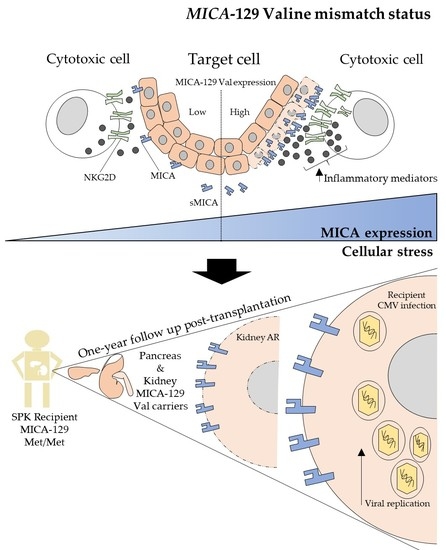
A Valine Mismatch at Position 129 of MICA Is an Independent Predictor of Cytomegalovirus Infection and Acute Kidney Rejection in Simultaneous Pancreas–Kidney Transplantation Recipients
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population and Outcome Parameters
4.2. MIC-A 454G/A and rs2596538A/G Genotyping
4.3. sMICA Measurement
4.4. Statistical Analysis
Supplementary Materials
Author contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AR | Acute rejection |
| bp | base pair |
| D | Donor |
| HIV | Human immunodeficiency virus |
| HLA | Human leukocyte antigen |
| LD | Linkage disequilibrium |
| Met | Methionine |
| MHC | Major histocompatibility complex |
| MICA | Major histocompatibility complex class I chain-related molecule A |
| NK | Natural killer |
| NKG2D | Natural-killer group 2 member D |
| PBS | Phosphate buffered saline |
| R | Recipient |
| SD | Standard deviation |
| sMICA | soluble MICA |
| SNP | Single nucleotide polymorphism |
| SPKT | Simultaneous pancreas-kidney transplantation |
| Val | Valine |
References
- Bazerbachi, F.; Selzner, M.; Boehnert, M.U.; Marquez, M.A.; Norgate, A.; McGilvray, I.D.; Schiff, J.; Cattral, M.S. Thymoglobulin versus basiliximab induction therapy for simultaneous kidney-pancreas transplantation: Impact on rejection, graft function, and long-term outcome. Transplantation 2011, 92, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Razonable, R.R.; Rivero, A.; Rodriguez, A.; Wilson, J.; Daniels, J.; Jenkins, G.; Larson, T.; Hellinger, W.C.; Spivey, J.R.; Paya, C.V. Allograft rejection predicts the occurrence of late-onset cytomegalovirus (CMV) disease among CMV-mismatched solid organ transplant patients receiving prophylaxis with oral ganciclovir. J. Infect. Dis. 2001, 184, 1461–1464. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, A.K.; Mehra, N.K. Major Histocompatibility complex class i chain-related A (MICA) molecules: Relevance in solid organ transplantation. Front. Immunol. 2017, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Halliwell, J.A.; Hayhurst, J.D.; Flicek, P.; Parham, P.; Marsh, S.G.E. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015, 43, D423–D431. [Google Scholar] [CrossRef] [PubMed]
