Molecular Genetics and Breeding for Nutrient Use Efficiency in Rice
Abstract
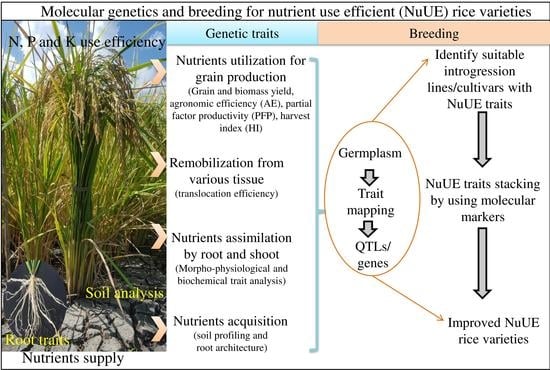
:1. Introduction
2. Screening Protocols and Breeding Efforts for Traits Related to Nutrient Use Efficiency
2.1. Phosphorus
2.2. Nitrogen
2.3. Potassium
3. Identification and Use of QTLs Related to Nutrient Use Efficiency
3.1. QTLs Related to Nitrogen Use Efficiency
3.2. Phosphorus Use Efficiency and Related QTLs
3.3. Potassium Use Efficiency and Related QTLs
4. Effect of Nutrient Use Efficiency across Medium- and Long-Duration Rice
5. QTLs for Both Low Nitrogen and Phosphorus Stress
6. Agronomic Efficiency and Partial Factor Productivity QTLs
7. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ladha, J.K.; Pathak, H.; Krupnik, T.J.; Six, J.; Kessel, C.V. Efficiency of fertilizer nitrogen in cereal production: Retrospects and prospects. Adv. Agron. 2005, 87, 85–156. [Google Scholar]
- Xu, X.; Liu, X.; He, P. Yield Gap, Indigenous Nutrient Supply and Nutrient Use Efficiency for Maize in China. PLoS ONE 2015, 10, e0140767. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.L. Improving Nutrient Use Efficiency. Turk. J. Agric. For. 2008, 32, 177–182. [Google Scholar]
- Liu, Z.; Zhu, C.; Jiang, Y.; Tian, Y.; Yu, J.; An, H.; Tang, W.; Sun, J.; Tang, J.; Chen, G.; et al. Association mapping and genetic dissection of nitrogen use efficiency-related traits in rice (Oryza sativa L.). Funct. Integr. Genom. 2016, 16, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Vinod, K.K.; Heuer, S. Approaches towards nitrogen- and phosphorus-efficient rice. AoB Plants 2012, 28, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.; Ladha, J.K.; Castillo, E.G.; Punzalam, G.; Tirol-Padre, A.; Duqueza, M. Genotypic variation in nitrogen use efficiency in medium and long-duration rice. Field Crops Res. 1998, 58, 35–53. [Google Scholar] [CrossRef]
- Han, M.; Okamoto, M.; Beatty, P.H.; Rothstein, S.J.; Good, A.G. The Genetics of Nitrogen Use Efficiency in Crop Plants. Annu Rev. Genet. 2015, 49, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Van Bueren, E.T.L.; Struik, P.C. Diverse concepts of breeding for nitrogen use efficiency, a review. Agron. Sustain. Dev. 2017, 37, 50. [Google Scholar] [CrossRef]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y.; et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J.; Singh, N.; Dwiyanti, M.S.; Wang, D.R.; Wright, M.H.; Perez, F.A.; DeClerck, G.; Chin, J.H.; Malitic-Layaoen, G.A.; Juanillas, V.M.; et al. Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications. Rice 2017, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Chen, K.; Cui, Y.; Wu, Z.; Zheng, T.; Zhu, Y.; Ali, J.; Wang, B.; Xu, J.; Zhang, W.; et al. Genetic Dissection and Simultaneous Improvement of Drought and Low Nitrogen Tolerances by Designed QTL Pyramiding in Rice. Front. Plant Sci. 2018, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Baligar, V.C.; Fageria, N.K.; Hea, Z.L. Nutrient Use Efficiency in Plants. Commun. Soil Sci. Plant Anal. 2001, 32, 7–8. [Google Scholar] [CrossRef]
- Ali, J.; Xu, J.L.; Gao, Y.M.; Fontanilla, M.A.; Li, Z.K. Green super rice (GSR) technology: An innovative breeding strategy-achievements & advances. In Proceedings of the 12th SABRAO Congress-Plant Breeding towards 2025: Challenges in a Rapidly Changing World, Chiang Mai, Thailand, 13–16 January 2012; pp. 16–17. [Google Scholar]
- Kole, C.; Muthamilarasan, M.; Henry, R.; Edwards, D.; Sharma, R.; Abberton, M.; Batley, J.; Bentley, A.; Blakeney, M.; Bryant, J.; et al. Application of genomics-assisted breeding for generation of climate resilient crops: Progress and prospects. Front. Plant Sci. 2015, 6, 563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, D.; Iwata, A.; Goicoechea, J.L.; Wei, S.; et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, F.E.; De Datta, S.K.; Laureles, E.V. Measurement of nitrogen utilization efficiency in rice genotypes. Agron. J. 1987, 79, 786–791. [Google Scholar] [CrossRef]
- Singh, U.; Cassman, K.G.; Ladha, J.K. Innovative Nitrogen Management Strategies for Lowland Rice Systems; Fragile Lives in Fragile Ecosystems, International Rice Research Institute, P.O. Box 933: Manila, Philippines, 1995; pp. 229–254. [Google Scholar]
- Xiao, J.H.; Li, J.M.; Yuan, L.P.; Yuan, S.R. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a sub-specific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Wisser, R.J.; Sun, Q.; Hulbert, S.H.; Kresovich, S.; Nelson, R.J. Identification and Characterization of Regions of the Rice Genome Associated with Broad-Spectrum, Quantitative Disease Resistance. Genetics 2015, 169, 2277–2293. [Google Scholar] [CrossRef] [PubMed]
- Bocianowski, J. Epistasis interaction of QTL effects as a genetic parameter influencing estimation of the genetic additive effect. Genet. Mol. Biol. 2013, 36, 93–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Liu, Z.; Fu, X.; Dai, Z.; Wang, S.; Zhang, G.; Zeng, R.; Liu, G. Detection and characterization of epistasis between QTLs on plant height in rice using single segment substitution lines. Breed. Sci. 2015, 65, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.S.; Pinson, R.M.; Stansel, J.W.; Park, D. Identification of two major genes and quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 1995, 91, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Cho, Y.G.; Tlee, Y.; Eun, M.Y.; Shim, J.U. QTL mapping of genes conferring days to heading, culm length and panicle length based on molecular map of rice (Oryza sativa L.). RDA J. Crop Sci. 1998, 40, 55–61. [Google Scholar]
- Yamamoto, T.; Yonemaru, J.; Yano, M. Towards the Understanding of Complex Traits in Rice: Substantially or Superficially? DNA Res. 2009, 16, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]
- Nongpiur, R.C.; Singla-Pareek, S.L.; Pareek, A. Genomics Approaches for Improving Salinity Stress Tolerance in Crop Plants. Curr. Genom. 2016, 17, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Dang, D.T.; Pham, C.V.; Bertin, P. QTL mapping for nitrogen use efficiency and related physiological and agronomical traits during the vegetative phase in rice under hydroponics. Euphytica 2016, 212, 473–500. [Google Scholar] [CrossRef]
