Recent Developments in Using Drosophila as a Model for Human Genetic Disease
Abstract
:1. Introduction
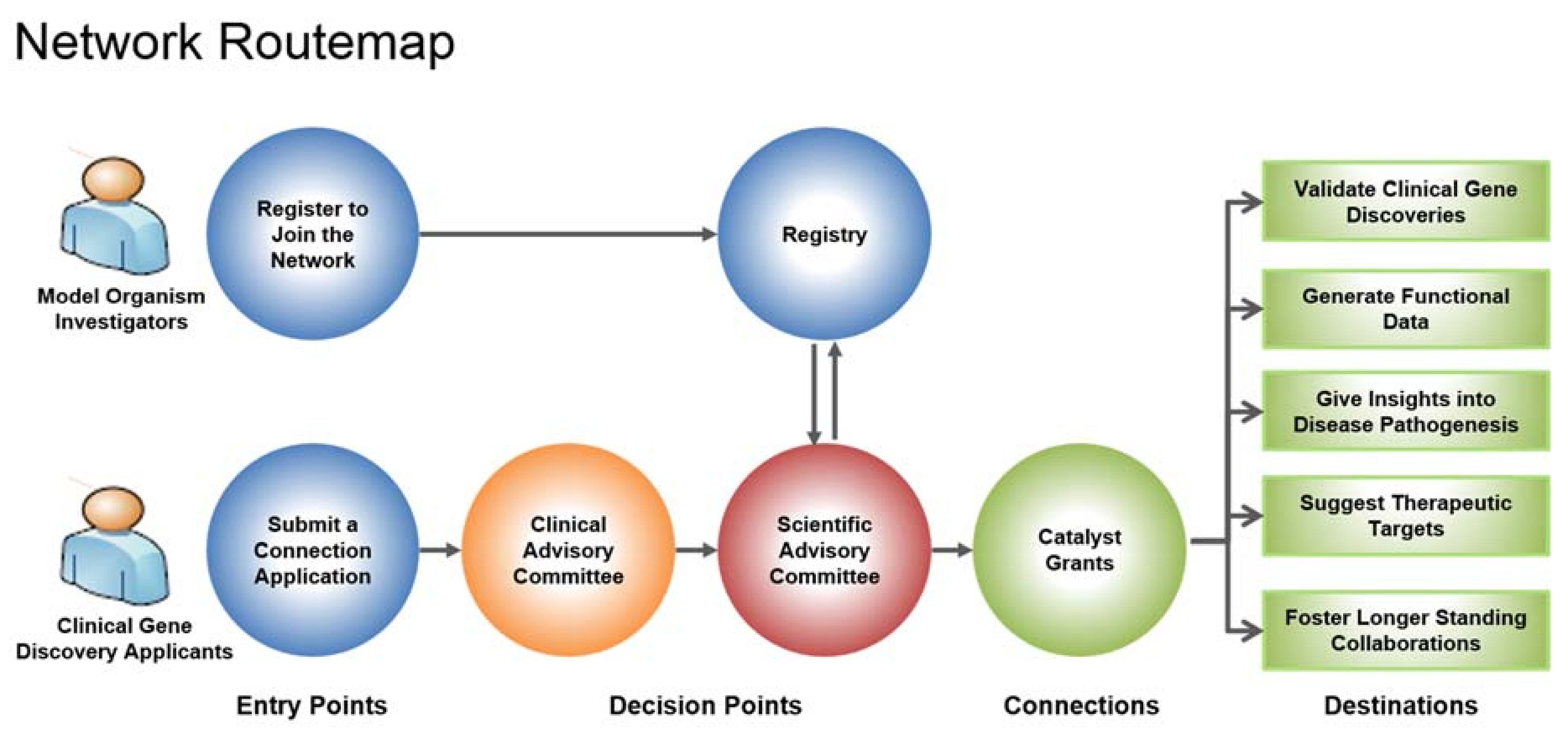
2. Rare Diseases: Models & Mechanisms (RDMM) Network
3. UDN and Centers for Mendelian Genomics
4. Solve-RD
5. Japanese RDMM
6. Concluding Thoughts
Funding
Conflicts of Interest
References
- Stark, M.B. An hereditary tumor in the fruit fly Drosophila. J. Cancer Res. 1918, 3, 279–301. [Google Scholar]
- Rubin, G.M.; Yandell, M.D.; Wortman, J.R.; Gabor Miklos, G.L.; Nelson, C.R.; Hariharan, I.K.; Fortini, M.E.; Li, P.W.; Apweiler, R.; Fleischmann, W.; et al. Comparative genomics of the eukaryotes. Science 2000, 287, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- Gramates, L.S.; Marygold, S.J.; Santos, G.D.; Urbano, J.M.; Antonazzo, G.; Matthews, B.B.; Rey, A.J.; Tabone, C.J.; Crosby, M.A.; Emmert, D.B.; et al. FlyBase at 25: Looking to the future. Nucleic Acids Res. 2017, 45, D663–D671. [Google Scholar] [CrossRef] [PubMed]
- Boycott, K.M.; Vanstone, M.R.; Bulman, D.E.; MacKenzie, A.E. Rare-disease genetics in the era of next-generation sequencing: Discovery to translation. Nat. Rev. Genet. 2013, 14, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Marmiesse, A.; Gouveia, S.; Couce, M.L. NGS technologies as a turning point in rare disease research, diagnosis and treatment. Curr. Med. Chem. 2018, 25, 404–432. [Google Scholar] [CrossRef] [PubMed]
- Wangler, M.F.; Yamamoto, S.; Bellen, H.J. Fruit flies in biomedical research. Genetics 2015, 199, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Wangler, M.F.; Yamamoto, S.; Chao, H.T.; Posey, J.E.; Westerfield, M.; Postlethwait, J.; Members of the Undiagnosed Diseases Network (UDN); Hieter, P.; Boycott, K.M.; Campeau, P.M.; et al. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 2017, 207, 9–27. [Google Scholar] [CrossRef] [PubMed]