- Risti, M.; Bicalho, M.D.G. MICA and NKG2D: Is There an Impact on Kidney Transplant Outcome? Front. Immunol. 2017, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Lu, J.; Wei, L.; Long, D.; Guo, J.Y.; Shan, J.; Li, F.S.; Lu, P.Y.; Li, P.Y.; Feng, L. The role of HIF-1 in up-regulating MICA expression on human renal proximal tubular epithelial cells during hypoxia/reoxygenation. BMC Cell Biol. 2010, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.H.Y.; Urabe, Y.; Kumar, V.; Tanikawa, C.; Koike, K.; Kato, N.; Miki, D.; Chayama, K.; Kubo, M.; Nakamura, Y.; et al. Identification of a functional variant in the MICA promoter which regulates MICA expression and increases HCV-related hepatocellular carcinoma risk. PLoS ONE 2013, 8, e61279. [Google Scholar] [CrossRef] [PubMed]
- Steinle, A.; Li, P.; Morris, D.L.; Groh, V.; Lanier, L.L.; Strong, R.K.; Spies, T. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics 2001, 53, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Isernhagen, A.; Schilling, D.; Monecke, S.; Shah, P.; Elsner, L.; Walter, L.; Multhoff, G.; Dressel, R. The MICA-129Met/Val dimorphism affects plasma membrane expression and shedding of the NKG2D ligand MICA. Immunogenetics 2016, 68, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Isernhagen, A.; Malzahn, D.; Bickeböller, H.; Dressel, R. Impact of the MICA-129Met/Val Dimorphism on NKG2D-Mediated Biological Functions and Disease Risks. Front. Immunol. 2016, 7, 588. [Google Scholar] [CrossRef] [PubMed]
- Boukouaci, W.; Busson, M.; Peffault de Latour, R.; Rocha, V.; Suberbielle, C.; Bengoufa, D.; Dulphy, N.; Haas, P.; Scieux, C.; Amroun, H.; et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood 2009, 114, 5216–5224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isernhagen, A.; Malzahn, D.; Viktorova, E.; Elsner, L.; Monecke, S.; von Bonin, F.; Kilisch, M.; Wermuth, J.M.; Walther, N.; Balavarca, Y.; et al. The MICA-129 dimorphism affects NKG2D signaling and outcome