Crosstalk of the Brassinosteroid Signalosome with Phytohormonal and Stress Signaling Components Maintains a Balance between the Processes of Growth and Stress Tolerance
Abstract
:1. Introduction
2. The BR Signalosome—An Update
3. BES1 and BZR1—The Major Transcription Factors Regulating BR-Dependent Gene Expression form a Hub in the Network of Coordinated Gene Expression
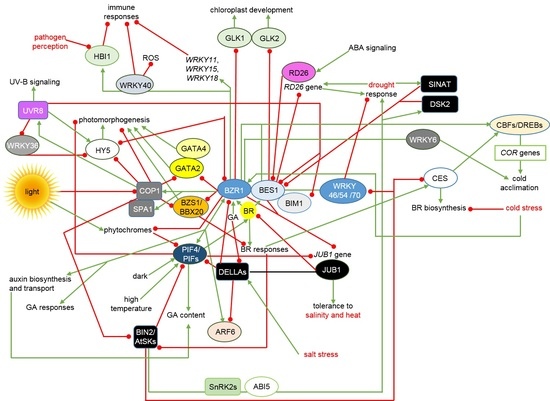
4. Interactions between the BRs Signalosome and Other Phytohormonal, Environmental and Stress Signaling Pathways Allow Maintenance of Physiological Homeostasis
5. BR Signaling in Cereal Crops—Novel and Specific Components
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Du, J.; Gao, Y.; Zhan, Y.; Zhang, S.; Wu, Y.; Xiao, Y.; Zou, B.; He, K.; Gou, X.; Li, G.; et al. Nucleocytoplasmic trafficking is essential for BAK1- and BKK1-mediated cell-death control. Plant J. 2016, 85, 520–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaiwanon, J.; Wang, W.; Zhu, J.-Y.; Oh, E.; Wang, Z.-Y. Information integration and communication in plant growth regulation. Cell 2016, 164, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.P.; Yu, J.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012, 17, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Vriet, C.; Russinova, E.; Reuzeau, C. Boosting crop yields with plant steroids. Plant Cell 2012, 24, 842–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruszka, D.; Janeczko, A.; Dziurka, M.; Pociecha, E.; Oklestkova, J.; Szarejko, I. Barley brassinosteroid mutants provide an insight into phytohormonal homeostasis in plant reaction to drought stress. Front. Plant Sci. 2016, 7, 1824. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, M. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016, 90, 856–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, I.; Srikanth, S.; Chen, Z. Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 2016, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, D. The brassinosteroid signaling pathway—New key players and interconnections with other signaling networks crucial for plant development and stress tolerance. Int. J. Mol. Sci. 2013, 14, 8740–8774. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Y.; Sae-Seaw, J.; Wang, Z.-Y. Brassinosteroid signalling. Development 2013, 140, 1615–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.-F.; Xiong, M.; Xu, P.; Huang, L.-C.; Zhang, C.-Q.; Liu, Q.-Q. Dissection of brassinosteroid-regulated proteins in rice embryos during germination by quantitative proteomics. Sci. Rep. 2016, 6, 34583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Wang, X.; Zhao, Z.; Wang, R.; Huang, X.; Zhu, Y.; Yuan, L.; Wang, Y.; Xu, X.; Burlingame, A.L.; et al. OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 2016, 170, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-F.; Lu, J.; Yu, J.-W.; Zhang, C.-Q.; He, J.-X.; Liu, Q.-Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta 2018, 1861, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Vukasinonic, N.; Russinova, E. BRexit: Possible brassinosteroid export and transport routes. Trends Plant Sci. 2018, 23, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Li, S.S.; Han, G.Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Kawamura, M.; Araki-Nakamura, S.; Fujimoto, H.; Ohmae-Shinohara, K.; Yamaguchi, M.; Fujii, A.; Sasaki, H.; Kasuga, S.; Sazuka, T. Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci. Rep. 2017, 7, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Deng, Z.; Wang, Z.Y. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr. Opin. Plant Biol. 2010, 13, 2727–2733. [Google Scholar] [CrossRef] [PubMed]
- Bojar, D.; Martinez, J.; Santiago, J.; Rybin, V.; Bayliss, R.; Hothorn, M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014, 78, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Belkhadir, Y.; Balsemao-Pires, E.; Dangl, J.L.; Chory, J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 2011, 108, 8503–8507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J. Direct involvement of leucine-rich repeats in assembling ligand-triggered receptor-coreceptor complexes. Proc. Natl. Acad. Sci. USA 2011, 108, 8073–8074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.L.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; Yin, H.; He, K.; Du, J.; Yi, J.; Xu, S.; Lin, H.; Clouse, S.D.; Li, J. Genetic evidence for an indispensable role of Somatic Embryogenesis Receptor Kinases in brassinosteroid signaling. PLoS Genet. 2012, 8, e1002452. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Sreeramulu, S.; Mostizky, Y.; Sunitha, S.; Shani, E.; Nahum, H.; Salomon, D.; Hayun, L.B.; Gruetter, C.; Rauh, D.; Ori, N.; et al. BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 2013, 74, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Guan, S.; Burlingame, A.S.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Mora-Garcia, S.; Vert, G.; Yin, Y.; Caño-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004, 18, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nam, K.H.; Vafeados, D.; Chory, J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001, 127, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Jonak, C.; Hirt, H. Glycogen synthase kinase3/SHAGGY-like kinases in plants: An emerging family with novel functions. Trends Plant Sci. 2002, 7, 457–461. [Google Scholar] [CrossRef]
- De Rybel, B.; Audenaert, D.; Vert, G.; Rozhon, W.; Mayerhofer, J.; Peelman, F.; Coutuer, S.; Denayer, T.; Jansen, L.; Nguyen, L.; et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 2009, 16, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhao, J.; Peng, P.; Chihara, R.K.; Li, J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009, 150, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhao, J.; Zhu, Y.; Asami, T.; Li, J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J. Biol. Chem. 2010, 285, 24646–24653. [Google Scholar] [CrossRef] [PubMed]