- Wang, H.; Qin, F. Genome-Wide Association Study Reveals Natural Variations Contributing to Drought Resistance in Crops. Front. Plant Sci. 2017, 30, 1110. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Aravindan, S.; Ngangkham, U.; Subudhi, H.N.; Bag, M.K.; Adak, T.; Munda, S.; Samantary, S.; Jena, M. Correction: Use of molecular markers in identification and characterization of resistance to rice blast in India. PLoS ONE 2017, 12, e0179467. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Ersoz, E.S.; Yu, J.; Buckler, E.S. Applications of linkage disequilibrium and association mapping in maize. Mol. Genet. Approach Maize Improv. 2009, 63, 173–195. [Google Scholar]
- Clark, R.M.; Schweikert, G.; Toomajian, C.; Ossowski, S.; Zeller, G.; Shinn, P.; Warthmann, N.; Hu, T.T.; Fu, G.; Hinds, D.A.; et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 2007, 317, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.M.; Zhu, H.T.; Ding, X.H.; Zeng, R.Z.; Zhang, Z.L.; Zhang, G. Detection of QTLs for Important Agronomic Traits and Analysis of Their Stabilities Using SSSLs in Rice. Agric. Sci. China 2007, 6, 769–778. [Google Scholar] [CrossRef]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yu, Y.; Ma, Y.; Gao, Q.; Cao, Y.; Chen, Z.; Ma, B.; Qi, M.; Li, Y.; Zhao, X.; et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 2017, 8, 15324. [Google Scholar] [CrossRef] [PubMed]
- McNally, K.L.; Childs, K.L.; Bohnert, R.; Davidson, R.M.; Zhao, K.; Ulat, V.J.; Zeller, G.; Clark, R.M.; Hoen, D.R.; Bureau, T.E.; et al. Genome wide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. USA 2009, 106, 12273–12278. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Ebana, K.; Yonemaru, J.; Fukuoka, S.; Iwata, H.; Kanamori, K.; Namiki, N.; Nagasaki, H.; Yano, M. Genetic structure revealed by a whole-genome single nucleotide polymorphism survey of 5 diverse accessions of cultivated Asian rice (Oryza sativa L.). Breed. Sci. 2010, 60, 390–397. [Google Scholar] [CrossRef]
- Zhao, K.; Aranzana, M.J.; Kim, S.; Lister, C.; Shindo, C.; Tang, C.; Toomajian, C.; Zheng, H.; Dean, C.; Marjoram, P.; et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007, 3, e4. [Google Scholar] [CrossRef] [PubMed]
- Agrama, H.A.; Yan, W.; Jia, M.; Fjellstrom, R.; McClung, A.M. Genetic structure associated with diversity and geographic distribution in the USDA rice world collection. Nat. Sci. 2010, 2, 247–291. [Google Scholar] [CrossRef]
- Ali, J.; Xu, J.; Ismail, A.M.; Fu, B.Y.; Vijaykumar, C.H.M.; Gao, Y.M.; Domingo, J.; Maghirang, R.; Yu, S.B.; Gregorio, G.; et al. Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Res. 2006, 97, 66–76. [Google Scholar] [CrossRef]
- MacDonald, G.K.; Bennett, E.M.; Potter, P.A.; Ramankutty, N. Agronomic phosphorus imbalances across the world's croplands. Proc. Natl. Acad. Sci. USA 2007, 108, 3086–3091. [Google Scholar] [CrossRef] [PubMed]
- Cordell, D.; Drangert, J.O.; White, S. The story of phosphorus global food security and food for thought. Glob. Environ. Chang. 2009, 19, 92–305. [Google Scholar] [CrossRef]
- Rose, T.J.; Wissuwa, M. Rethinking internal phosphorus utilization efficiency: A new approach is needed to improve PUE in grain crops. Adv. Agron. 2012, 116, 185–217. [Google Scholar]
- Krishnamurthy, P.; Sreedevi, B.; Ram, T.; Padmavathi, G.; Kumar, R.M.; Rao, P.R; Rani, N.S.; Latha, P.C.; Singh, S.P. Evaluation of rice genotypes for phosphorus use efficiency under soil mineral stress conditions. Oryza 2010, 47, 29–33. [Google Scholar]
- Fageria, N.K.; Morais, O.P.; Baligar, V.C.; Wrigh, R.J. Response of rice cultivars to phosphorus supply on an Oxisol. Fertilizer Res. 1988, 16, 195–206. [Google Scholar] [CrossRef]
- Saito, K.; Vandamme, E.; Segda, Z.; Fofana, M.; Ahouanton, K. A screening protocol for vegetative-stage tolerance to phosphorus deficiency in upland rice. Crop Sci. 2015, 55, 1223–1229. [Google Scholar] [CrossRef]
- Li, Z.K.; Fu, B.Y.; Gao, Y.M.; Xu, J.L.; Ali, J.; Lafitte, H.R.; Jiang, Y.Z.; Rey, J.D.; Vijayakumar, C.H.; Maghirang, R.; et al. Genome-wide ILs and Their Use in Genetic and Molecular Dissection of Complex Phenotypes in Rice (Oryza sativa L.). Plant Mol. Biol. 2005, 59, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, M.; Rao, D.N.; Sarla, N. Molecular mechanisms in response to phosphate starvation in rice. Biotechnol. Adv. 2009, 27, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Wissuwa, M.; Kretzschmar, T.; Rose, T.J. From promise to application: Root traits for enhanced nutrient capture in rice breeding. J. Exp. Bot. 2016, 67, 3605–3615. [Google Scholar] [CrossRef] [PubMed]
- Vejchasarn, P.; Lynch, J.P.; Brown, K.M. Genetic Variability in Phosphorus Responses of Rice Root Phenotypes. Rice 2016, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Aluwihare, Y.C.; Ishan, M.; Chamikara, M.D.M.; Weebadde, C.K.; Sirisena, D.N.; Samarasinghe, W.L.G.; Sooriyapathirana, S.D.S.S. Characterization and Selection of Phosphorus Deficiency Tolerant Rice Genotypes in Sri Lanka. Rice Sci. 2016, 23, 184–195. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K.; Singh, O.N. Traits-related QTLs and genes and their potential applications in rice improvement under low phosphorus condition. Arch. Agron. Soil Sci. 2017, 64, 449–464. [Google Scholar] [CrossRef]
- Yugandhar, P.; Veronica, N.; Panigrahy, M.; Nageswara Rao, D.; Subrahmanyam, D.; Voleti, S.R.; Mangrauthia, S.K.; Sharma, R.P.; Sarla, N. Comparing Hydroponics, Sand, and Soil Medium to Evaluate Contrasting Rice Nagina 22 Mutants for Tolerance to Phosphorus Deficiency. Crop Sci. 2017, 57, 1–9. [Google Scholar] [CrossRef]
- Chithrameenal, K.; Vellaikumar, S.; Ramalingam, J. Identification of rice (Oryza sativa L.) genotypes with high phosphorus use efficiency (PUE) under field and hydroponic conditions. Indian Res. J. Genet. Biotechnol. 2017, 9, 23–37. [Google Scholar]
