS | AF035630 |
| PcEF1FwRt PcEF1RwRt | CTCCATTGGGTCGTTTTGCT GGTCACCTTGGCACCAGTTG | 60 | 101 | EF1α | X14449 |
| PcFPSFwRt PcFPSRwRt | GCAAAGCAGTACAGGCAGTGC TCCCAATGGGAGAATGAAGTTC | 60 | 101 | LeFPS1 | AF048747 |
| TDC-F TDC-R | ACTGTTAGCTCCGCTGCGTTT CCATTTCCAACTCCGTGCAT | 60 | 105 | TDC | AA824781 |
| ArgE-F ArgE-R | ACAACCCACCGGATCTTATCC TGATGATCAATGCTCCGCC | 60 | 102 | ArgE | AW737876 |
| ACoC-F ACoC-R | CAAAGAGGCGGAAGTTCACAAA CCAAAGTTTCACCTCCCACTCA | 60 | 114 | ACoC | AW928749 |
| ES895666–F ES895666-R | TACACAGTTTGGCCCGGTACA CAGGAGCGGAGAGTTGGATAGT | 60 | 103 | ES895666 | ES895666 |
| PAL-F PAL-R | GCTGAGCAACACAACCAAGATG TGGCAAAGAGCCACGAGATAG | 60 | 116 | PAL | M83314 |
| BF097728-F BF097728-R | GGTTTTATGGTGCTGCTTCACT GCAAATCGGAAAAGCCCAC | 60 | 172 | BF097728 | BF097728 |
| RO-F RO-R | CAGAAGCTGTAATGGAGCCAGG GCAAAGTTCATGCCTTCCCAG | 60 | 102 | RO | BP881050 |
| ACS-F ACS-R | CGACGAAATATACGCTGGCAC TGCCAACTCTGAAACCTGGAA | 60 | 161 | ACS | M83322 |
| AY348872-F AY348872-R | TGCTGGTGCACGCTTTGTT CGCAGTAAGCCAATTCGTGAG | 60 | 102 | AY348872 | AY348872 |
| PXD-F PXD-R | CCCCGGCATTGTTTCTTGT GGCATCTCTTCTGCCCAATTTT | 60 | 88 | PXD | AK320453 |
| F35H-F F35H-R | ACGTTCGTGCCAATGAGCTAG CCATCGCAAACGTTAACACATC | 60 | 103 | F35H | EU626067 GQ904194 |
| ACS3-R ACS3-F | CGCAAAAAAGCGCAACCTT GTGAATGCCTTTTTCGTCGATG | 60 | 109 | ACS3 | U17972.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coppola, M.; Cascone, P.; Bossi, S.; Corrado, G.; Garonna, A.P.; Maffei, M.; Rao, R.; Guerrieri, E. TPS Genes Silencing Alters Constitutive Indirect and Direct Defense in Tomato. Int. J. Mol. Sci. 2018, 19, 2748. https://doi.org/10.3390/ijms19092748
Coppola M, Cascone P, Bossi S, Corrado G, Garonna AP, Maffei M, Rao R, Guerrieri E. TPS Genes Silencing Alters Constitutive Indirect and Direct Defense in Tomato. International Journal of Molecular Sciences. 2018; 19(9):2748. https://doi.org/10.3390/ijms19092748
Chicago/Turabian StyleCoppola, Mariangela, Pasquale Cascone, Simone Bossi, Giandomenico Corrado, Antonio Pietro Garonna, Massimo Maffei, Rosa Rao, and Emilio Guerrieri. 2018. "TPS Genes Silencing Alters Constitutive Indirect and Direct Defense in Tomato" International Journal of Molecular Sciences 19, no. 9: 2748. https://doi.org/10.3390/ijms19092748