of hematopoietic stem cell transplantation. EMBO Mol. Med. 2015, 7, 1480–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carapito, R.; Jung, N.; Kwemou, M.; Untrau, M.; Michel, S.; Pichot, A.; Giacometti, G.; Macquin, C.; Ilias, W.; Morlon, A.; et al. Matching for the nonconventional MHC-I MICA gene significantly reduces the incidence of acute and chronic GVHD. Blood 2016, 128, 1979–1986. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, D.; Neuchel, C.; Niederwieser, D.; Bunjes, D.; Gramatzki, M.; Wagner, E.; Wulf, G.; Glass, B.; Pfreundschuh, M.; Einsele, H.; et al. Matching for the MICA-129 polymorphism is beneficial in unrelated hematopoietic stem cell transplantation. Blood 2016, 128, 3169–3176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askar, M.; Sobecks, R.; Wang, T.; Haagenson, M.; Majhail, N.; Madbouly, A.; Thomas, D.; Zhang, A.; Fleischhauer, K.; Hsu, K.; et al. MHC Class I Chain-Related Gene A (MICA) Donor-Recipient Mismatches and MICA-129 Polymorphism in Unrelated Donor Hematopoietic Cell Transplantations Has No Impact on Outcomes in Acute Lymphoblastic Leukemia, Acute Myeloid Leukemia, or Myelodysplastic Syndrome: A center of international blood and marrow transplant research study. Biol. Blood Marrow Transplant. 2017, 23, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.T.; Stephens, H.A.F.; Fernando, R.; Karasu, A.; Harber, M.; Howie, A.J.; Powis, S.; Zou, Y.; Stastny, P.; Madrigal, J.A.; et al. Major histocompatibility complex class I-related chain A allele mismatching, antibodies, and rejection in renal transplantation. Hum. Immunol. 2011, 72, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Tonnerre, P.; Gerard, N.; Chatelais, M.; Poli, C.; Allard, S.; Cury, S.; Bressollette, C.; Cesbron-Gautier, A.; Charreau, B. MICA variant promotes allosensitization after kidney transplantation. J. Am. Soc. Nephrol. 