- Rozhon, W.; Mayerhofer, J.; Petutschnig, E.; Fujioka, S.; Jonak, C. ASKtheta, a group III Arabidopsis GSK3, functions in the brassinosteroid signaling pathway. Plant J. 2010, 62, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, L.; Yin, Y. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J. Integr. Plant Biol. 2011, 53, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Nakano, T.; Gendron, J.M.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002, 2, 505–513. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Chory, J. Downstream nuclear events in brassionsteroid signaling. Nature 2006, 441, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14–3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef] [PubMed]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14–3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007, 13, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Chu, C. Brassinosteroid signaling and application in rice. J. Genet. Genomics 2012, 39, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fan, X.Y.; Cao, D.M.; He, K.; Tang, W.; Zhu, J.Y.; He, J.X.; Bai, M.Y.; Zhu, S.; Oh, E.; et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Gudesblat, G.E.; Russinova, E. Plants grow on brassinosteroids. Curr. Opin. Plant. Biol. 2011, 14, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, L.; Aluru, M.; Aluru, S.; Yin, Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 2013, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, X.; Thompson, A.; Guo, M.; Yoshida, S.; Asami, T.; Chory, J.; Yin, Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009, 58, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Hategan, L.; Godza, B.; Kozma-Bognar, L.; Bishop, G.J.; Szekeres, M. Differential expression of the brassinosteroid receptor-encoding BRI1 gene in Arabidopsis. Planta 2014, 239, 989–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goda, H.; Shimada, Y.; Asami, T.; Fujioka, S.; Yoshida, S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002, 130, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Mockler, T.C.; Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004, 2, e258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, T.; Morinaka, Y.; Inukai, Y.; Kitano, H.; Fujioka, S. Auxin signal transcription factor regulates expression of brassinosteroid receptor gene in rice. Plant J. 2013, 73, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; Lv, X.; Chen, T.; Li, R.; Xue, Y.; Jiang, J.; Jin, B.; Baluska, F.; Samaj, J.; et al. Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol. Plant 2015, 8, 1334–1349. [Google Scholar] [CrossRef] [PubMed]
- Bücherl, C.A.; van Esse, G.W.; Kruis, A.; Luchtenberg, J.; Westphal, A.H.; Aker, J.; van Hoek, A.; Albrecht, C.; Borst, J.W.; de Vries, S.C. Visualization of BRI1 and BAK1 (SERK3) membrane receptor hetero-oligomers during brassinosteroid signaling. Plant Physiol. 2013, 162, 1911–1925. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Henzler, C.; Hothorn, M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 2013, 341, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Han, Z.; Tang, J.; Hu, Z.; Chai, C.; Zhou, B.; Chai, J. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013, 23, 1326–1329. [Google Scholar] [CrossRef] [PubMed]
- Hutten, S.J.; Hamers, D.S.; den Toorn, M.A.; van Esse, W.; Nolles, A.; Bücherl, C.A.; de Vries, S.C.; Hohlbein, J.; Borst, J.W. Visualization of BRI1 and SERK3/BAK1 nanoclusters in Arabidopsis roots. PLoS ONE 2017, 12, e0169905. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Goshe, M.B.; Soderblom, E.J.; Phinney, B.S.; Kuchar, J.A.; Li, J.; Asami, T.; Yoshida, S.; Huber, S.C.; Clouse, S.D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 2005, 17, 1685–1703. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Meisenhelder, J.; Hunter, T.; Yoshida, S.; Asami, T.; Chory, J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 2005, 8, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Hothorn, M.; Belkhadir, Y.; Dabi, T.; Nimchuk, Z.L.; Meyerowitz, E.M.; Chory, J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011, 25, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.; Wang, J.; Chen, L.; Fan, S.-L.; Wu, J.-W.; Wang, X.; Wang, Z.-X. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014, 24, 1328–1341. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.L.; Platre, M.P.; Marques-Bueno, M.M.; Armengot, L.; Stanislas, T.; Bayle, V.; Caillaud, M.C.; Jaillais, Y. A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signaling in plants. Nat. Plants 2016, 2, 16089. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Vert, G. Brassinosteroid signaling and BRI1 dynamics went underground. Curr. Opin. Plant Biol. 2016, 33, 92–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–472. [Google Scholar] [CrossRef] [PubMed]
- She, J.; Han, Z.; Kim, T.W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Yang, M.; Wang, Z.Y.; Chai, J. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Yang, C.; Wang, H.; Wu, Z.; Jiang, J.; Liu, J.; He, Z.; Chang, F.; Ma, H.; Wang, X. BKI1 regulates plant architecture through coordinated inhibition of the brassinosteroid and ERECTA signaling pathways in Arabidopsis. Mol. Plant 2017, 10, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Shpak, E.D.; Berthiaume, C.T.; Hill, E.J.; Torii, K.U. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 2004, 131, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Van Zanten, M.; Snoek, L.B.; Proveniers, M.C.; Peeters, A.J. The many functions of ERECTA. Trends Plant Sci. 2009, 14, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Shpak, E.D. Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 2013, 55, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Chen, X.; Mang, H.; Liu, C.; Yu, X.; Gao, X.; Torii, K.U.; He, P.; Shan, L. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 2015, 25, 2361–2372. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, M.; Yuan, M.; Oses-Prieto, J.; Cai, X.; Sun, Y.; Burlingame, A.L.; Wang, Z.-Y.; Tang, W. The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B’ subunits. Mol. Plant 2016, 9, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Wang, X.; Li, X.; Kamiya, Y.; Otegui, M.S.; Chory, J. Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 2011, 172, ra29. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Dohmann, E.M.; Cayrel, A.; Johnson, A.; Fischer, W.; Pojer, F.; Satiat-Jeunemaître, B.; Jaillais, Y.; Chory, J.; Geldner, N.; et al. Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 2015, 6, 6151. [Google Scholar] [CrossRef] [PubMed]
- Di Rubbo, S.; Irani, N.G.; Kim, S.Y.; Xu, Z.-Y.; Gadeyne, A.; Dejonghe, W.; Vanhoutte, I.; Persiau, G.; Eeckhout, D.; Simon, S.; et al. The clathrin adaptor complex AP-2 mediates endocytosis of BRASSINOSTEROID INSENSITIVE1 in Arabidopsis. Plant Cell 2013, 25, 2986–2997. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Hyman, D.L.; Wang, X.; Schumacher, K.; Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007, 21, 1598–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irani, N.G.; Di Rubbo, S.; Mylle, E.; Van den Begin, J.; Schneider-Pizoń, J.; Hniliková, J.; Šíša, M.; Buyst, D.; Vilarrasa-Blasi, J.; Szatmári, A.M.; et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 2012, 8, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Yamagami, A.; Saito, C.; Nakazawa, M.; Fujioka, S.; Uemura, T.; Matsui, M.; Sakuta, M.; Shinozaki, K.; Osada, H.; Nakano, A.; et al. Evolutionarily conserved BIL4 suppresses the degradation of brassinosteroid receptor BRI1 and regulates cell elongation. Sci. Rep. 2017, 7, 5739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Liu, D.; Wang, P.; Ma, X.; Lin, W.; Chen, S.; Mishev, K.; Lu, D.; Kumar, R.; Vanhoutte, I.; et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 2018, 115, 1906–1915. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, D.; Cao, Y.; Sun, X.; Espinoza, C.; Nguyen, C.T.; Liang, Y.; Stacey, G. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017, 214, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ahn, I.-P.; Ning, Y.; Park, C.-H.; Zeng, L.; Whitehill, J.G.A.; Lu, H.; Zhao, Q.; Ding, B.; Xie, Q.; et al. The U-box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 2012, 159, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lu, D.; Xu, G.; Finlayson, S.A.; He, P.; Shan, L. The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering and senescence. J. Exp. Bot. 2015, 66, 3353–3366. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.I.; Wong, A.; Marondedze, C.; Groen, A.J.; Kwezi, L.; Freihat, L.; Vyas, J.; Raji, M.A.; Irving, H.R.; Gehring, C. The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant J. 2017, 91, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 2006, 45, 700–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isner, J.C.; Nuhse, T.; Maathuis, F.J. The cyclic nucleotide cGMP is involved in plant hormone signaling and alters phosphorylation of Arabidopsis thaliana root proteins. J. Exp. Bot. 2012, 63, 3199–3205. [Google Scholar] [CrossRef] [PubMed]
- Facette, M.R.; Shen, Z.; Bjornsdottir, F.R.; Briggs, S.P.; Smith, L.G. Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell 2013, 25, 2798–2812. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Groen, A.; Thomas, L.; Lilley, K.S.; Gehring, C. A quantitative phosphoproteome analysis of cGMP-dependent cellular responses in Arabidopsis thaliana. Mol. Plant 2016, 9, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qi, Z.; Berkowitz, G.A. Teaching an old hormone new tricks: Cytosolic Ca2+ elevation involvement in plant brassinosteroid signal transduction cascades. Plant Physiol. 2013, 163, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Bouche, N.; Yellin, A.; Snedden, W.A.; Fromm, H. Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 2005, 56, 435–466. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signaling. Biochem. J. 2009, 425, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Kim, H.S.; Wu, X.; Clouse, S.D.; Zielinski, R.E.; Huber, S.C. Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signaling. Biochem. J. 2012, 443, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Poovaiah, B.W. Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 2005, 437, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lv, M.; Feng, Z.; Campbell, T.; Liscum, E.; Li, J. TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol. Plant 2016, 9, 582–592. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Li, L.; Luan, S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004, 134, 1248–1267. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. Immunophilins: For the love of proteins. Cell Mol. Life Sci. 2006, 63, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Bailly, A. Tete-a-tete: The function of FKBPs in plant development. Trends Plant Sci. 2007, 12, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Bailly, A.; Zwiewka, M.; Henrichs, S.; Azzarello, E.; Mancuso, S.; Maeshima, M.; Friml, J.; Schulz, A.; Geisler, M. Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell 2013, 25, 202–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.X.; Ye, H.B.; Xuan, Y.H.; Yao, D.N. Overexpression of a SNARE protein AtBS14b alters BR response in Arabidopsis. Bot. Stud. 2014, 55, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Shi, Q.M.; Yang, X.H.; Xu, Z.H.; Xue, H.W. Membrane steroid-binding protein 1 (MSBP1) negatively regulates brassinosteroid signaling by enhancing the endocytosis of BAK1. Cell Res. 2009, 19, 864–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Q.M.; Yang, X.; Song, L.; Xue, H.W. Arabidopsis MSBP1 is activated by HY5 and HYH and is involved in photomorphogenesis and brassinosteroid sensitivity regulation. Mol. Plant 2011, 4, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Chen, L.; Li, S.; Zhang, Y.; Xu, R.; Liu, Z.; Liu, W.; Kong, J.; Huang, X.; Wang, Y.; et al. BRI1 and BAK1 interact with G proteins and regulate sugar-responsive growth and development in Arabidopsis. Nat. Commun. 2018, 9, 1522. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, M.; Laxmi, A. Multiple interactions between glucose and brassinosteroid signal transduction pathways in Arabidopsis are uncovered by whole-genome transcriptional profiling. Plant Physiol. 2015, 168, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Kwon, Y.; Lee, B.; Nam, K.H. Overexpression of miR172 suppresses the brassinosteroid signaling defects of bak1 in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 447, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Lauter, N.; Kampani, A.; Carlson, S.; Goebel, M.; Moose, S.P. MicroRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 2005, 102, 9412–9417. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Kutuzov, M.A.; Andreeva, A.V. Protein Ser/Thr phosphatases with Kelch-like repeat domains. Cell. Signal. 2002, 14, 745–750. [Google Scholar] [CrossRef]
- Maselli, G.A.; Slamovits, C.H.; Bianchi, J.I.; Vilarrasa-Blasi, J.; Cano-Delgado, A.I.; Mora-Garcia, S. Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 2014, 164, 1527–1541. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-J.; Youn, J.-H.; Park, C.-H.; Kim, T.-W.; Guan, S.; Xu, S.; Burlingame, A.L.; Kim, Y.-P.; Kim, S.-K.; Wang, Z.-Y.; et al. Oligomerization between BSU1 family members potentiates brassinosteroid signaling in Arabidopsis. Mol. Plant 2016, 9, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-J.; Lee, S.-H.; Park, C.-H.; Kim, T.-W. Functional role of BSL1 subcellular localization in brassinosteroid signaling. J. Plant Biol. 2018, 61, 40–49. [Google Scholar] [CrossRef]