- Duronio, R.J.; O’Farrell, P.H.; Sluder, G.; Su, T.T. Sophisticated lessons from simple organisms: Appreciating the value of curiosity-driven research. Dis. Models Mech. 2017, 10, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Housden, B.E.; Muhar, M.; Gemberling, M.; Gersbach, C.A.; Stainier, D.Y.; Seydoux, G.; Mohr, S.E.; Zuber, J.; Perrimon, N. Loss-of-function genetic tools for animal models: Cross-species and cross-platform differences. Nat. Rev. Genet. 2017, 18, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Dietzl, G.; Chen, D.; Schnörrer, F.; Su, K.C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Perkins, L.A.; Holderbaum, L.; Tao, R.; Hu, Y.; Sopko, R.; McCall, K.; Yang-Zhou, D.; Flockhart, I.; Binari, R.; Shim, H.S.; et al. The transgenic RNAi project at Harvard Medical School: Resources and validation. Genetics 2015, 201, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Heigwer, F.; Port, F.; Boutros, M. RNA interference (RNAi) screening in Drosophila. Genetics 2018, 208, 853–874. [Google Scholar] [CrossRef] [PubMed]
- Jenett, A.; Rubin, G.M.; Ngo, T.T.; Shepherd, D.; Murphy, C.; Dionne, H.; Pfeiffer, B.D.; Cavallaro, A.; Hall, D.; Jeter, J.; et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012, 4, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Kvon, E.Z.; Kazmar, T.; Stampfel, G.; Yáñez-Cuna, J.O.; Pagani, M.; Schernhuber, K.; Dickson, B.J.; Stark, A. Genome-scale functional characterization of Drosophila developmental enhancers. Nature 2014, 512, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Jeibmann, A.; Schulz, J.; Eikmeier, K.; Johann, P.D.; Thiel, K.; Tegeder, I.; Ambrée, O.; Frühwald, M.C.; Pfister, S.M.; Kopl, M.; et al. SMAD dependent signaling plays a detrimental role in a fly model of SMARCB1-deficiency and the biology of atypical teratoid/rhabdoid tumors. J. Neurooncol. 2017, 131, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Lochmüller, H.; Torrent I Farnell, J.; Le Cam, Y.; Jonker, A.H.; Lau, L.P.; Baynam, G.; Kaufmann, P.; Dawkins, H.J.; Lasko, P.; Austin, C.P.; et al. IRDiRC Consortium Assembly. The International Rare Diseases Research Consortium: Policies and guidelines to maximize impact. Eur. J. Hum. Genet. 2017, 25, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Rare Diseases Models and Mechanisms Network. Available online: http://rare-diseases-catalyst-network.ca (accessed on 18 May 2018).
- MICYRN: Better Health for Mothers and Children. Available online: www.micyrn.ca (accessed on 18 May 2018).