2013, 24, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Alvarez, B.; López-Vázquez, A.; Díaz-Molina, B.; Bernardo-Rodríguez, M.J.; Alvarez-López, R.; Pascual, D.; Astudillo, A.; Martínez-Borra, J.; Lambert, J.L.; González, S.; et al. The predictive value of soluble major histocompatibility complex class I chain-related molecule A (MICA) levels on heart allograft rejection. Transplantation 2006, 82, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, A.K.; Goswami, S.; Bhat, D.K.; Kaur, G.; Agarwal, S.K.; Mehra, N.K. Soluble Major Histocompatibility Complex Class I related Chain A (sMICA) levels influence graft outcome following Renal Transplantation. Hum. Immunol. 2018, 79, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Ashiru, O.; López-Cobo, S.; Fernández-Messina, L.; Pontes-Quero, S.; Pandolfi, R.; Reyburn, H.T.; Valés-Gómez, M. A GPI anchor explains the unique biological features of the common NKG2D-ligand allele MICA*008. Biochem. J. 2013, 454, 295–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidel, E.; Le, V.T.K.; Bar-On, Y.; Tsukerman, P.; Enk, J.; Yamin, R.; Stein, N.; Schmiedel, D.; Oiknine Djian, E.; Weisblum, Y.; et al. Dynamic co-evolution of host and pathogen: HCMV downregulates the prevalent allele MICA∗008 to escape elimination by NK cells. Cell Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Chalupny, N.J.; Rein-Weston, A.; Dosch, S.; Cosman, D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 2006, 346, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ashiru, O.; Bennett, N.J.; Boyle, L.H.; Thomas, M.; Trowsdale, J.; Wills, M.R. NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J. Virol. 2009, 83, 12345–12354. [Google Scholar] [CrossRef] [PubMed]
- Moenkemeyer, M.; Heiken, H.; Schmidt, R.E.; Witte, T. Higher risk of cytomegalovirus reactivation in human immunodeficiency virus–1–infected patients homozygous for MICA5.1. Hum. Immunol. 2009, 70, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Cerboni, C.; Neri, F.; Casartelli, N.; Zingoni, A.; Cosman, D.; Rossi, P.; Santoni, A.; Doria, M. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J. Gen. Virol. 2007, 88, 242–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodier, M.R.; Jonjić, S.; Riley, E.M.; Juranić Lisnić, V. CMV and natural killer cells: Shaping the response to vaccination. Eur. J. Immunol. 2018, 48, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Odom, C.I.; Gaston, D.C.; Markert, J.M.; Cassady, K.A. Human herpesviridae methods of natural killer cell evasion. Adv. Virol. 2012, 2012, 359869. [Google Scholar] [CrossRef] [PubMed]