- Gudesblat, G.E.; Schneider-Pizon, J.; Betti, C.; Mayerhofer, J.; Vanhoutte, I.; van Dongen, W.; Boeren, S.; Zhiponova, M.; de Vries, S.; Jonak, C. SPEECHLESS integrates brassinosteroid and stomata signaling pathways. Nat. Cell Biol. 2012, 14, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Saidi, Y.; Hearn, T.J.; Coates, J.C. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 2012, 17, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-H.; Kim, T.-W.; Kim, E.-J.; Bu, S.; Kim, S.-K.; Wang, Z.-Y.; Kim, T.-W. Structural and functional characterization of Arabidopsis GSK3-like kinase AtSK12. Mol. Cells 2013, 36, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wang, H.; Qiao, S.; Leng, L.; Wang, X. Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl. Acad. Sci. USA 2016, 113, 10418–10423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-Y.; Li, Y.; Cao, D.-M.; Yang, H.; Oh, E.; Bi, Y.; Zhu, S.; Wang, Z.-Y. The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 2017, 66, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Truernit, E.; Bauby, H.; Belcram, K.; Barthelemy, J.; Palauqui, J.-C. OCTOPUS, a polarly localised membrane-associated protein regulates phloem differentiation entry in Arabidopsis thaliana. Development 2012, 139, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Anne, P.; Azzopardi, M.; Gissot, L.; Beaubiat, S.; Hematy, K.; Palauqui, J.-C. OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr. Biol. 2015, 25, 2584–2590. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.-J.; Li, J.; Zhu, D.; Deng, X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA 2017, 114, 3539–3544. [Google Scholar] [CrossRef] [PubMed]
- Samakovli, D.; Margaritopoulou, T.; Prassinos, C.; Milioni, D.; Hatzopoulos, P. Brassinosteroid nuclear signaling recruits HSP90 activity. New Phytol. 2014, 203, 743–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Lachowiec, J.; Lemus, T.; Thomas, J.H.; Murphy, P.J.; Nemhauser, J.L.; Queitsch, C. The protein chaperone HSP90 can facilitate the divergence of gene duplicates. Genetics 2013, 193, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, T.; Zaizen, Y.; Asami, T.; Yoshida, S.; Nakamura, Y.; Okamoto, S.; Matsuo, T.; Sugimoto, Y. Molecular evidence of the involvement of heat shock protein 90 in brassinosteroid signaling in Arabidopsis T87 cultured cells. Plant Cell Rep. 2014, 33, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudesblat, G.E.; Betti, C.; Russinova, E. Brassinosteroids tailor stomatal production to different environments. Trends Plant Sci. 2012, 17, 685–687. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.; Rozhon, W.; Bigeard, J.; Pflieger, D.; Husar, S.; Pitzschke, A.; Teige, M.; Jonak, C.; Hirt, H.; Poppenberger, B. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J. Biol. Chem. 2013, 288, 7519–7527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ye, H.; Guo, H.; Johnson, A.; Zhang, M.; Lin, H.; Yin, Y. Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 2014, 77, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, L.; Guo, H.; Yin, Y. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 20142–20147. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-M.; Shang, J.-X.; Chen, Q.-X.; Oses-Prieto, J.A.; Bai, M.-Y.; Yang, Y.; Yuan, M.; Zhang, Y.-L.; Mu, C.-C.; Deng, Z.; et al. Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol. Cell. Proteomics 2013, 12, 3653–3665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.; Xu, Y.; Li, H.; Li, S.; Zhang, D.; Mao, Z.; Guo, S.; Yang, C.; Weng, Y.; et al. The cyclophilin CYP20-2 modulates the conformation of BRASSINAZOLE-RESISTANT1, which binds the promoter of FLOWERING LOCUS D to regulate flowering in Arabidopsis. Plant Cell 2013, 25, 2504–2521. [Google Scholar] [CrossRef] [PubMed]
- Shimada, S.; Komatsu, T.; Yamagami, A.; Nakazawa, M.; Matsui, M.; Kawaide, H.; Natsume, M.; Osada, H.; Asami, T.; Nakano, T. Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell 2015, 27, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Du, Y.; Hou, Y.J.; Zhao, Y.; Hsu, C.C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.P.; Zhu, J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, D.; Janeczko, A.; Dziurka, M.; Pociecha, E.; Fodor, J. Non-enzymatic antioxidant accumulations in BR-deficient and BR-insensitive barley mutants under control and drought conditions. Physiol. Plant. 2018, 163, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, J.-Y.; Roh, J.; Marchive, C.; Kim, S.-K.; Meyer, C.; Sun, Y.; Wang, W.; Wang, Z.-Y. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 2016, 26, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 2017, 41, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.; Wang, X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 2013, 27, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Jeong, Y.J.; Corvalan, C.; Fujioka, S.; Cho, S.; Park, T.; Choe, S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014, 77, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, C.; Cai, Z.; Hu, Y.; Nolan, T.; Yu, F.; Yin, Y.; Xie, Q.; Tang, G.; Wang, X. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 2017, 41, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R. E3 ubiquitin ligases: Key regulators of hormone signaling in plants. Mol. Cell Proteomics 2018, 17, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, P.; Wang, W.; Wang, S.; Caruana, J.C.; Yang, H.-Q.; Lian, H. Arabidopsis G-protein β subunit AGB1 interacts with BES1 to regulate brassinosteroid signaling and cell elongation. Front. Plant Sci. 2018, 8, 2225. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Assmann, S.M. Plants: The latest model system for G-protein research. EMBO Rep. 2004, 5, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Mei, S.; Shi, C.; Yang, Y.; Peng, Y.; Ma, L.; Wang, F.; Li, X.; Huang, X.; Yin, Y.; et al. UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell 2018, 44, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Zhu, Z. Illuminating the nucleus: UVR8 interacts with more. Trends Plant Sci. 2018, 23, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Skvortsova, M.Y.; Loubéry, S.; Ulm, R. COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. USA 2016, 113, 4415–4422. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Song, J.-H.; Park, S.-U.; Jeong, Y.-S.; Kim, S.-H. Brassinosteroid-induced transcriptional repression and dephosphorylation-dependent protein degradation negatively regulate BIN2-interacting AIF2 (a BR signaling-negative regulator) bHLH transcription factor. Plant Cell Physiol. 2017, 58, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, Y.; Fujioka, S.; Asami, T.; Li, J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009, 21, 3781–3791. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 2006, 61, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. ATBS1 Interacting factors negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal. Behav. 2013, 8, e23448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-Y.; Bai, M.-Y.; Wu, J.; Zhu, J.-Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Bai, M.Y.; Wang, Z.Y. The brassinosteroid signaling network—A paradigm of signal integration. Curr. Opin. Plant. Biol. 2014, 21, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.-Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Guo, M.; Chory, J.; Yin, Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 7618–7623. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Cui, X.; Zhang, S.; Jenuwein, T.; Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011, 43, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Germann, S.; Blus, B.J.; Khorasanizadeh, S.; Gaudin, V.; Jacobsen, S.E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007, 14, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; Colot, V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007, 3, e86. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, H.; Guo, H.; Yin, Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 3918–3923. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J.; Xie, Z.; Liu, S.; Nolan, T.; Ye, H.; Zhang, M.; Guo, H.; Schnable, P.S.; Li, Z.; et al. Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol. Plant 2014, 7, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Sui, P.; Jin, J.; Ye, S.; Mu, C.; Gao, J.; Feng, H.; Shen, W.H.; Yu, Y.; Dong, A. H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 2012, 70, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Ryu, H.; Hwang, I.; Wang, Z.Y. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 2014, 5, 4140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Jing, Y.; Jiang, Z.; Lin, R. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 2014, 26, 2472–2485. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D. Brassinosteroid/abscisic acid antagonism in balancing growth and stress. Dev. Cell 2016, 38, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Zheng, S.; Liu, C.; Shen, J.; Li, J.; Li, L. OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 2016, 38, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Chen, J.; Yin, Y. Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 2017, 474, 2641–2661. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Dai, C.; Lee, M.M.; Kwak, J.M.; Nam, K.H. BRI1-associated receptor kinase 1 regulates guard cell ABA signaling mediated by open stomata 1 in Arabidopsis. Mol. Plant 2016, 9, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Mustilli, A.-C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.R.; Jeon, B.W.; Zhang, W.; Assmann, S.M. Open stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 2013, 200, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Shang, Y.; Nam, K.H. Brassinosteroids modulate ABA-induced stomatal closure in Arabidopsis. J. Exp. Bot. 2016, 67, 6297–6308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, C.; Boutrot, F.; Segonzac, C.; Schwessinger, B.; Gimenez-Ibanez, S.; Chinchilla, D.; Rathjen, J.P.; de Vries, S.C.; Zipfel, C. Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 2012, 109, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Belkhadir, Y.; Jaillais, Y.; Epple, P.; Balsemão-Pires, E.; Dangl, J.L.; Chory, J. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2012, 109, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Lu, D.; Gao, X.; Jiang, S.; Ma, X.; Wang, Z.; Mengiste, T.; He, P.; Shan, L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA. 2013, 110, 12114–12119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, M.; Wang, X.; Wang, D.; Xu, F.; Ding, X.; Zhang, Z.; Bi, D.; Cheng, Y.T.; Chen, S.; Li, X.; et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 2009, 6, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Liebrand, T.W.; van den Burg, H.A.; Joosten, M.H. Two for all: Receptor-associated kinases SOBIR and BAK1. Trends Plant Sci. 2014, 19, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Ferreras, A.; Kiss-Papp, M.; Jehle, A.K.; Felix, G.; Chinchilla, D. An overdose of the Arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR OF BIR1-1-dependent manner. Plant Physiol. 2015, 168, 1106–1121. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Shang, Y.; Joo, S.-H.; Kim, S.-K.; Nam, K.H. Overexpression of BAK1 causes salicylic acid accumulation and deregulation of cell death control genes. Biochem. Biophys. Res. Commun. 2017, 484, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringe effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Aceti, D.J.; Sabat, G.; Song, J.; Makino, S.; Fox, B.G.; Bent, A.F. Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog. 2013, 9, e1003313. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [PubMed]
- Segonzac, C.; Macho, A.P.; Sanmartin, M.; Ntoukakis, V.; Sanchez-Serrano, J.J.; Zipfel, C. Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 2014, 33, 2069–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couto, D.; Niebergall, R.; Liang, X.; Bücherl, C.A.; Sklenar, J.; Macho, A.P.; Ntoukakis, V.; Derbyshire, P.; Altenbach, D.; Maclean, D.; et al. The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog. 2016, 12, e1005811. [Google Scholar] [CrossRef] [PubMed]
- Laluk, K.; Luo, H.; Chai, M.; Dhawan, R.; Lai, Z.; Mengiste, T. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 2011, 23, 2831–2849. [Google Scholar] [CrossRef] [PubMed]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Shen, Q.; Qi, Y.; Yan, H.; Nie, H.; Chen, Y.; Zhao, T.; Katagiri, F.; Tang, D. BR-SIGNALING KINASE 1 physically associates with FLAGELLIN SENSING 2 and regulates plant innate immunity in Arabidopsis. Plant Cell 2013, 25, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yan, H.; Li, J.; Tang, D. BSK1, a receptor-like cytoplasmic kinase, involved in both BR signaling and innate immunity in Arabidopsis. Plant Signal. Behav. 2013, 8, 24996. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhao, Y.; Shi, H.; Li, J.; Wang, Y.; Tang, D. BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 2018, 176, 2991–3002. [Google Scholar] [CrossRef] [PubMed]
- De Bruyne, L.; Höfte, M.; De Vleesschauwer, D. Connecting growth and defense: The emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 2014, 7, 943–959. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Rozhon, W.; Khan, M.; Husar, S.; Adam, G.; Luschnig, C.; Fujioka, S.; Sieberer, T. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 2011, 30, 1149–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Tang, J.; Liu, J.; Hu, J.; Liu, J.; Chen, Y.; Cai, Z.; Wang, X. Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 2018, 11, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, J.; Wang, H.; Yang, C.; Chen, Y.; Li, Y.; Pan, S.; Dong, R.; Tang, G.; Barajas-Lopez Jde, D.; et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 9651–9656. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE 2 interacts with ABSCISIC ACID INSENSITIVE 5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cai, Z.; Wang, X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 4543–4548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dal Santo, S.; Stampfl, H.; Krasensky, J.; Kempa, S.; Gibon, Y.; Petutschnig, E.; Rozhon, W.; Heuck, A.; Clausen, T.; Jonak, C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 2012, 24, 3380–3392. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-H.; Kim, T.-W. Functional insights of plant GSK3-like kinases: Multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 2015, 8, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.