- Liu, W.; Yan, B.; An, D.; Xiao, J.; Hu, F.; Zhou, D. Sporadic periventricular nodular heterotopia: Classification, phenotype and correlation with Filamin A mutations. Epilepsy Res. 2017, 133, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Farhan, S.M.K.; Nixon, K.C.J.; Everest, M.; Edwards, T.N.; Long, S.; Segal, D.; Knip, M.J.; Arts, H.H.; Chakrabarti, R.; Wang, J.; et al. Identification of a novel synaptic protein, TMTC3, involved in periventricular nodular heterotopia with intellectual disability and epilepsy. Hum. Mol. Gen. 2017, 26, 4278–4289. [Google Scholar] [CrossRef] [PubMed]
- Howlett, I.C.; Rusan, Z.M.; Parker, L.; Tanouye, M.A. Drosophila as a model for intractable epilepsy: Gilgamesh suppresses seizures in parabss1 heterozygote flies. G3 2013, 3, 1399–1407. [Google Scholar] [CrossRef] [PubMed]
- Reuter, C.M.; Brimble, E.; DeFilippo, C.; Dries, A.M.; Undiagnosed Diseases Network; Enns, G.M.; Ashley, E.A.; Bernstein, J.A.; Fisher, P.G.; Wheeler, M.T. A new approach to rare diseases of children: The Undiagnosed Diseases Network. J. Pediatr. 2018, 196, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Jaiswal, M.; Charng, W.-L.; Gambin, T.; Karaca, E.; Mirzaa, G.; Wiszniewski, W.; Sandoval, H.; Haelterman, N.A.; Xiong, B.; et al. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 2014, 159, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Şentürk, M.; Bellen, H.J. Genetic strategies to tackle neurological diseases in fruit flies. Curr. Opin. Neurobiol. 2018, 50, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Al-Ouran, R.; Hu, Y.; Kim, S.Y.; Wan, Y.W.; Wangler, M.F.; Yamamoto, S.; Chao, H.T.; Comjean, A.; Mohr, S.E.; et al. MARRVEL: Integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 2017, 100, 843–853. [Google Scholar] [CrossRef] [PubMed]
- MARRVEL. Available online: marrvel.org (accessed on 18 May 2018).
- Chao, H.T.; Davids, M.; Burke, E.; Pappas, J.G.; Rosenfeld, J.A.; McCarty, A.J.; Davis, T.; Wolfe, L.; Toro, C.; Tifft, C.; et al. A syndromic neurodevelopmental disorder caused by de novo variants of EBF3. Am. J. Hum. Genet. 2017, 100, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Usui, T.; Satoh, D.; Moriyama, S.; Shimono, K.; Itoh, T.; Shirahige, K.; Uemura, T. Sensory-neuron subtype-specific transcriptional programs controlling dendrite morphogenesis: Genome-wide analysis of Abrupt and Knot/Collier. Dev. Cell 2013, 27, 530–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crozatier, M.; Valle, D.; Dubois, L.; Ibnsouda, S.; Vincent, A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 1996, 6, 707–718. [Google Scholar] [CrossRef]
- Liu, N.; Schoch, K.; Luo, X.; Pena, L.; Bhavana, V.H.; Kukolich, M.K.; Stringer, S.; Powis, Z.; Radtke, K.; Mroske, C.; et al. Functional variants in TBX2 are associated with a syndromic cardiovascular and skeletal developmental disorder. Hum. Mol. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Harrelson, Z.; Kelly, R.G.; Goldin, S.N.; Gibson-Brown, J.J.; Bollag, R.J.; Silver, L.M.; Papaioannou, V.E. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 2004, 131, 5041–5052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manning, L.; Ohyama, K.; Saeger, B.; Hatano, O.; Wilson, S.A.; Logan, M.; Placzek, M. Regional morphogenesis in the hypothalamus: A BMP-Tbx2 pathway coordinates fate and proliferation through Shh downregulation. Dev. Cell 2006, 11, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Behesti, H.; Papaioannou, V.E.; Sowden, J.C. Loss of Tbx2 delays optic vesicle invagination leading to small optic cups. Dev. Biol. 2009, 333, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Borke, J.L.; Chen, J.R.; Yu, J.C.; Bollag, R.L.; Orellana, M.F.; Isales, C.M. Negative transcriptional regulation of connexin 42 by Tbx2 in rat immature coronal sutures and ROS 17/2.8 cells in culture. Cleft Palate Craniofac. J. 2003, 40, 284–290. [Google Scholar] [CrossRef]
