A Functional Mutation in KIAA1462 Promoter Decreases Glucocorticoid Receptor Affinity and Affects Egg-Laying Performance in Yangzhou Geese
Abstract
:1. Introduction
2. Results
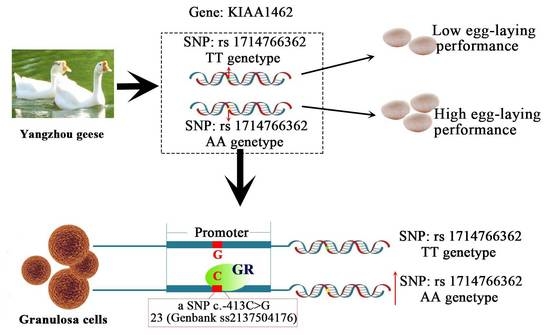
2.1. SNP (Single Nucleotide Polymorphisms) Genotyping and Association Analysis
2.2. Sequence Characterization and Phylogenetic Relationships among Species of Yangzhou Geese KIAA1462 Gene
2.3. KIAA1462 mRNA Expression Profile in Yangzhou Geese Tissues
2.4. KIAA1462 mRNA Level Differs in Individuals with Different Genotypes
2.5. The mRNA Level of Caspase-3 Is Negative Regulated by KIAA1462
2.6. Direct Sequencing of the 5′ Flanking Region of KIAA1462
2.7. The c.-413C>G Mutation Causes Allele-Specific Binding of GR
2.8. GR mRNA Expression Profile in Yangzhou Geese Tissues
2.9. Glucocorticoid Promotes the Transcription Activity of the C But Not of the G Allele
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals and Samples Preparation
4.3. Genotyping and Association Analysis
4.4. RNA Isolation and First-Strand cDNA Synthesis
4.5. Cloning of KIAA1462 Gene Coding Sequence in Yangzhou Geese
4.6. Construction of pEGFP-KIAA1462-N1 Expression Vector
4.7. Quantitative Real-Time PCR Analysis
4.8. Granulosa Cell Culture and KIAA1462 Gene Overexpression
4.9. siRNA Preparation and Transfection
4.10. Mutation Detection in KIAA1462 Promoter Region
4.11. Electrophoretic Mobility Shift Assay (EMSA)
4.12. Dual Luciferase Reporter Assay
4.13. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | Adenosine 5′-monophosphate (AMP)-activated protein kinase |
| AS-PCR | Allele-Specific PCR |
| CDS | Coding Sequence |
| DMEM | Dulbecco’s modified Eagle’s medium |
| EMSA | Electrophoretic Mobility Shift Assay |
| GR | Glucocorticoid Receptor |
| HFD | High-Fat Diet |
| JCAD | Junctional Protein Associated with Coronary Artery Disease |
| ORF | Open Reading Frame |
| Q-PCR | Quantitative Real Time Polymerase Chain Reaction |
| RAD sequencing | Restriction Site-Associated Sequencing |
| SNP | Single-Nucleotide Polymorphism |
| TOR | Target of Rapamycin |
References
- Qin, Q.; Sun, A.; Guo, R.; Lei, M.; Ying, S.; Shi, Z. The characteristics of oviposition and hormonal and gene regulation of ovarian follicle development in Magang geese. Reprod. Biol. Endocrinol. 2013, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, H.; Pan, Z.; Xia, L.; Dong, X.; Li, L.; Xu, F.; He, H.; Wang, J. Molecular cloning, expression profile and transcriptional modulation of two splice variants of very low density lipoprotein receptor during ovarian follicle development in geese (Anser cygnoide). Anim. Reprod. Sci. 2014, 149, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Akashi, M.; Higashi, T.; Masuda, S.; Komori, T.; Furuse, M. A coronary artery disease-associated gene product, JCAD/KIAA1462, is a novel component of endothelial cell-cell junctions. Biochem. Biophys. Res. Commun. 2011, 413, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Chu, W.; Zhang, L.; Han, H.; Zhao, R.; Wu, W.; Zhu, J.; Dodson, M.V.; Wei, W.; Liu, H.; et al. Identification of laying-related SNP markers in geese using RAD sequencing. PLoS ONE 2015, 10, e0131572. [Google Scholar] [CrossRef] [PubMed]