- Racusen, L.C.; Solez, K.; Colvin, R.B. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999, 55, 713–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nückel, H.; Switala, M.; Sellmann, L.; Horn, P.A.; Dürig, J.; Dührsen, U.; Küppers, R.; Grosse-Wilde, H.; Rebmann, V. The prognostic significance of soluble NKG2D ligands in B-cell chronic lymphocytic leukemia. Leukemia 2010, 24, 1152–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Variables | AR (n = 19) | No AR (n = 31) | p |
|---|---|---|---|
| Age (median: 25–75%) | 50 (35.0–56.5) | 43.0 (34.5–53.0) | 0.289 |
| Recipient gender (male/female) | 10/9 | 19/12 | 0.547 |
| Donor gender (male/female) | 11/8 | 14/17 | 0.382 |
| BMI (median: 25–75%) | 24.2 (21.8–26.43) | 25.0 (20.8–27.8) | 0.818 |
| Urea, mg/dL (median: 25–75%) | 18.6 (16.1–27.2) | 18.5 (15.4–20.5) | 0.431 |
| Creatinine, mg/dL (median: 25–75%) | 1.36 (1.09–1.61) | 1.30 (1.02–1.55) | 0.516 |
| Glucose, mg/dL (median: 25–75%) | 104 (90.0–119.0) | 97.5 (88.0–116.0) | 0.382 |
| HbA1C, % (median: 25–75%) | 5.85 (5.33–6.70) | 5.70 (5.3–6.4) | 0.693 |
| Kidney graft cold ischemia time, min (±SD) | 816.7 (158.4) | 798.4 (173.9) | 0.772 |
| Pancreas graft cold ischemia time, min (±SD) | 677.4 (133.4) | 682.6 (137.2) | 0.697 |
| HLA-A and -B mismatches (median: 25%–75%) | 3.0 (3.0–4.0) | 3.0 (2.0–3.0) | 0.267 |
| HLA–DR mismatch (median: 25–75%) | 2.0 (1.5–2.0) | 2.0 (1.0–2.0) | 0.225 |
| Anti–MICA pretransplantation | 5 (26.3) | 1 (3.4) | 0.030 |
| Anti–MICA post-transplantation | 2 (11.1) | 4 (14.8) | 1.000 |
| Anti–MHC I post-transplantation | 5 (27.8) | 5 (18.5) | 0.489 |
| Anti–MHC II post-transplantation | 4 (22.3) | 6 (22.2) | 1.000 |
| Cytomegalovirus D+ | 11 (57.9) | 11 (35.5) | 0.121 |
| Cytomegalovirus R+ | 5 (26.3) | 19 (61.3) | 0.016 |
| Cytomegalovirus, R−/D+ | 8 (42.1) | 5 (16.1) | 0.042 |