- Chin, J.H.; Gamuyao, R.; Dalid, C.; Bustamam, M.; Prasetiyono, J.; Moeljopawiro, S.; Wissuwa, M.; Heuer, S. Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application. Plant Physiol. 2011, 156, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Sirisena, D.N.; Wanninayake, W.M.N. Identification of promising rice varieties for low fertile soils in the low country intermediate zone in Sri Lanka. Ann. Sri Lanka Dep. Agric. 2014, 14, 95–105. [Google Scholar]
- Cancellier, E.L.; Brandao, D.R.; Silva, J.; Santos, M.M.; Fidelis, R.R. Phosphorus use efficiency of upland rice cultivars on Cerrado soil. Ambience 2012, 8, 307–318. [Google Scholar]
- Fageria, N.K.; Knupp, A.M.; Moraes, M.F. Phosphorus Nutrition of Lowland Rice in Tropical Lowland Soil. Commun. Soil Sci. Plant Anal. 2013, 44, 2932–2940. [Google Scholar] [CrossRef]
- Panigrahy, M.; Nageswara Rao, D.; Yugandhar, P.; Sravan Raju, N.; Krishnamurthy, P.; Voleti, S.R.; Ashok Reddy, G.; Mohapatra, T.; Robin, A.; Singh, A.K.; et al. Hydroponic experiment for identification of tolerance traits developed by rice Nagina 22 mutants to low-phosphorus in field condition. Arch. Agron. Soil Sci. 2014, 60, 565–576. [Google Scholar] [CrossRef]
- Wu, P.; Ma, L.; Hou, X. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003, 132, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, Y.; Dai, A.; Liu, L.; Li, Z. Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using ILs of rice. J. Genet. Genom. 2009, 36, 173–183. [Google Scholar] [CrossRef]
- Li, L.; Qiu, X.; Li, X.; Wang, S.; Zhang, Q.; Lian, X.M. Transcriptomic analysis of rice responses to low phosphorus stress. Chin. Sci. Bull. 2010, 55, 251–258. [Google Scholar] [CrossRef]
- Wissuwa, M.; Yano, M.; Ae, N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 777–783. [Google Scholar] [CrossRef]
- Wissuwa, M.; Ae, N. Further characterization of two QTLs that increase phosphorus uptake of rice (Oryza sativa L.) under phosphorus deficiency. Plant Soil. 2001, 237, 275–286. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, W.; Shi, Q.; Liang, Y.; Chen, F.; He, H.; Liang, K. Screening methodology for rice (Oryza sativa) genotypes with high phosphorus use efficiency at their seedling stage. J. Appl. Ecol. 2002, 13, 1587–1591. [Google Scholar]
- Yuan, H.; Liu, D. Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 2008, 50, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.H.; Lu, X.; Haefele, S.M.; Gamuyao, R.; Ismail, A.; Wissuwa, M.; Heuer, S. Development and application of gene-based markers for the major rice QTL Phosphate uptake 1. Theor. Appl. Genet. 2010, 120, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Ramaekers, L.; Remans, R.; Rao, I.M.; Blair, M.W.; Vanderleyden, J. Strategies for improving phosphorus acquisition efficiency of crop plants. Field Crops Res. 2010, 117, 169–176. [Google Scholar] [CrossRef]
- Wissuwa, M.; Mazzola, M.; Picard, C. Novel approaches in plant breeding for rhizosphere-related traits. Plant Soil 2009, 321, 409–430. [Google Scholar] [CrossRef]
- Chen, L.; Lin, L.; Cai, G.; Sun, Y.; Huang, T.; Wang, K.; Deng, J. Identification of Nitrogen, Phosphorus, and Potassium Deficiencies in Rice Based on Static Scanning Technology and Hierarchical Identification Method. PLoS ONE 2014, 9, e113200. [Google Scholar] [CrossRef] [PubMed]
- Place, G.A.; Sims, J.L.; Hall, U.L. Effects of nitrogen and phosphorous on the growth yield and cooking characteristics of rice. Agron. J. 1970, 62, 239–241. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Pham, C.V.; Bertin, P. The effect of nitrogen concentration on nitrogen use efficiency and related parameters in cultivated rices (Oryza sativa L. subsp. indica and japonica and O. glaberrima Steud.) in hydroponics. Euphytica 2014, 198, 137–151. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Naylor, R.; Crews, T.; David, M.B.; Drinkwater, L.E.; Holland, E.; Johnes, P.J.; Katzenberger, J.; Martinelli, L.A.; Matson, P.A.; et al. Agriculture. Nutrient imbalances in agricultural development. Science 2009, 324, 1519–1520. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.F.; Boumans, L.J.M.; Batjes, N.H. Emissions of N2O and NO from fertilised fields: Summary of available measurement data. Glob. Biogeochem. Cycles 2002, 16, 6-1–6-13. [Google Scholar] [CrossRef]
- Samborski, S.; Kozak, M.; Azevedo, R.A. Does nitrogen uptake affect nitrogen uptake efficiency or vice versa? Acta Physiol. Plant. 2008, 30, 419–420. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Ren, B.; Shen, Q.; Guo, S. Why nitrogen use efficiency decreases under high nitrogen supply in rice (Oryza sativa L.) seedlings. J. Plant Growth Regul. 2012, 31, 47–52. [Google Scholar] [CrossRef]
- Singh, H.; Verma, A.; Ansari, M.A.; Shukla, A. Physiological response of rice (Oryza sativa L.) genotypes to elevated nitrogen applied under field conditions. Plant Signal. Behav. 2015, 9, e29015. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, I. Effect of Nitrogen Fertilizers on Growth, Yield and Quality of Hybrid Rice (Oryza sativa L.). J. Cent. Eur. Agric. 2005, 6, 611–618. [Google Scholar]
- Manzoor, Z.; Awan, T.H.; Zahid, M.A.; Faiz, F.A. Response of rice crop (Super Basmati) to different nitrogen levels. J. Anim. Plant Sci. 2006, 16, 1–2. [Google Scholar]
- Swamy, K.N.; Kondamudi, R.; Kiran, T.V.; Vijayalakshmi, P.; Rao, Y.V.; Rao, P.R.; Subrahmanyam, D.; Voleti, S.R. Screening for nitrogen use efficiency with their root characteristics in rice (Oryza spp.) genotypes. Ann. Biol. Sci. 2015, 3, 8–11. [Google Scholar]
- Haque, M.A.; Haque, M.M. Growth, Yield and Nitrogen Use Efficiency of New Rice Variety under Variable Nitrogen Rates. Am. J. Plant Sci. 2016, 7, 612–622. [Google Scholar] [CrossRef]
- Kumagai, E.; Araki, T.; Kubota, F. Effects of nitrogen supply restriction on gas exchange and photosystem 2 function in flag leaves of a traditional low-yield cultivar and a recently improved high-yield cultivar of rice (Oryza sativa L.). Photosynthetica 2007, 45, 489–495. [Google Scholar] [CrossRef]
- Maske, N.S.; Borkar, S.L.; Rajgire, H.J. Effects of Nitrogen Levels on Growth, Yield and Grain Quality of Rice. J. Soil Crop 1997, 7, 83–86. [Google Scholar]