- Nissim, S.; Allard, P.; Bandyopadhyay, A.; Harfe, B.D.; Tabin, C.J. Characterization of a novel ectodermal signaling center regulating Tbx2 and Shh in the vertebrate limb. Dev. Biol. 2007, 304, 9–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pflugfelder, G.O.; Roth, H.; Poeck, B.; Kerscher, S.; Schwarz, H.; Jonschker, B.; Heisenberg, M. The lethal(1)optomotor-blind gene of Drosophila melanogaster is a major organizer of optic lobe development: Isolation and characterization of the gene. Proc. Natl. Acad. Sci. USA 1992, 89, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Oláhová, M.; Yoon, W.H.; Thompson, K.; Jangam, S.; Fernandez, L.; Davidson, J.M.; Kyle, J.E.; Grove, M.E.; Fisk, D.G.; Kohler, J.N.; et al. Biallelic mutations in ATP5F1D, which encodes a subunit of ATP synthase, cause a metabolic disorder. Am. J. Hum. Genet. 2018, 102, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Rosenfeld, J.A.; Yamamoto, S.; Harel, T.; Zuo, Z.; Hall, M.; Wierenga, K.J.; Pastore, M.T.; Bartholomew, D.; Delgado, M.R.; et al. Clinically severe CACNA1A alleles affect synaptic function and neurodegeneration differentially. PLoS Genet. 2017, 13, e1006905. [Google Scholar] [CrossRef] [PubMed]
- Pietrobon, D. CaV2.1 channelopathies. Pflugers Arch. 2010, 460, 375–393. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Gala, U.; Zhang, Y.; Shang, W.; Jaiswal, S.N.; diRonza, A.; Jaiswal, M.; Yamamoto, S.; Sandoval, H.; Duraine, L.; et al. A voltage-gated calcium channel regulates lysosomal fusion with endosomes and autophagosomes and is required for neuronal homeostasis. PLoS Biol. 2015, 13, e1002013. [Google Scholar] [CrossRef] [PubMed]
- Marcogliese, P.C.; Shashi, V.; Spillmann, R.C.; Stong, N.; Rosenfeld, J.A.; Koenig, M.K.; Martinez-Agosto, J.A.; Herzog, M.; Chen, A.H.; Dickson, P.I.; et al. Loss-of-function in IRF2BPL is associated with neurological phenotypes. Am. J. Hum. Genet. 2018. [Google Scholar] [CrossRef]
- Tan, K.L.; Haelterman, N.A.; Kwartler, C.S.; Regalado, E.S.; Lee, P.T.; Nagarkar-Jaiswal, S.; Guo, D.C.; Duraine, L.; Wangler, M.F.; University of Washington Center for Mendelian Genomics; et al. Ari-1 regulates myonuclear organization together with parkin and is associated with aortic aneurysms. Dev. Cell 2018, 45, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Bone, C.R.; Starr, D.A. Nuclear migration events throughout development. J. Cell Sci. 2016, 129, 1951–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucking, C.B.; Durr, A.; Bonafati, V.; Vaughan, J.; DeMichele, G.; Gasser, T.; Harhangi, B.S.; Meco, G.; Denefle, P.; Wood, N.W.; et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N. Engl. J. Med. 2000, 342, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Sobreira, N.; Schiettecatte, F.; Valle, D.; Hamosh, A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015, 36, 928–930. [Google Scholar] [CrossRef] [PubMed]
- Harel, T.; Yoon, W.H.; Garone, C.; Gu, S.; Coban-Akdemir, Z.; Eldomery, M.K.; Posey, J.E.; Jhangiani, S.N.; Rosenfeld, J.A.; Cho, M.T.; et al. Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 2016, 99, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Gilquin, B.; Taillebourg, E.; Cherradi, N.; Hubstenberger, A.; Gay, O.; Merle, N.; Assard, N.; Fauvarque, M.O.; Tomohiro, S.; Kuge, O.; et al. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol. Cell. Biol. 2010, 30, 1984–1996. [Google Scholar] [CrossRef] [PubMed]
- Yoon, W.H.; Sandoval, H.; Nagarkar-Jaiswal, S.; Jaiswal, M.; Yamamoto, S.; Haelterman, N.A.; Putluri, N.; Putluri, V.; Sreekumar, A.; Tos, T.; et al. Loss of Nardilysin, a mitochondrial co-chaperone for a-Ketoglutarate Dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron 2017, 93, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Solve-RD: Solving the Unsolved Rare Diseases. Available online: www.solve-rd.eu (accessed on 18 May 2018).
- Japanese Rare Diseases Models and Mechanisms Network. Available online: irudbeyond.nig.ac.jp/en (accessed on 9 July 2018).
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oriel, C.; Lasko, P. Recent Developments in Using Drosophila as a Model for Human Genetic Disease. Int. J. Mol. Sci. 2018, 19, 2041. https://doi.org/10.3390/ijms19072041
Oriel C, Lasko P. Recent Developments in Using Drosophila as a Model for Human Genetic Disease. International Journal of Molecular Sciences. 2018; 19(7):2041. https://doi.org/10.3390/ijms19072041
Chicago/Turabian StyleOriel, Christine, and Paul Lasko. 2018. "Recent Developments in Using Drosophila as a Model for Human Genetic Disease" International Journal of Molecular Sciences 19, no. 7: 2041. https://doi.org/10.3390/ijms19072041