- Murdock, D.G.; Bradford, Y.; Schnetz-Boutaud, N.; Mayo, P.; Allen, M.J.; D’Aoust, L.N.; Liang, X.; Mitchell, S.L.; Zuchner, S.; Small, G.W.; Gilbert, J.R.; et al. KIAA1462, a coronary artery disease associated gene, is a candidate gene for late onset Alzheimer disease in APOE carriers. PLoS ONE 2013, 8, e82194. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.; Bois, P.R.; Feingold, E.; Sherman, S.L.; Cheung, V.G. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009, 5, e1000648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, J.; Luo, B.; Peri, S.; Wirchansky, B.; Hughes, L.; Forsythe, C.; Wu, H. Whole exome sequence analysis of serous borderline tumors of the ovary. Gynecol. Oncol. 2013, 130, 560–564. [Google Scholar] [CrossRef] [PubMed]
- El-lethey, H.; Jungi, T.W.; Huber-Eicher, B. Effects of feeding corticosterone and housing conditions on feather pecking in laying hens (Gallus gallus domesticus). Physiol. Behav. 2001, 73, 243–251. [Google Scholar] [CrossRef]
- Williams, J.B.; Etches, R.J.; Rzasa, J. Induction of a pause in laying by corticosterone infusion or dietary alterations: Effects on the reproductive system, food consumption and body weight. Br. Poult. Sci. 1985, 26, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, X.; Jiao, H.; Zhao, J.; Lin, H. Glucocorticoids inhibited hypothalamic target of rapamycin in high fat diet-fed chicks. Poult. Sci. 2015, 94, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Song, Z.; Jiao, H.; Lin, H. Glucocorticoids increase NPY gene expression via hypothalamic AMPK signaling in broiler chicks. Endocrinology 2014, 155, 2190–2198. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, G.S.; Al-Harbi, S.; Almasan, A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol. Biol. 2015, 1219, 1–9. [Google Scholar] [PubMed]
- Hrabia, A.; Leśniak-Walentyn, A.; Sechman, A.; Gertler, A. Chicken oviduct-the target tissue for growth hormone action: Effect on cell proliferation and apoptosis and on the gene expression of some oviduct-specific proteins. Cell Tissue Res. 2014, 357, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Jiang, Z.; Jiang, S.; Li, L.; Lin, X.; Gou, Z.; Fan, Q. Dietary vitamin A supplementation improved reproductive performance by regulating ovarian expression of hormone receptors, caspase-3 and Fas in broiler breeders. Poult. Sci. 2016, 95, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Werner, T. Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol. Biol. 2000, 132, 337–349. [Google Scholar] [PubMed]
- Balkovetz, D.F. Opening Pandora’s box in the tight junction. J. Am. Soc. Nephrol. 2007, 18, 1624–1625. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, A.; Tschernutter, M.; Bainbridge, J.W.; Balaggan, K.S.; Mowat, F.; West, E.L.; Munro, P.M.; Thrasher, A.J.; Matter, K.; Balda, M.S.; et al. The tight junction associated signalling proteins ZO-1 and ZONAB regulate retinal pigment epithelium homeostasis in mice. PLoS ONE 2010, 5, e15730. [Google Scholar] [CrossRef] [PubMed]
- Flores, I.; Jones, D.R.; Merida, I. Changes in the balance between mitogenic and antimitogenic lipid second messengers during proliferation, cell arrest, and apoptosis in T-lymphocytes. FASEB J. 2000, 14, 1873–1875. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Tsutomi, Y.; Akahane, K.; Araki, T.; Miura, M. Resistance to Fas-mediated apoptosis: Activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 1998, 17, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Yakovlev, A.G.; Ota, K.; Wang, G.; Movsesyan, V.; Bao, W.L.; Yoshihara, K.; Faden, A.I. Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J. Neurosci. 2001, 21, 7439–7446. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; Woods, D.C. Dynamics of avian ovarian follicle development: Cellular mechanisms of granulosa cell differentiation. Gen. Comp. Endocrinol. 