- Peng, S.; Cassman, K.G.; Virmani, S.S.; Sheehy, J.; Khush, G.S. Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci. 1999, 39, 1552–1559. [Google Scholar] [CrossRef]
- Lawlor, D.W. Carbon and nitrogen assimilation in relation to yield: Mechanisms are the key to understanding production systems. J. Exp. Bot. 2002, 53, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Peng, S.; Zhang, Z.; Wang, Z.; Visperas, R.M.; Zhu, Q. Grain and dry matter yields and portioning of assimilates in Japonica/Indica hybrid rice. Crop Sci. 2002, 42, 766–772. [Google Scholar] [CrossRef]
- Ahmed, M.; Islam, M.M.; Paul, S.K.; Khulna, B. Effect of Nitrogen on Yield and Other Plant Characters of Local, T. Aman Rice Var. Jatai. Res. J. Agric. Biol. Sci. 2005, 1, 158–161. [Google Scholar]
- Hamaoka, N.; Uchida, Y.; Tomita, M.; Kumagai, E.; Araki, T.; Ueno, O. Genetic variations in dry matter production, nitrogen uptake, and nitrogen use efficiency in the AA genome Oryza species grown under different nitrogen conditions. Plant Prod. Sci. 2013, 16, 107–116. [Google Scholar] [CrossRef]
- Yogendra, N.D.; Kumara, B.H.; Chandrashekar, N.; Prakash, N.B.; Anantha, M.S.; Shashidhar, H.E. Real-time nitrogen management in aerobic rice by adopting leaf color chart (LCC) as influenced by silicon. J. Plant Nutr. 2017, 40, 1277–1286. [Google Scholar] [CrossRef]
- Mandal, N.N.; Chaudhry, P.P.; Sinha, D. Nitrogen, phosphorus and potash uptake of wheat (var. Sonalika). Environ. Ecol. 1992, 10, 297. [Google Scholar]
- Wang, Y.; Wu, W.H. Genetic approaches for improvement of the crop potassium acquisition and utilization efficiency. Curr. Opin. Plant Biol. 2015, 25, 46–52. [Google Scholar] [CrossRef] [PubMed]
- FAO. Current World Fertilizer Trends and Outlook to 2016; Food and Agriculture Organization of the United Nations: Rome, Italy, 2012. [Google Scholar]
- Dobermann, A.; Cassman, K.G.; Mamaril, C.P.; Sheehy, J.E. Management of phosphorus, potassium and sulfur in intensive irrigated lowland rice. Field Crops Res. 1998, 56, 113–138. [Google Scholar] [CrossRef]
- Xiaoe, Y.; Romheld, V.; Marschner, H.; Baligar, V.C.; Martens, D.C. Shoot photosynthesis and root growth of hybrid and conventional rice cultivars as affected by N and K levels in the root zone. Pedosphere 1997, 7, 35–42. [Google Scholar]
- Epstein, E.; Bloom, A.J. Mineral Nutrition of Plants: Principles and Perspectives, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2005. [Google Scholar]
- Fageria, N.K.; Dos Santos, A.B.; Moreira, A.; Moraes, M.F. Potassium soil test calibration for lowland rice on an inceptisol. Commun. Soil Sci. Plant Anal. 2010, 41, 2595–2601. [Google Scholar] [CrossRef]
- Grzebisz, W.; Gransee, A.; Szczepaniak, W. The effects of potassium fertilization on water-use efficiency in crop plants. J. Pant Nutr. Soil Sci. 2013, 176, 355–374. [Google Scholar] [CrossRef] [Green Version]
- Mehdi, S.M.; Sarfraz, M.; Hafeez, M. Response of rice advanced line PB-95 to potassium in saline sodic soil. Pak. J. Biol. Sci. 2007, 10, 2938–2939. [Google Scholar]
- Fageria, N.K.; Dos Santos, A.B.; De Moraes, M.F. Yield, Potassium Uptake, and Use Efficiency in Upland Rice Genotypes. Commun. Soil Sci. Plant Anal. 2010, 41, 2676–2684. [Google Scholar] [CrossRef]
- Arif, M.; Arshad, M.; Asghar, H.N.; Basara, S.M.A. Response of rice (Oryza sativa) genotypes varying in K use efficiency to various levels of potassium. Int. J. Agric. Biol. 2010, 12, 926–930. [Google Scholar]
- Kalita, U.; Ojha, N.J.; Talukdar, M.C. Effect of levels and time of potassium application on yield and yield attributes of upland rice. J. Potassium Res. 1993, 11, 203–206. [Google Scholar]
- Dunn, D.; Stevens, G. Rice potassium nutrition research progress (Missouri). Better Crops 2005, 89, 15–17. [Google Scholar]
- Awan, T.H.; Manzoor, Z.; Safdar, M.E.; Ahmad, M. Yield response of rice to dynamic use of potassium in traditional rice growing area of Punjab. Pak. J. Agric. Sci. 2007, 44, 130–135. [Google Scholar]
- Bahmaniar, M.A.; Ranjbar, G.A. Effects of nitrogen and potassium fertilizers on rice (Oryza sativa L.) genotypes processing characteristics. Pak. J. Biol. Sci. 2007, 10, 1829–1834. [Google Scholar] [PubMed]
- Sarkar, R.K.; Malik, G.C. Effect of foliar spray of KNO3 and Ca (NO3)2 on grass pea (Lathyrus sativus L.) grown in rice fallows. Lathyrus Lathyrism Newslett. 2001, 2, 47–48. [Google Scholar]
- Islam, A.; Saha, P.K.; Biswas, J.C.; Saleque, M.A. Potassium Fertilization in Intensive Wetland Rice System: Yield, Potassium Use Efficiency and Soil Potassium Status. Int. J. Agric. Pap. 2016, 1, 7–21. [Google Scholar]
- De, D.S.K.; Broadbent, F.E. Development changes related to nitrogen-use efficiency in rice. Field Crops Res. 1993, 34, 47–56. [Google Scholar]
- William, R.R.; Johnson, G.V. Improving nitrogen use efficiency for cereal production. Agron. J. 1999, 91, 357–363. [Google Scholar]
- Gregard, A.; Gelanger, G.; Michaud, R. Nitrogen use efficiency and morphological characteristics of timothy populations selected for low and high forage nitrogen concentrations. Crop Sci. 2000, 40, 422–429. [Google Scholar] [CrossRef]
- Anil, K.; Nidhi, G.; Atul, K.G.; Vikram, S.G. Identification of Biomarker for Determining Genotypic Potential of Nitrogen-Use-Efficiency and Optimization of the Nitrogen Inputs in Crop Plants. J. Crop Sci. Biotechnol. 2009, 12, 183–194. [Google Scholar]
- Pingali, P.L. Green revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef] [PubMed]
- Beatty, P.H.; Shrawat, A.K.; Carroll, R.T.; Zhu, T.; Good, A.G. Transcriptome analysis of nitrogen-efficient rice over-expressing alanine aminotransferase. Plant Biotechnol. J. 2009, 7, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Bi, Y.; Rothstein, S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Kabir, G. Genetic approaches of increasing nutrient use efficiency especially nitrogen in cereal crops—A review. J. Bio-Sci. 2014, 22, 111–125. [Google Scholar] [CrossRef]