| Cytomegalovirus infection, first-year post-transplantation | 3 (15.8) | 4 (12.9) | 1.000 |
| Pancreas and kidney graft AR | 1 (5.2) | ND | |
| Pancreas graft AR | 4 (21.1) | ND | |
| Kidney graft AR | 16 (84.2) | ND | |
| Immunosuppressive drugs (n (%)) | |||
| ATG | 19 (100) | 31 (100) | |
| Steroids | 19 (100) | 31 (100) | |
| Tacrolimus | 18 (94.7) | 30 (96.8) | |
| Mycophenolic acid | 19 (100) | 30 (96.8) | |
| Cyclosporine A | 1 (5.3) | 1 (3.2) | |
| Azathioprine | ND | 1 (3.2) | |
| Simulect | ND | 1 (3.2) | |
| Rituximab | ND | 1 (3.2) |
| Variables | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Univariate analysis (n = 7) | ||||
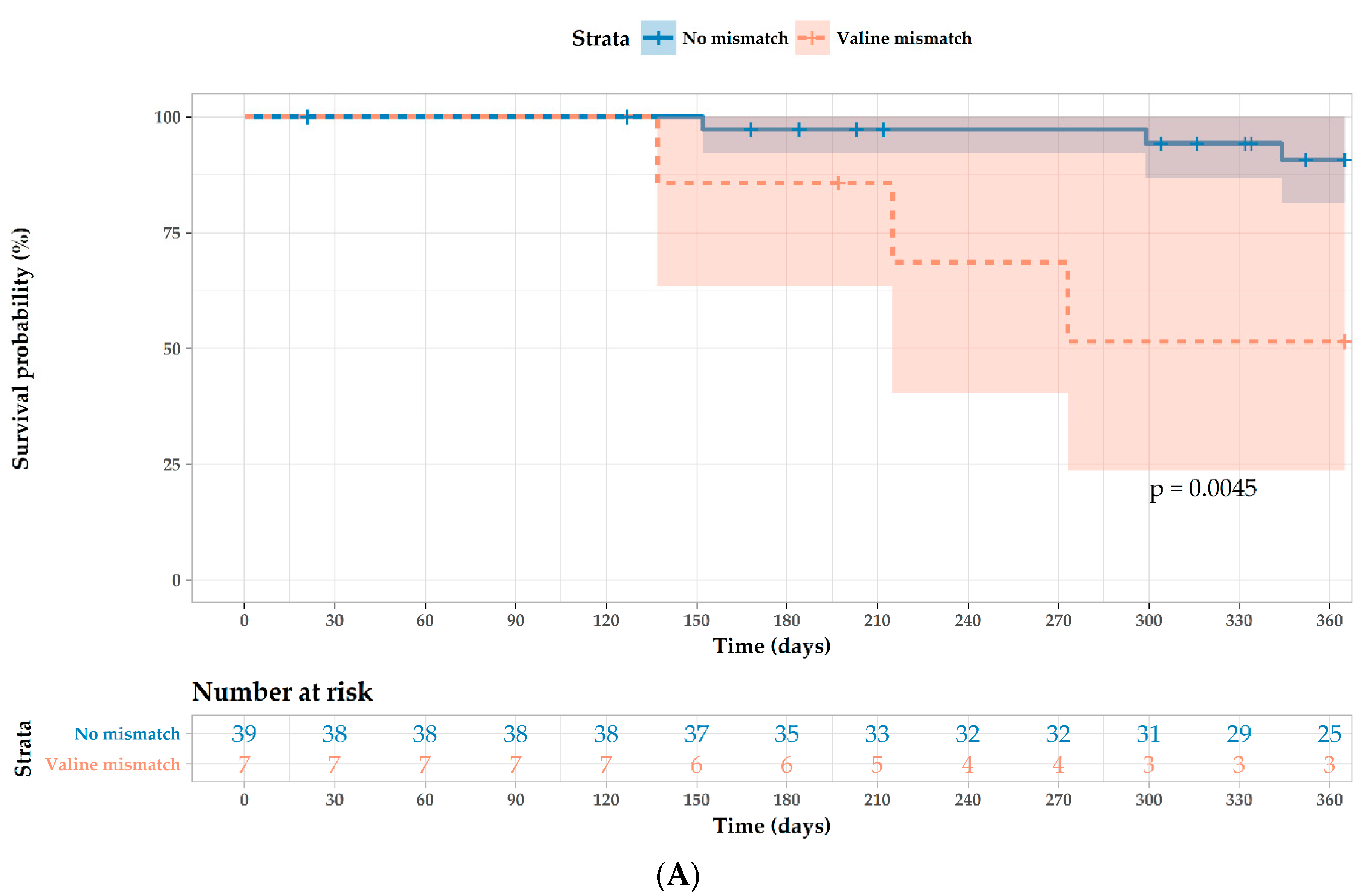
| Valine mismatch 1 | 7.37 (1.47–36.9) | 0.015 | 5.32 (1.0–28.1) | 0.049 |
| MICA antibodies pretransplantation 2 | 0.04 (0.00–1381.6) | 0.546 | ||
| HLA mismatch 3 | ||||
| 4/6 | 1.39 (0.12–15.3) | 0.790 | ||
| 5/6 | 0.80 (0.07–8.76) | 0.851 | ||
| 6/6 | 1.74 (0.16–19.2) | 0.651 | ||
| Cytomegalovirus R−/D+ 4 | 4.29 (0.96–19.6) | 0.057 | 4.89 (0.86–27.9) | 0.074 |
| Kidney graft ischemia time | 1.00 (1.00–1.00) | 0.860 | ||
| Kidney graft AR | 1.61 (0.36–7.20) | 0.532 | ||
| Variables | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Univariate analysis (n = 16) | ||||
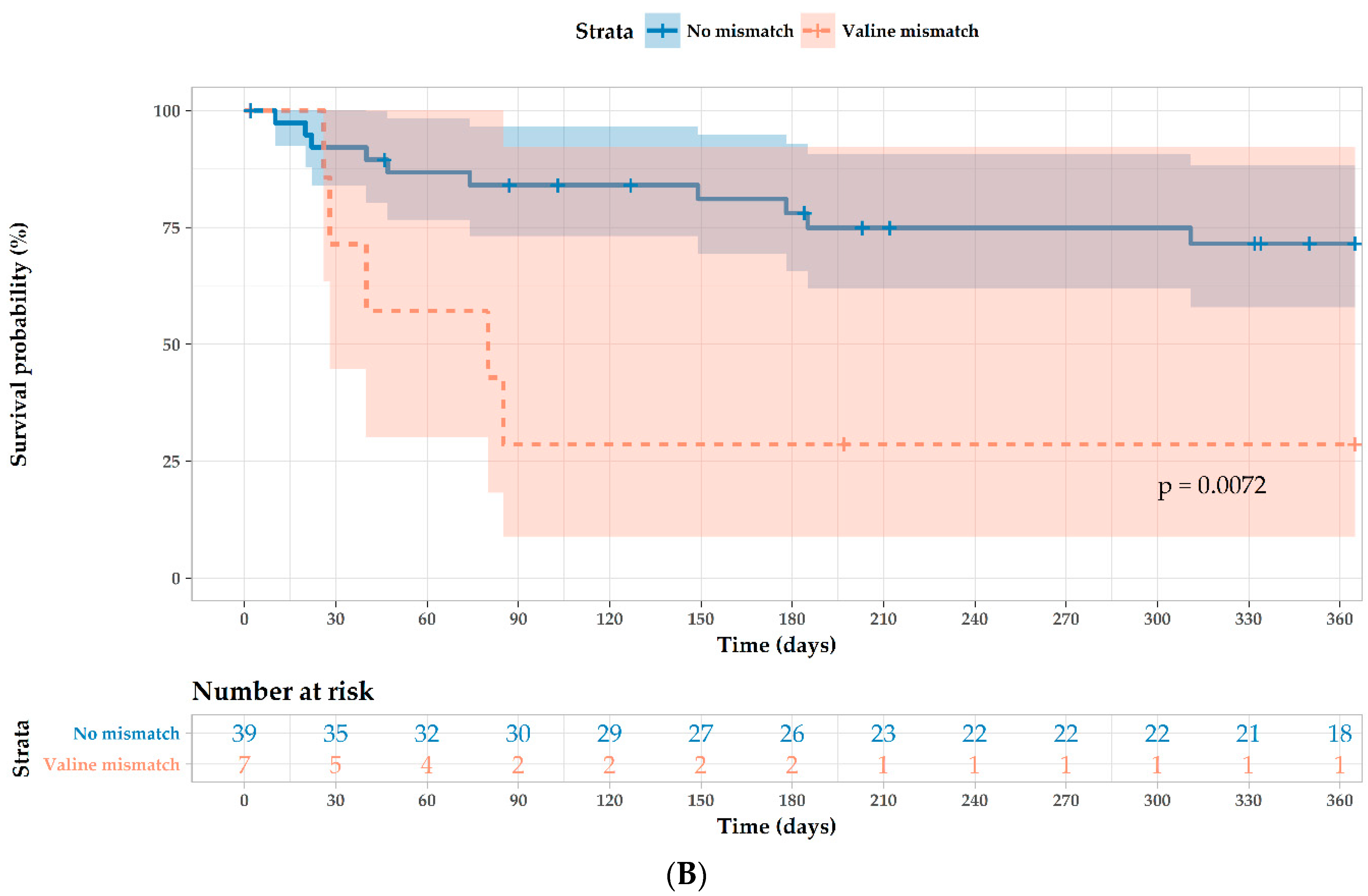
| Valine mismatch 1 | 4.00 (1.4–12.0) | 0.012 | 6.04 (1.68–21.7) | 0.006 |
| MICA antibodies pretransplantation 2 | 2.80 (0.89–8.7) | 0.077 | 3.40 (0.84–13.8) | 0.086 |
| HLA mismatch 3 | ||||
| 4/6 | 2.91 (0.32–26.0) | 0.340 | 3.65 (0.39–34.0) | 0.256 |
| 5/6 | 4.02 (0.49–32.8) | 0.193 | 5.34 (0.64–44.7) | 0.122 |
| 6/6 | 4.03 (0.45–36.1) | 0.212 | 3.11 (0.32–30.6) | 0.330 |
| HLA-DR mismatch 4 | 1.82 (0.58–5.64) | 0.300 | ||
| Gender mismatch 5 | 1.47 (0.51–4.20) | 0.480 | ||
| Cytomegalovirus infection 6 | 1.40 (0.40–4.90) | 0.590 | ||
| Kidney graft ischemia time | 1.00 (1.00–1.00) | 0.860 | ||
| Measurements, Days after Transplantation (± SD) | Val/Val | Val/Met | Met/Met | p * |
|---|---|---|---|---|
| Val129Met | ||||
| 1st, 0.58 (0.90) 1 | 0.87 (0.75–1.44) | 1.22 (0.60–1.75) | <0.02 | 0.265 |