-L.; Liu, C.; Liu, N.; Zhao, J.-L.; Ai, L.-F.; Chi, H.; Li, K.L.; Chien, C.-W.; Burlingame, A.L.; Zhang, S.-W.; et al. Immunopurification and mass spectrometry identifies protein phosphatase 2A (PP2A) and BIN2/GSK3 as regulators of AKS transcription factors in Arabidopsis. Mol. Plant 2017, 10, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Ebisu, Y.; Kinoshita, T.; Doi, M.; Okuma, E.; Murata, Y.; Shimazaki, K. bHLH transcription factors that facilitate K(+) uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 2013, 6, ra48. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Ryu, H.; Rho, S.; Hill, K.; Smith, S.; Audenaert, D.; Park, J.; Han, S.; Beeckman, T.; Bennett, M.J.; et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 2014, 16, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-S.; Chen, Q.-S.; Xin, D.-W.; Qi, Z.-M.; Zhang, C.; Li, S.-N.; Jin, Y.-M.; Li, M.; Mei, H.Y.; Su, A.-Y.; et al. Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots. J. Integr. Agric. 2018, 17, 60345–60347. [Google Scholar] [CrossRef]
- Vanstraelen, M.; Benkova, E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; He, J.X. Mechanisms of signaling crosstalk between brassinosteroids and gibberellins. Plant Signal. Behav. 2013, 8, e24686. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Rozhon, W.; Papacek, M. Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 2015, 27, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Shang, J.X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.P.; Wang, Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolome, J.; Minguet, E.G.; Grau-Enguix, F.; Abbas, M.; Locascio, A.; Thomas, S.G.; Alabadi, D.; Blazquez, M.A. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci USA 2012, 109, 13446–13451. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Wang, C.; Jiang, L.; Li, S.; Sun, S.S.; He, J.X. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 2012, 5, ra72. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 2011, 21, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Liao, L.; Jiang, C.; Desnos, T.; Bartlett, J.; Fu, X.; Harberd, N. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007, 143, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sawa, S.; Asami, T.; Fujioka, S.; Shimada, Y.; Yoshida, S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004, 134, 1555–1573. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Nakajima, N.; Goda, H.; Shimada, Y.; Hayashi, K.; Nozaki, H.; Asami, T.; Yoshida, S.; Fujioka, S. Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 2006, 45, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.Y.; Song, L.; Xue, H.W. Brassinosteroids regulate the differential growth of Arabidopsis hypocotyls through auxin signaling components IAA19 and ARF7. Mol. Plant 2013, 6, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-H.; Kim, M.K.; Kim, E.J.; Son, S.H.; Lee, J.E.; Jang, M.S.; Kim, T.-W.; Kim, S.K. ARF7 increases the endogenous contents of castasterone through suppression of BAS1 expression in Arabidopsis thaliana. Phytochemistry 2016, 122, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3, e03031. [Google Scholar] [CrossRef] [PubMed]
- Favero, D.S.; Le, K.N.; Neff, M.M. Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J. 2017, 89, 1133–1145. [Google Scholar] [PubMed]
- Favero, D.S.; Jacques, C.N.; Iwase, A.; Le, K.N.; Zhao, J.; Sugimoto, K.; Neff, M.M. SUPPRESSOR OF PHYTOCHROME B4-#3 represses genes associated with auxin signaling to modulate hypocotyl growth. Plant Physiol. 2016, 171, 2701–2716. [Google Scholar] [PubMed]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Bai, Y.; Shang, J.; Xin, R.; Tang, W. The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT 1. Plant Cell Environ. 2016, 39, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Schumaker, K.S.; Guo, Y. Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 12822–12827. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely non-overlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sasaki, E.; Akiyama, K.; Maruyama-Nakashita, A.; Nakabayashi, K.; Li, W.; Ogawa, M.; Yamauchi, Y.; Preston, J.; Aoki, K.; et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008, 55, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Tian, H.; Zhang, F.; Liu, J.; Lu, S.; Bai, M.; Li, C. Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 2018, 14, e1007144. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Chae, H.S.; Kieber, J.J. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009, 57, 606–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.Y.; Sun, Y.; Cao, D.M.; Bai, M.Y.; Luo, X.M.; Yang, H.J.; Wei, C.Q.; Zhu, S.W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Vert, G. Brassinosteroids, gibberellins and light-mediated signalling are the three-way controls of plant sprouting. Nat. Cel. Biol. 2012, 14, 788–790. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Moylan, E.C.; Langdale, J.A. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008, 56, 432–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.-M.; Lin, W.-H.; Zhu, S.; Zhu, J.-Y.; Sun, Y.; Fan, X.-Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M.; et al. Integration of light and brassinosteroid signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 2010, 19, 872–883. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, C.; Delker, C.; Martinez, C.; Burstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Lucas, M.; Prat, S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014, 202, 1126–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Xu, S.L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014, 344, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Liu, B.; Zhao, C.; Pepper, M.; Lin, C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011, 16, 684–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, O.S.; Deng, X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Zhang, D.; Wang, X.; Tang, W.; Wang, W.; Huai, J.; Xu, G.; Chen, D.; Li, Y.; Lin, R. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 2013, 25, 242–256. [Google Scholar] [CrossRef] [PubMed]
- De Lucas, M.; Daviere, J.M.; Rodriguez-Falcon, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blazquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Locascio, A.; Blazquez, M.A.; Alabadi, D. Genomic analysis of DELLA protein activity. Plant Cell Physiol. 2013, 54, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; He, J.X. BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 2016, 9, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Oravecz, A.; Baumann, A.; Máté, Z.; Brzezinska, A.; Molinier, J.; Oakeley, E.J.; Ádám, E.; Schäfer, E.; Nagy, F.; Ulm, R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 2006, 18, 1975–1990. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ouyang, X.; Deng, X.W. Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 2014, 21, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Ulm, R.; Baumann, A.; Oravecz, A.; Máté, Z.; Adám, E.; Oakeley, E.J.; Schäfer, E.; Nagy, F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1397–1402. [Google Scholar] [CrossRef] [PubMed]