- Rose, T.J.; Kretzschmar, T.; Waters, D.L.E.; Balindong, J.L.; Wissuwa, M. Prospects for Genetic Improvement in Internal Nitrogen Use Efficiency in Rice. Agronomy 2017, 7, 70. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, Y.; Tang, D.; Wang, J.; Zhong, J.; Wang, Y.; Yuan, Q.; Yu, X.; Zhang, Y. Identification of QTL Associated with Nitrogen Uptake and Nitrogen Use Efficiency Using High Throughput Genotyped CSSLs in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1166. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Sevanthi, V.A.M.; Chaudhary, S.; Tyagi, P.; Venkadesan, S.; Rani, M.; Mandal, P.K. Transcriptome Analysis of Two Rice Varieties Contrasting for Nitrogen Use Efficiency under Chronic N Starvation Reveals Differences in Chloroplast and Starch Metabolism-Related Genes. Genes (Basel) 2018, 11, E206. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.H.; Zhang, Y.L.; Ye, L.T.; Fan, X.R.; Xu, G.H.; Shen, Q.R. Responses of Rice Cultivars with Different Nitrogen Use Efficiency to Partial Nitrate Nutrition. Ann. Bot. 2007, 99, 1153–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Xie, D.; Chen, J.; Lu, H.; Xu, Y.; Ma, C.; Xu, G. Over-expression of OsPTR6 in rice increased plant growth at different nitrogen supplies but decreased nitrogen use efficiency at high ammonium supply. Plant Sci. 2014, 227, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; Qian, Q.; Li, J. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5,a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shomura, A.; Sasaki, T.; Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 2001, 98, 7922–7927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under shortday conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Oikawa, T.; Sugiyama, N.; Tanisaka, T.; Yano, M.; Shimamoto, K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002, 16, 2006–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Ookawa, T.; Hobo, T.; Yano, M.; Murata, K.; Ando, T.; Miura, H.; Asano, K.; Ochiai, Y.; Ikeda, M.; Nishitani, R.; et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010, 1, 132. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; Yano, M. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Hattori, U.Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; Matsuoka, M. The ethylene response factors SNOKEL1 and SNOKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Koyama, E.; Kono, I.; Ando, T.; Yano, M.; Ma, J.F. Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in rice. Plant Cell Physiol. 2009, 50, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhou, A.; Sang, T. Rice domestication by reducing shattering. Science 2006, 311, 1936–1939. [Google Scholar] [CrossRef] [PubMed]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; Sun, C. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Romero, L.E.; Lozano, I.; Garavito, A.; Carabali, S.J.; Triana, M.; Villareal, N.; Reyes, L.; Duque, M.C.; Martinez, C.P. Major QTLs Control Resistance to Rice Hoja Blanca Virus and Its Vector Tagosodes orizicolus. G3 (Bethesda) 2014, 4, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Yamamoto, E.; Kanno, N.; Kawai, S.; Mizubayashi, T.; Fukuoka, S. A major QTL controlling deep rooting on rice chromosome 4. Sci. Rep. 2013, 3, 3040. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.J.; Wu, P.; Senadhira, D.; Huang, N. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 1361–1369. [Google Scholar] [CrossRef]
- Wissuwa, M.; Wegner, J.; Ae, N.; Yano, M. Substitution mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet. 2002, 105, 890–897. [Google Scholar] [PubMed]
- Shimizu, A.; Yanagihara, S.; Kawasaki, S.; Ikehashi, H. Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor. Appl. Genet. 2004, 109, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Lang, N.T.; Buu, B.C. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Omonrice 2006, 14, 1–9. [Google Scholar]
- Shimizu, A.; Kato, K.; Komatsu, A.; Motomura, K.; Ikehashi, H. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.): Fine QTL mapping and multivariate analysis of related traits. Theor. Appl. Genet. 2008, 117, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Chao, X.; Jie, R.; Xiu-qin, Z.; Zai-song, D.; Jing, Z.; Chao, W.; Jun-wei, Z.; Joseph, C.A.; Qiang, Z.; et al. Genetic Dissection of Low Phosphorus Tolerance Related Traits Using Selected Introgression Lines in Rice. Rice Sci. 2015, 22, 264–274. [Google Scholar] [CrossRef]
- Wang, K.; Cui, K.; Liu, G.; Xie, W.; Yu, H.; Pan, J.; Huang, J.; Nie, L.; Shah, F. Identification of quantitative trait loci for phosphorus use efficiency traits in rice using a high density SNP map. BMC Genet. 2014, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Xianwu, Z.; Guohua, M.; He, P.; Zhu, L.; Zhang, F. Identification of quantitative trait loci affecting tolerance to low phosphorus in rice (Oryza sativa L.). Chin. Sci. Bull. 2000, 45, 519–525. [Google Scholar]
- Fang, P.; Wu, P. QTL x N-level interaction for plant height in rice (Oriza sativa L.). Plant Soil 2001, 236, 237–242. [Google Scholar] [CrossRef]
- Ishimaru, K.; Kobayashi, N.; Ono, K.; Yano, M.; Ohsugi, R. Are contents of Rubisco, soluble protein and nitrogen in flag leaves of rice controlled by the same genetics? J. Exp. Bot. 2001, 52, 1827–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, S.; Valencia, M.O.; Lorieux, M.; Arbelaez, J.D.; McCouch, M.; Ishitani, M.; Selvaraj, M.G. Identification of QTLs associated with agronomic performance under nitrogen-deficient conditions using chromosome segment substitution lines of a wild rice relative, Oryza rufipogon. Acta Physiol. Plant. 2016, 38, 103. [Google Scholar] [CrossRef]
- Obara, M.; Kajiura, M.; Fukuta, Y.; Yano, M.; Hayashi, M.; Yamaya, T.; Sato, T. Mapping of QTLs associated with cytosolic glutamine synthetase and NADH- glutamate synthase in rice (Oryza sativa L.). J. Exp. Bot. 2001, 52, 1209–1217. [Google Scholar] [PubMed]
- Senthilvel, S.; Govindaraj, P.; Arumugachamy, S.; Latha, R.; Malarvizhi, P.; Gopalan, A.; Maheswaran, M. Mapping genetic loci associated with nitrogen use efficiency in rice (Oryza sativa L.). In Proceedings of the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004. [Google Scholar]