| 2nd, 98.5 (46.0) 2 | 0.42 (0.35–0.90) | 0.55 (0.53–0.88) | <0.02 | 0.007 |
| 3rd, 238.9 (72.5) 3 | 0.66 (0.40–0.75) | 0.50 (0.45–1.08) | <0.02 | 0.035 |
| Median values at one year post-transplantation 4 | 0.69 (0.49–0.87) | 1.04 (0.71–1.19) | <0.02 | 0.005 |
| Healthy individuals ** | 1.16 (0.88–1.54) | 0.55 (0.44–0.62) | 0.00 (0.00–0.04) | <0.001 |
| rs2596538GA | GG | GA | AA | |
| First, 0.58 (0.9) 1 | 0.87 (0.65–1.59) | 0.87 (0.81–1.64) | <0.02 | 0.241 |
| Second, 98.5 (46.0) 2 | 0.49 (0.42–0.76) | 0.55 (0.45–0.72) | 0.00 (0.00–0.45) | 0.103 |
| Third, 238.9 (72.5) 3 | 0.58 (0.32–1.17) | 0.63 (0.47–0.89) | <0.02 | 0.036 |
| Median values at one year post-transplantation 4 | 0.76 (0.60–1.17) | 0.71 (0.47–0.97) | <0.02 | 0.075 |
| Healthy individuals ** | 1.37 (0.91–1.55) | 0.58 (0.43–0.83) | 0.00 (0.00–0.06) | <0.001 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michita, R.T.; Chies, J.A.B.; Schramm, S.; Horn, P.A.; Heinemann, F.M.; Wunsch, A.; Viebahn, R.; Schenker, P.; Rebmann, V. A Valine Mismatch at Position 129 of MICA Is an Independent Predictor of Cytomegalovirus Infection and Acute Kidney Rejection in Simultaneous Pancreas–Kidney Transplantation Recipients. Int. J. Mol. Sci. 2018, 19, 2618. https://doi.org/10.3390/ijms19092618
Michita RT, Chies JAB, Schramm S, Horn PA, Heinemann FM, Wunsch A, Viebahn R, Schenker P, Rebmann V. A Valine Mismatch at Position 129 of MICA Is an Independent Predictor of Cytomegalovirus Infection and Acute Kidney Rejection in Simultaneous Pancreas–Kidney Transplantation Recipients. International Journal of Molecular Sciences. 2018; 19(9):2618. https://doi.org/10.3390/ijms19092618
Chicago/Turabian StyleMichita, Rafael Tomoya, José Artur Bogo Chies, Sabine Schramm, Peter A. Horn, Falko M. Heinemann, Andreas Wunsch, Richard Viebahn, Peter Schenker, and Vera Rebmann. 2018. "A Valine Mismatch at Position 129 of MICA Is an Independent Predictor of Cytomegalovirus Infection and Acute Kidney Rejection in Simultaneous Pancreas–Kidney Transplantation Recipients" International Journal of Molecular Sciences 19, no. 9: 2618. https://doi.org/10.3390/ijms19092618