- Favory, J.J.; Stec, A.; Gruber, H.; Rizzini, L.; Oravecz, A.; Funk, M.; Albert, A.; Cloix, C.; Jenkins, G.I.; Oakeley, E.J.; et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009, 28, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liang, T.; Zhang, L.; Shao, K.; Gu, X.; Shang, R.; Shi, N.; Li, X.; Zhang, P.; Liu, H. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants 2018, 4, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signaling. Nat. Plants 2016, 2, 16013. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Wu, R.; Wee, C.W.; Xie, F.; Wei, X.; Chan, P.M.Y.; Tham, C.; Duan, L.; Dinneny, J.R. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 2013, 25, 2132–2154. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Fridman, Y.; Friedlander-Shani, L.; Tarkowska, D.; Strnad, M.; Savaldi-Goldstein, S. Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol. 2014, 166, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liao, H.; Lucas, W.J. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 2014, 56, 192–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaiwanon, J.; Wang, Z.Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 2015, 25, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Divi, U.K.; Krishna, P. Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. N. Biotechnol. 2009, 26, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, B.H.; Lim, C.J.; Lim, C.O.; Nam, K.H. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol. Plant. 2010, 138, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Unterholzner, S.J.; Rathnayake, A.I.; Castellanos, M.; Khan, M.; Kugler, K.G.; May, S.T.; Mayer, K.F.X.; Rozhon, W.; Poppenberger, B. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 2016, 113, 5982–5991. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Rozhon, W.; Unterholzner, S.J.; Chen, T.; Eremina, M.; Wurzinger, B.; Bachmair, A.; Teige, M.; Sieberer, T.; Isono, E.; et al. Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 2014, 5, 4687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2015, 56, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yin, Y.; Fei, S. Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 2015, 234, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Northey, J.G.B.; Liang, S.; Jamshed, M.; Deb, S.; Foo, E.; Reid, J.B.; McCourt, P.; Samuel, M.A. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat. Plants 2016, 2, 16114. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk between drought and brassinosteroid signaling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathways. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Kwon, S.I.; Choe, S. Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Mol. Cells 2014, 37, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 transcriptions factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Macho, A.P.; Boutrot, F.; Segonzac, C.; Somssich, I.E.; Zipfel, C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2013, 2, e00983. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Zipfel, C. Trade-off between growth and immunity: Role of brassinosteroids. Trends Plant Sci. 2015, 20, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Bai, M.Y.; Kim, J.G.; Wang, T.; Oh, E.; Chen, L.; Park, C.H.; Son, S.H.; Kim, S.K.; Mudgett, M.B.; et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Malinovsky, F.G.; Batoux, M.; Schwessinger, B.; Youn, J.H.; Stransfeld, L.; Win, J.; Kim, S.K.; Zipfel, C. Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol. 2014, 164, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Yang, F.; Li, L.; Chen, H.; Chen, S.; Zhang, J. The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant Physiol. 2015, 167, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Marcos, R.; Izquierdo, Y.; Vellosillo, T.; Kulasekaran, S.; Cascon, T.; Hamberg, M.; Castresana, C. 9-Lipoxygenase-derived oxylipins activate brassinosteroid signaling to promote cell wall-based defense and limit pathogen infection. Plant Physiol. 2015, 169, 2324–2334. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Chono, M.; Honda, I.; Zeniya, H.; Yoneyama, K.; Saisho, D.; Takeda, K.; Takatsuto, S.; Hoshino, T.; Watanabe, Y. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 2003, 133, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Morinaka, Y.; Sakamoto, T.; Inukai, Y.; Agetsuma, M.; Kitano, H.; Ashikari, M.; Matsuoka, M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006, 141, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Li, H.-Y. Oryza sativa BRASSINOSTEROID UPREGULATED1 LIKE1 induces the expression of a gene encoding a small leucine-rich-repeat protein to positively regulate lamina inclination and grain size in rice. Front. Plant Sci. 2017, 8, 1253. [Google Scholar] [CrossRef] [PubMed]
- Ferrero-Serrano, A.; Assmann, S.M. The a-subunit of the rice heterotrimeric G protein, RGA1, regulates drought tolerance during the vegetative phase in the dwarf rice mutant d1. J. Exp. Bot. 2016, 67, 3433–3443. [Google Scholar] [CrossRef] [PubMed]
- Corvalan, C.; Choe, S. Identification of brassinosteroid genes in Brachypodium distachyon. BMC Plant Biol. 2017, 17, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.-J.; Lin, W.-H.; Fu, F.-F.; Xu, Z.-H.; Xue, H.-W. Receptor-like protein ELT1 promotes brassinosteroid signaling through interacting with and suppressing the endocytosis-mediated degradation of receptor BRI1. Cell Res. 2017, 27, 1182–1185. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Je, B.I.; Piao, H.L.; Park, S.J.; Park, S.H.; Kim, C.M.; Xuan, Y.H.; Park, S.H.; Huang, J.; Do Choi, Y.; An, G.; et al. RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 2010, 22, 1777–1791. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Tanaka, A.; Mori, M. Brassinosteroid signaling in rice. In Brassinosteroids: A Class of Plant Hormone; Hayat, S., Ahmad, A., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 83–117. ISBN 978-94-007-0189-2. [Google Scholar]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, Y.; Guo, S.; Zhu, J.; Huan, Q.; Liu, H.; Wang, L.; Luo, G.; Wang, X.; Chong, K. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 2012, 8, e1002686. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Qian, Q.; Xu, T.; Zhang, Y.; Dong, G.; Gao, T.; Xie, Q.; Xue, Y. The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G alpha subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet. 2013, 9, e1003391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Song, X.; Guo, H.; Wu, Y.; Chen, X.; Fang, R. A small G protein as a novel component of the rice brassinosteroid signal transduction. Mol. Plant 2016, 9, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signaling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bai, M.Y.; Chong, K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruszka, D.; Szarejko, I.; Maluszynski, M. New allele of HvBRI1 gene encoding brassinosteroid receptor in barley. J. Appl. Genet. 2011, 52, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Dockter, C.; Gruszka, D.; Braumann, I.; Druka, A.; Druka, I.; Franckowiak, J.; Gough, S.P.; Janeczko, A.; Kurowska, M.; Lundqvist, J.; et al. Induced variations in brassinosteroid genes define barley height and sturdiness, and expand the green revolution genetic toolkit. Plant Physiol. 2014, 166, 1912–1927. [Google Scholar] [CrossRef] [PubMed]