- Tong, H.H.; Mei, H.W.; Yu, X.Q.; Xu, X.Y.; Li, M.S.; Zhang, S.Q.; Luo, L.J. Identification of Related QTLs at Late Developmental Stage in Rice (Oryza sativa L.) Under Two Nitrogen Levels. Acta Genet. Sin. 2006, 33, 458–467. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.J.; Chen, D.Y.; Yu, S.B. Analysis of Quantitative Trait Loci in Response to Nitrogen and Phosphorus Deficiency in Rice Using Chromosomal Segment Substitution Lines. Acta Agron. Sin. 2009, 35, 580–587. [Google Scholar] [CrossRef]
- Laza, M.R.; Kondo, M.; Ideta, O.; Barleen, E.; Imbe, T. Identification of quantitative trait loci for d13C and productivity in irrigated lowland rice. Crop Sci. 2006, 46, 763–773. [Google Scholar] [CrossRef]
- MacMillan, K.; Emrich, K.; Piepho, H.P.; Mullins, C.E.; Price, A.H. Assessing the importance of genotype × environment interaction for root traits in rice using a mapping population II: Conventional QTL analysis. Theor. Appl. Genet. 2006, 113, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.I.; Jiang, W.Z.; Chin, J.H.; Piao, Z.; Cho, Y.G.; McCouch, S.; Koh, H.J. Identification of QTLs associated with physiological nitrogen use efficiency in rice. Mol. Cell 2007, 23, 72–79. [Google Scholar]
- Senthilvel, S.; Vinod, K.K.; Malarvizhi, P.; Maheswaran, M. QTL and QTL × environment effects on agronomic and nitrogen acquisition traits in rice. J. Integr. Plant Biol. 2008, 50, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Piao, Z.; Li, M.; Li, P.; Zhang, C.; Wang, H.; Luo, Z.; Lee, J.; Yang, R. Bayesian dissection for genetic architecture of traits associated with nitrogen utilization efficiency in rice. Afr. J. Biotechnol. 2009, 8, 6834–6839. [Google Scholar]
- Srividya, A.; Vemireddy, L.R.; Hariprasad, A.S.; Jayaprada, M.; Sakile, S.; Puram, V.R.R.; Anuradha, G.; Siddiq, E.A. Identification and mapping of landrace derived QTL associated with yield and its components in rice under different nitrogen levels and environments. Int. J. Plant Breed. Genet. 2010, 4, 210–227. [Google Scholar] [CrossRef]
- Tong, H.H.; Chen, L.; Li, W.P.; Mei, H.; Xing, Y.; Yu, X.; Xu, X.; Zhang, S.; Luo, L. Identification and characterization of quantitative trait loci for grain yield and its components under different nitrogen fertilization levels in rice (Oryza sativa L.). Mol. Breed. 2011, 28, 495–509. [Google Scholar] [CrossRef]
- Yue, F.; Rong-rong, Z.; Ze-chuan, L.; Li-yong, C.; Xing-hua, W.; Shi-hua, C. Quantitative trait locus analysis for rice yield traits under two nitrogen levels. Rice Sci. 2015, 22, 108–115. [Google Scholar] [CrossRef]
- Wei, D.; Cui, K.; Ye, G.; Pan, J.; Xiang, J.; Huang, J.; Nie, L. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice. Plant Soil 2012, 359, 281–295. [Google Scholar] [CrossRef]
- Wu, P.; Ni, J.J.; Luo, A.C. QTLs underlying Rice Tolerance to Low-Potassium Stress in Rice Seedlings. Crop Sci. 1998, 38, 1458–1462. [Google Scholar] [CrossRef]
- Senaratne, R.; Ratnasinghe, D.S. Nitrogen fixation and beneficial effects of some grain legumes and green-manure crops on rice. Biol. Fer. Soils 1995, 19, 49–54. [Google Scholar] [CrossRef]
- Lin, X.Q.; Zhou, W.J.; Zhu, D.F.; Zhang, Y. Effect of water management on photosynthetic rate and water use efficiency of leaves in paddy rice. Chin. J. Rice Sci. 2004, 18, 333–338, (in Chinese with English abstract). [Google Scholar]
- Peng, S.; Cassman, K.G. Upper thresholds of nitrogen uptake rates and associated nitrogen fertilizer efficiencies in irrigated rice. Agron. J. 1998, 90, 178–185. [Google Scholar] [CrossRef]
- Yamaya, T.; Obara, M.; Nakajima, H.; Sasaki, S.; Hayakawa, T.; Sato, T. Genetic manipulation and quantitative trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 2002, 53, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Xing, Y.; Yan, H.; Xu, C.; Li, X.; Zhang, Q. QTLs for low nitrogen tolerance at seedling stage identified using a recombinant inbred line population derived from an elite rice hybrid. Theor. Appl. Genet. 2005, 112, 85–96. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Velk, P.L.G. The role of Azolla cover in improving the nitrogen use efficiency of lowland rice. Plant Soil 2004, 263, 311–321. [Google Scholar]
- Ladha, J.K.; Kirk, G.J.D.; Bennett, J.; Peng, S.; Reddy, C.K.; Reddy, P.M.; Singh, U. Opportunities for increased nitrogen use efficiency from improved lowland rice germplasm. Field Crops Res. 1998, 56, 41–71. [Google Scholar] [CrossRef]
- Obara, M.; Sato, T.; Sasaki, S.; Kashiba, K.; Nagano, A.; Nakamura, I.; Ebitani, T.; Yano, M.; Yamaya, T. Identification and characterization of a QTL on chromosome 2 for cytosolic glutamine synthetase content and panicle number in rice. Theor. Appl. Genet. 2004, 110, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.H.; Wang, Y.; Pan, X.B. Mapping of QTLs for nitrogen use efficiency and related traits in rice (Oryza sativa L.). Agric. Sci. China 2005, 4, 721–727. [Google Scholar]
- Dong, G.C.; Wang, Y.L.; Zhang, Y.F.; Chen, P.; Yang, L.; Huang, J.; Zuo, B. Characteristics of yield and yield components in conventional indica rice cultivars with different nitrogen use efficiencies for grain output. Acta Agron. Sin. 2006, 32, 1511–1518. [Google Scholar]
- Dong, G.C.; Wang, Y.; Yu, X.F. Differences of nitrogen uptake and utilization of conventional rice varieties with different growth duration. Sci. Agric. Sin. 2011, 44, 4570–4582. [Google Scholar]
- Sanchez, P.A.; Salinas, J.G. Low-input technology for managing oxisols and ultisols in tropical America. Adv. Agron. 1981, 34, 279–406. [Google Scholar]
- Dobermann, A.; Fairhurst, T. Rice: Nutrient Disorders & Nutrient Management; Potash & Phosphate Institute, Potash & Phosphate Institute of Canada, and International Rice Research Institute: Singapore; Los Baños, Philippines, 2000. [Google Scholar]
- Su, J.Y.; Xiao, Y.M.; Li, M.; Liu, Q.; Li, B.; Tong, Y.; Jia, J.; Li, Z. Mapping QTLs for phosphorus-deficiency tolerance at wheat seedling stage. Plant Soil 2006, 281, 25–36. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Z.C.; Mi, G.H.; Zhang, H.L.; Mu, P.; Wang, X. Screen and identification for tolerance to low-phosphorus stress of rice germplasm (Oryza sativa L.). Acta Agron. Sin. 2005, 31, 238–242, (in Chinese with English abstract). [Google Scholar]
- Ping, M.U.; Huang, C.; Li, J.X.; Liu, L.F.; Li, Z.C. Yield Trait Variation and QTL Mapping in a DH Population of Rice Under Phosphorus Deficiency. Acta Agron. Sin. 2008, 34, 1137–1142. [Google Scholar]
- Liu, L.J.; Chang, E.H.; Fan, M.M.; Wang, Z.Q.; Yang, J.C. Effects of Potassium and Calcium on Root Exudates and Grain Quality During Grain Filling. Acta Agron. Sin. 2011, 37, 661–669. [Google Scholar]
- Torkashv, M.; Vahed, S. The efficiency of potassium fertilization methods on the growth of rice (Oryza sativa L.) under salinity stress. Afr. J. Biotechnol. 2011, 10, 15946–15952. [Google Scholar]
- Ali, J.; Franje, N.J.; Revilleza, J.E.; Acero, B. Breeding for Low-Input Responsive Green Super Rice (GSR) Varieties for Rainfed Lowlands of Asia and Africa. University Library; University of the Philippines at Los Baños: Los Baños, Philippines, 2016. [Google Scholar]
- Yorobe, J.M.; Ali, J.; Pede, V.; Rejesus, R.M.; Velarde, O.P.; Wang, W. Yield and income effects of rice varieties with tolerance of multiple abiotic stresses: The case of green super rice (GSR) and flooding in the Philippines. Agric. Econ. 2016, 47, 1–11. [Google Scholar] [CrossRef]
- Wu, L.; Yuan, S.; Huang, L.; Sun, F.; Zhu, G.; Li, G.; Fahad, S.; Peng, S.; Wang, F. Physiological Mechanisms Underlying the High-Grain Yield and High-Nitrogen Use Efficiency of Elite Rice Varieties under a Low Rate of Nitrogen Application in China. Front. Plant Sci. 2016, 7, 1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Peng, S. Yield potential and nitrogen use efficiency of China’s super rice. J. Integr. Agric. 2017, 16, 1000–1008. [Google Scholar] [CrossRef]
- Ali, J.; Xu, J.L.; Gao, Y.; Fontanilla, M.; Li, Z.K. Breeding for yield potential and enhanced productivity across different rice ecologies through green super rice (GSR) breeding strategy. In International Dialogue on Perception and Prospects of Designer Rice; Muralidharan, K., Siddiq, E.A., Eds.; Society for the Advancement of Rice Research, Directorate of Rice Research: Hyderabad, India, 2013; pp. 60–68. [Google Scholar]
- Mortvedt, J.J.; Murphy, L.S.; Follett, R.H. Fertilizer Technology and Application; Meister Publishing Co.: Willoughby, OH, USA, 2001. [Google Scholar]
- Yadav, R.L. Assessing on-farm efficiency and economics of fertilizer N., P and K in rice-wheat systems of India. Field Crops Res. 2003, 18, 39–51. [Google Scholar] [CrossRef]
- Cassman, K.G.; Gines, G.C.; Dizon, M.A.; Samson, M.I.; Alceantara, J.M. Nitrogen use efficiency in tropical lowland rice systems: Contributions from indigenous and applied nitrogen. Fields Crops Res. 1996, 47, 1–12. [Google Scholar] [CrossRef]
- Dobermann, A.R. Nitrogen Use Efficiency-State of the Art; Agronomy-Faculty Publications: Lincoln, NE, USA, 2005; p. 316. [Google Scholar]
- Wen-xia, X.; Guang-huo, W.; Qi-chun, Z.; Guo, H.C. Effects of nitrogen fertilization strategies on nitrogen use efficiency in physiology, recovery, and agronomy and redistribution of dry matter accumulation and nitrogen accumulation in two typical rice cultivars in Zhejiang. China J. Zhejiang Univ. Sci. B 2007, 8, 208–216. [Google Scholar]
- Yoshida, S. Fundamentals of Rice Crop Science; IRRI: Los Baños, Laguna, Philippines, 1981; 269p. [Google Scholar]
- Amanullah; Muhammad, A.; Almas, L.K.; Amanullaj, J.; Zahir, S.; Rahman, H.; Khalil, S.K. Agronomic Efficiency and Profitability of P-Fertilizers Applied at Different Planting Densities of Maize in Northwest. Pak. J. Plant Nutr. 2012, 35, 331–341. [Google Scholar] [CrossRef]
- Rao, T.N. Improving nutrient use efficiency: The role of beneficial management practices. In Better Crops-India; IPNI–India Program 133: Gurgaon, India, 2007; Volume 1, pp. 6–7. [Google Scholar]
- Singh, D.P. Vermiculture biotechnology and biocomposting. In Environmental Microbiology and Biotechnology; Singh, D.P., Dwivedi, S.K., Eds.; New Age International (P) Limited Publishers: Darya Ganj, New Delhi, 2004; pp. 97–112. [Google Scholar]


| S. No. | Traits | Name of QTL | Encoded Protein | Nature of Allele Suitable for Use in Breeding Programs | References |
|---|---|---|---|---|---|
| 1 | Grain number | Gn1a | Cytokinin oxidase | Low expression | [122] |
| 2 | Grain number and strong culm | dep1 | PEBP-like domain protein | Loss of function | [123] |
| 3 | Grain number | WFP | OsSPL14 | High expression | [124] |
| 4 | Grain number, low tiller number, and strong culm | Ipa | OsSPL14 | High and ectopic expression | [125] |
| 5 | Grain size | gs3 | Transmembrane protein | Loss of function | [126] |
| 6 | Grain size and filling | gw2 | RING-type ubiquitin E3 ligase | Loss of function | [127] |
| 7 | Grain size | qSW5/GW5 | Unknown | Loss of function | [128] |
| 8 | Grain filling | GIF1 | Cell wall invertase | Restricted expression in the ovular vascular trace | [129] |
| 9 | Heading date | Hd1 | CONSTANS-like protein | Loss-of-function allele leads to late heading | [130] |
| 10 | Heading date | Hd6 | Subunit of protein kinase | Loss–of-function allele leads to early heading | [131] |
| 11 | Heading date | Hd3a | FT-like | Low expression leads to late heading | [132,133,134] |
| 12 | Heading date | Ehd1 | B-type response regulator | Loss-of-function allele leads to late heading | [135] |
| 13 | Grain number, plant height and heading date | Ghd7 | CCT domain protein | Functional allele | [136] |
| 14 | Days to heading | DTH8 | CCT domain protein | Functional allele | [137] |
| 15 | Plant height | sd1 | Gibberellin 20 oxidase | Loss of function | [138] |
| 16 | Lodging resistance | SCM2 | F-box protein | High expression | [139] |
| 17 | Disease resistance | pi21 | Proline-rich protein | Loss of function | [140] |
| 18 | Disease resistance | Pb1 | CC-NBS-LRR protein | Functional allele | [141] |
| 19 | Salt tolerance | SKC1 | HKT-type transporter | Gain of function | [142] |
| 20 | Cold tolerance | qLTG3-1 | GRP and LTP domain | Functional allele | [143] |
| 21 | Submerge tolerance | Sub1A | ERF-related factor | Gain of function | [144] |