- Thole, V.; Peraldi, A.; Worland, B.; Nicholson, P.; Doonan, J.H.; Vain, P. T-DNA mutagenesis in Brachypodium distachyon. J. Exp. Bot. 2012, 63, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Kir, G.; Ye, H.; Nelissen, H.; Neelakandan, A.K.; Kusnandar, A.S.; Luo, A.; Inze, D.; Sylvester, A.W.; Yin, Y.; Becraft, P.W. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 2015, 169, 826–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Breja, P.; Khurana, J.P.; Khurana, P. Wheat Brassinosteroid-Insensitive1 (TaBRI1) interacts with members of TaSERK gene family and cause early flowering and seed yield enhancement in Arabidopsis. PLoS ONE 2016, 11, e0153273. [Google Scholar] [CrossRef] [PubMed]
- Bittner, T.; Nadler, S.; Schulze, E.; Fischer-Iglesias, C. Two homolog wheat Glycogen Synthase Kinase 3/SHAGGY-like kinases are involved in brassinosteroid signaling. BMC Plant Biol. 2015, 15, 247. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Khurana, P. Ectopic expression of Triticum aestivum SERK genes (TaSERKs) control plant growth and development in Arabidopsis. Sci. Rep. 2017, 7, 12368. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Guo, H.; Zhang, G.; Wu, Y.; Wang, G.; Chen, X. OsPRA2 fine-tunes rice brassinosteroid receptor. Plant Signal. Behav. 2017, 12, e1257455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Xu, Y.-Y.; Ma, Q.-B.; Li, D.; Xu, Z.-H.; Chong, K. Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Res. 2006, 16, 916–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oki, K.; Inaba, N.; Kitagawa, K.; Fujioka, S.; Kitano, H.; Fujisawa, Y.; Kato, H.; Iwasaki, Y. Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol. 2009, 50, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Chen, J.G.; Young, J.C.; Im, K.H.; Sussman, M.R.; Jones, A.M. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 2001, 292, 2066–2069. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Chen, J.G.; Wang, S.; Jones, A.M. Role of Heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 2002, 129, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, S.; Asami, T.; Chen, J.G. Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein alpha subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant Cell Physiol. 2008, 49, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Yasuda, A.; Yamaoka, K.; Ueda, M.; Nakayama, A.; Takatsuto, S.; Honda, I. Brachytic 1 of barley (Hordeum vulgare L.) encodes the α subunit of heterotrimeric G protein. J. Plant Physiol. 2017, 213, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Braumann, I.; Dockter, C.; Beier, S.; Himmelbach, A.; Lok, F.; Lundqvist, U.; Skadhauge, B.; Stein, N.; Zakhrabekova, S.; Zhou, R.; et al. Mutations in the gene of the Gα subunit of the heterotrimeric G protein are the cause for the brachytic1 semi-dwarf phenotype in barley and applicable for practical breeding. Hereditas 2018, 155, 10. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kitano, H.; Fujioka, S. An E3 ubiquitin ligase, ERECT LEAF1, functions in brassinosteroid signaling of rice. Plant Signal. Behav. 2013, 8, e27117. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zheng, J.; Huang, R.; Huang, Y.; Wang, H.; Jiang, L.; Fang, X. Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep. 2016, 35, 2423–2433. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, D.; Zhang, G.; Tong, H.; Chu, C. Brassinosteroids regulate OFP1, a DLT interacting protein, to modulate plant architecture and grain morphology in rice. Front. Plant Sci. 2017, 8, 1698. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chang, Y.; Guo, J.; Chen, J.G. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007, 50, 858–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, A.J.; Begcy, K.; Sarath, G.; Walia, H. Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 2015, 241, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shen, W.; He, Y.; Tian, Z.; Li, J. OVATE Family Protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PLoS Genet. 2016, 12, e1006118. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol. 2018, 176, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Nakagawa, H.; Tomita, C.; Shimatani, Z.; Ohtake, M.; Nomura, T.; Jiang, C.J.; Dubouzet, J.G.; Kikuchi, S.; Sekimoto, H.; et al. BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009, 151, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Yuan, P.; Zhang, C.; Li, T.Y.; Xuan, Y.H. RAVL1, an upstream component of brassinosteroid signalling and biosynthesis, regulates ethylene signalling via activation of EIL1 in rice. Plant Biotechnol. J. 2018, 16, 1399–1401. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, X.; Zhou, W.; Ren, Y.; Wang, Z.; Liu, Z.; Tang, J.; Tong, H.; Fang, J.; Bu, Q. Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol. 2017, 175, 1337–1349. [Google Scholar] [CrossRef] [PubMed]


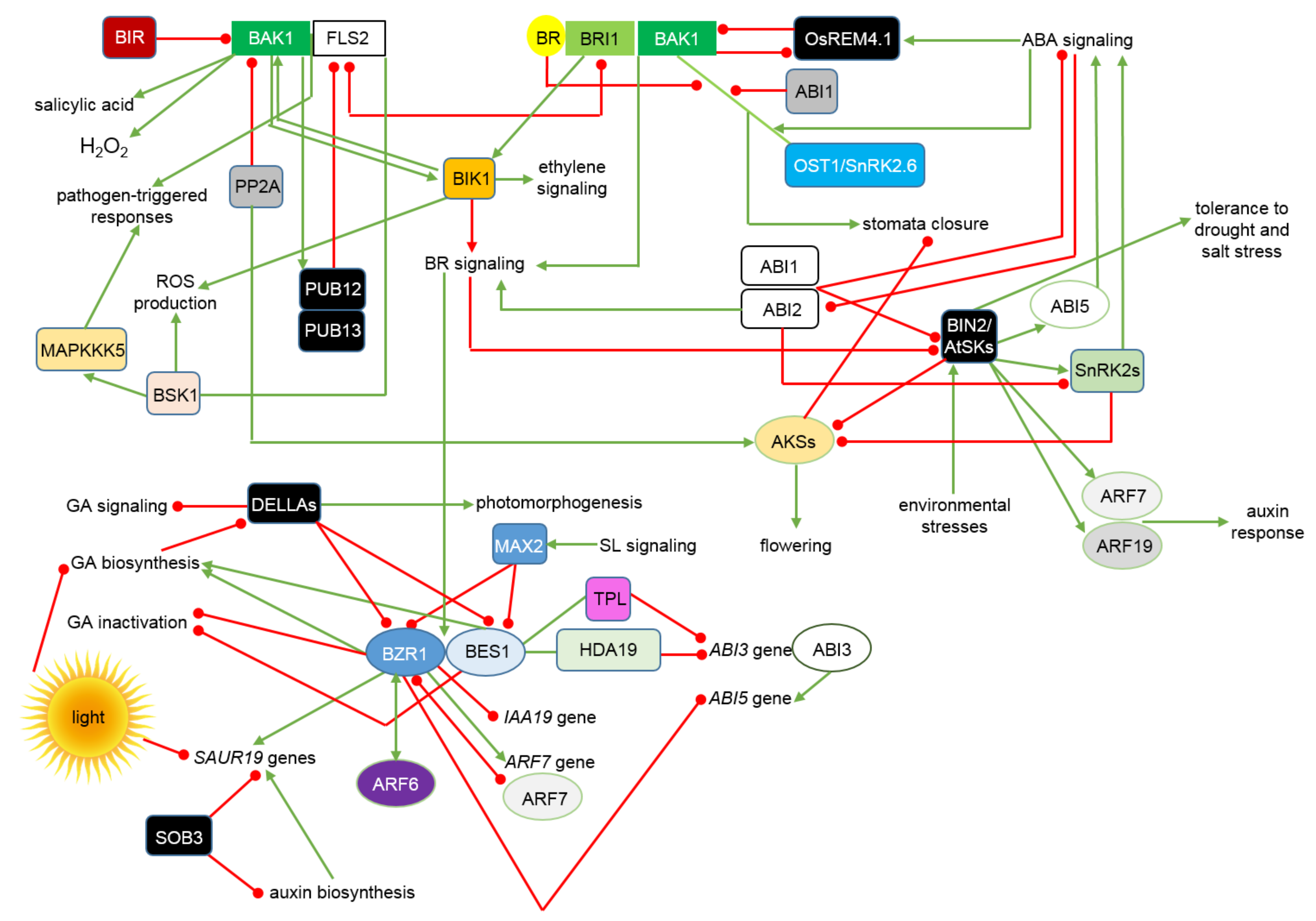
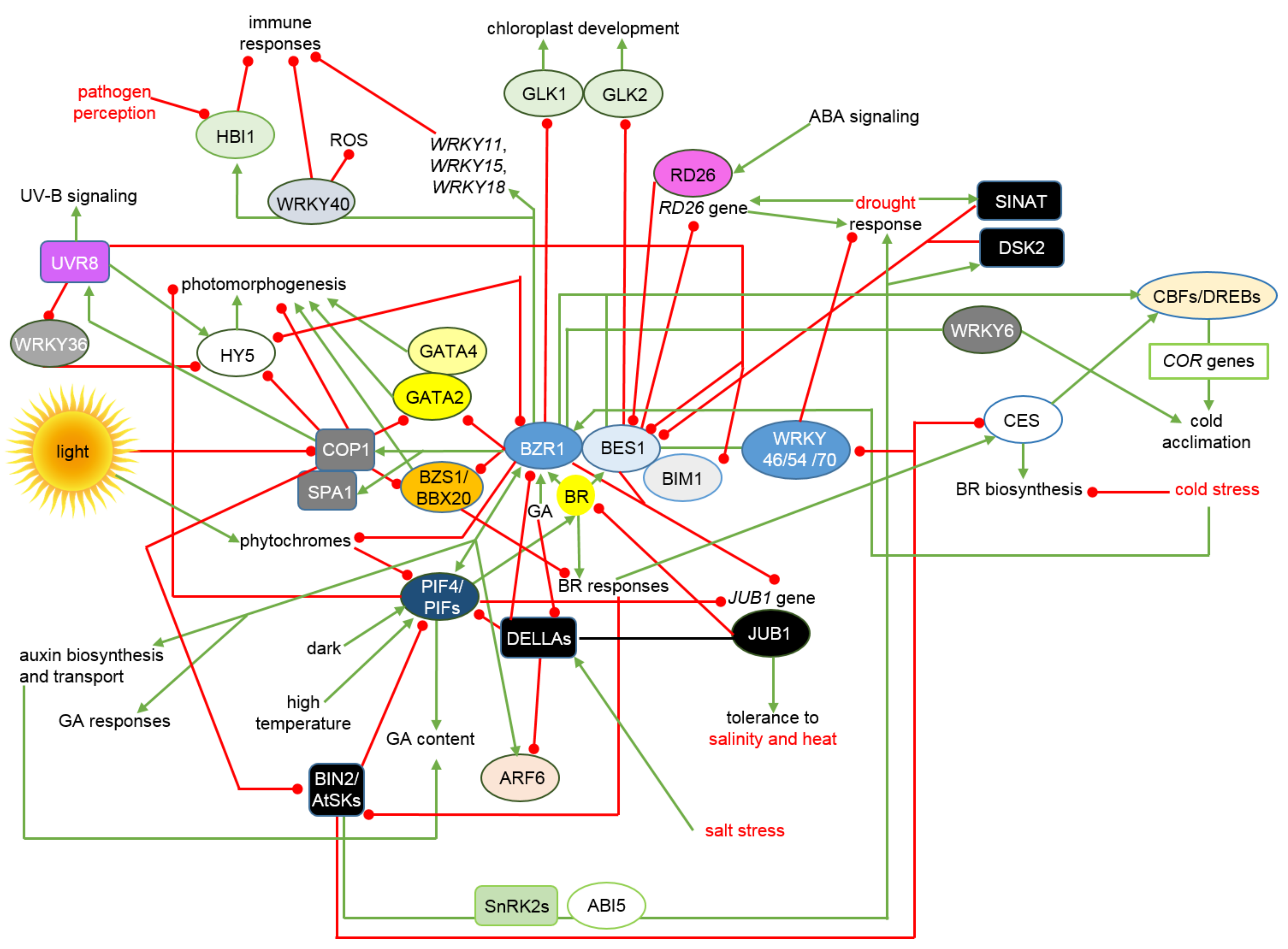
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruszka, D. Crosstalk of the Brassinosteroid Signalosome with Phytohormonal and Stress Signaling Components Maintains a Balance between the Processes of Growth and Stress Tolerance. Int. J. Mol. Sci. 2018, 19, 2675. https://doi.org/10.3390/ijms19092675
Gruszka D. Crosstalk of the Brassinosteroid Signalosome with Phytohormonal and Stress Signaling Components Maintains a Balance between the Processes of Growth and Stress Tolerance. International Journal of Molecular Sciences. 2018; 19(9):2675. https://doi.org/10.3390/ijms19092675
Chicago/Turabian StyleGruszka, Damian. 2018. "Crosstalk of the Brassinosteroid Signalosome with Phytohormonal and Stress Signaling Components Maintains a Balance between the Processes of Growth and Stress Tolerance" International Journal of Molecular Sciences 19, no. 9: 2675. https://doi.org/10.3390/ijms19092675