| 22 | Internode elongation under submergence conditions | SK2 | ERF-related factor | Gain of function | [145] |
| 23 | Cadmium accumulation | OsHMA3 | Putative heavy metal transporter | Functional allele | [146] |
| 24 | Seed shattering | sh4 | Myb3 transcription factor | Loss of function | [147] |
| 25 | Seed shattering | qSH1 | BEL1-like homeobox protein | Low expression in abscission layer between panicle and spikelet | [148] |
| 26 | Prostrate growth | PROG1 | Zinc finger transcription factor | Loss of function | [149,150] |
| 27 | Disease resistance | RHBV | NS3 protein | Favorable gene or QTL alleles | [151] |
| 28 | Phosphorus uptake | Pup1 | OsPupK46-2 | High expression | [57] |
| 29 | Deep rooting | DRO1 | Auxin signaling pathway | Functional allele | [152] |
| Entry | Phosphorus | |||||
|---|---|---|---|---|---|---|
| S. No. | Traits | Population | Cross | No. of QTLs | Reference | |
| M | E | |||||
| 1 | Phosphorus uptake, plant dry weight, tiller number; phosphorus use efficiency | NILs | Nipponbare/Kasalath | 8 | - | [65] |
| 2 | Relative tillering ability, relative shoot dry weight, relative root dry weight | RILs | IR20/IR55178 | 4 | - | [153] |
| 3 | Phosphorus uptake, tiller number | NIL | Nipponbare/Kasalath | 1 (Pup) | - | [154] |
| 4 | Root elongation, shoot dry weight, relative phosphorus content, relative Fe content | F8 | Gimbozu/Kasalath | 6 | - | [155] |
| 5 | Relative root length, relative shoot length, relative shoot dry weight, relative root dry weight | BILs | OM2395/AS996 | 1 | - | [156] |
| 6 | Root elongation under phosphorus deficiency | CSSLs | Nipponbare/Kasalath CSSL29 | 1 | - | [157] |
| 7 | Plant height, maximum root length, root number, root volume, root fresh weight, root dry weight, shoot dry weight, total dry weight, root/shoot dry weight ratio | ILs | Yuefa/IRAT109 | 24 | 29 | [63] |
| 8 | Relative root length, relative root dry weight, relative shoot dry weight, relative total dry weight, relative root-shoot ratio of dry weight | BC2F4 | Shuhui 527/Minghui 86 | 48 | - | [158] |
| 9 | Total aboveground biomass, harvest index, P use efficiency for grain yield based on P accumulation in grains, P harvest index, P translocation, P translocation efficiency, P total aboveground P uptake, P use efficiency for biomass accumulation, P use efficiency for grain yield, P use efficiency for straw dry weight based on P accumulation in straw | RILs | Zhenshan 97/Minghui 63 | 36 | - | [159] |
| 10 | Root dry weight, relative shoot dry weight, relative total dry weight | DHs | ZYQ8/JX17 | 6 | - | [160] |
| Nitrogen | ||||||
| 1 | Plant height | DHs | IR64/Azucena | 10 | - | [161] |
| 2 | Rubisco, total leaf nitrogen, soluble protein content | BILs | Nipponbare/Kasalath | 15 | - | [162] |
| 3 | N uptake (NUP), grain yield, biomass yield, N use efficiency (NUE) | CSSLs | 9311/Nipponbare | 13 | [118] | |
| 4 | Toot system architecture, NDT, and morphological and physiological traits | CSSLs | Curinga/IRGC105491 | 13 | [163] | |
| 5 | Twelve physiological and agronomic traits | RILs | IR64/Azucena | 63 | [27] | |
| 6 | Glutamine synthetase, glutamate synthase | BILs | Nipponbare/Kasalath | 13 | - | [164] |
| 7 | Glutamine synthetase, panicle number per plant, panicle weight | NILs | Koshihikari/Kasalath | 1 | - | [164] |
| 8 | Total grain nitrogen, total shoot nitrogen, nitrogen uptake, nitrogen use efficiency, nitrogen translocation efficiency | F3 | Basmati370/ASD16 | 43 | - | [165] |
| 9 | Root dry weight, shoot dry weight, biomass | RILs | Zhenshan97/Minghui 63 | 52 | 103 | [166] |
| 10 | Plant height, panicle number per plant, chlorophyll content, shoot dry weight | CSSLs | Teqing/Lemont | 31 | - | [167] |
| 11 | Total grain number, total leaf nitrogen, total shoot nitrogen, nitrogen uptake, specific leaf nitrogen | RILs | IR69093-4-3-2/IR72 | 32 | - | [168] |
| 12 | Root length, root thickness, root biomass, biomass, etc. | RILs | Bala/Azucena | 17 | - | [169] |
| 13 | Relative root dry weight, spikelet number per panicle, spikelet fertility, 1000-grain weight | ILs | Shuhui 527 × Minghui 86 | 48 | [170] | |
| 14 | Total grain number, total leaf nitrogen, total shoot nitrogen, physiological nitrogen-use efficiency, biomass | RILs | Dasanbyeo/TR22183 | 20 | 58 | [170] |
| 15 | Total plant nitrogen, nitrogen-use efficiency | DHs | IR64/Azucena | 16 | - | [171] |
| 16 | Total plant nitrogen, nitrogen dry matter production efficiency, nitrogen grain production efficiency, total grain number | RIL | Dasanbyeo/TR22183 | 28 | 23 | [172] |
| 17 | Grain yield per plant, biomass, harvest index, etc. | RILs | IR64/INRC10192 | 46 | - | [173] |
| 18 | Plant height, root dry weight, shoot dry weight, chlorophyll content, root length, biomass | RILs | R9308/Xieqingzao B | 7 | - | [161] |
| 19 | Grain yield per plant, grain number per panicle | RILs | Zhenshan 97/HR5 | 19 | 11 | [174] |
| 20 | Number of panicles per plant, number of spikelets per panicle, number of filled grains per panicle, grain density per panicle | RILs | Xieqingzao B/Zhonghui 9308 | 52 | - | [175] |
| 21 | Nitrogen deficiency tolerance and nitrogen-use efficiency | RILs | Zhenshan 97 and Minghui 63 | 12 | [176] | |
| Potassium | ||||||
| 1 | Plant height, tiller number, shoot and root oven-dry weight | DHs | IR64/Azucena. | 4 | - | [177] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, J.; Jewel, Z.A.; Mahender, A.; Anandan, A.; Hernandez, J.; Li, Z. Molecular Genetics and Breeding for Nutrient Use Efficiency in Rice. Int. J. Mol. Sci. 2018, 19, 1762. https://doi.org/10.3390/ijms19061762
Ali J, Jewel ZA, Mahender A, Anandan A, Hernandez J, Li Z. Molecular Genetics and Breeding for Nutrient Use Efficiency in Rice. International Journal of Molecular Sciences. 2018; 19(6):1762. https://doi.org/10.3390/ijms19061762
Chicago/Turabian StyleAli, Jauhar, Zilhas Ahmed Jewel, Anumalla Mahender, Annamalai Anandan, Jose Hernandez, and Zhikang Li. 2018. "Molecular Genetics and Breeding for Nutrient Use Efficiency in Rice" International Journal of Molecular Sciences 19, no. 6: 1762. https://doi.org/10.3390/ijms19061762