2009, 163, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.A. Follicle selection in the avian ovary. Reprod. Domest. Anim. 2012, 47, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Anti-inflammatory actions of glucocorticoids: Molecular mechanisms. Clin. Sci. 1998, 94, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Tsurufuji, S.; Sugio, K.; Takemasa, F. The role of glucocorticoid receptor and gene expression in the anti-inflammatory action of dexamethasone. Nature 1979, 280, 408–410. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.D.; Schote, A.B.; Macedo, J.A.; Pelascini, L.P.; Muller, C.P. Tissue specific glucocorticoid receptor expression, a role for alternative first exon usage? Biochem. Pharmacol. 2006, 72, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Itani, O.A.; Liu, K.Z.; Cornish, K.L.; Campbell, J.R.; Thomas, C.P. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E971–E979. [Google Scholar] [CrossRef] [PubMed]
- McKay, L.I.; Cidlowski, J.A. Molecular control of immune/inflammatory responses: Interactions between nuclear factor-kappa B and steroid receptor-signaling pathways. Endocr. Rev. 1999, 20, 435–459. [Google Scholar] [PubMed]
- Gilbert, A.B.; Evans, A.J.; Perry, M.M.; Davidson, M.H. A method for separating the granulosa cells, the basal lamina and the theca of the preovulatory ovarian follicle of the domestic fowl (Gallus domesticus). J. Reprod. Fertil. 1977, 50, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Bustos, A.D.; Rubio, P.; Jouve, N. Molecular characterisation of the inactive allele of the gene Glu-A1 and the development of a set of AS-PCR markers for HMW glutenins of wheat. Theor. Appl. Genet. 2000, 100, 1085–1094. [Google Scholar] [CrossRef]
- Pauciullo, A.; Gallo, D.; Colimoro, L. Genotyping at the CSN1S1 locus by PCR-RFLP and AS-PCR in a Neapolitan goat population—Small Ruminant Research. Small Rumin. Res. 2008, 74, 84–90. [Google Scholar]
- Alsiddig, M.A.; Yu, S.G.; Pan, Z.X.; Widaa, H.; Badri, T.M.; Chen, J.; Liu, H.L. Association of single nucleotide polymorphism in melatonin receptor 1A gene with egg production traits in Yangzhou geese. Anim. Genet. 2017, 48, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Guo, J.R.; Yang, H.M.; Zhou, R.J.; Liu, J.X.; Li, S.Z.; Dong, C.Y. Differential expression profiling of ovarian genes in prelaying and laying geese. Poult. Sci. 2009, 88, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.S.; Barbosa, L.T.; Brito, C.; Fernandes, R.P.; Mann, R.S.; Pinto, A.P.; Oliveira, H.C.; Dodson, M.V.; Guimaraes, S.E.; Duarte, M.S. Identification of suitable reference genes for real time quantitative polymerase chain reaction assays on pectoralis major muscle in chicken (Gallus gallus). PLoS ONE 2015, 10, e0127935. [Google Scholar] [CrossRef] [PubMed]
- Schybli, M.; Sigrist, B.; Hess, M.; van Leerdam, B.; Hoop, R.K.; Vogtlin, A. Development of a new real-time polymerase chain reaction assay to detect Duck adenovirus A DNA and application to samples from Swiss poultry flocks. J. Vet. Diagn. Investig. 2014, 26, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.C.; Faller, D.V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991, 19, 2499. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.H.; Urabe, Y.; Kumar, V.; Tanikawa, C.; Koike, K.; Kato, N.; Miki, D.; Chayama, K.; Kubo, M.; Nakamura, Y.; et al. Identification of a functional variant in the MICA promoter which regulates MICA expression and increases HCV-related hepatocellular carcinoma risk. PLoS ONE 2013, 8, e61279. [Google Scholar] [CrossRef] [PubMed]
- Attarzadeh-Yazdi, G.; Shipston, M.J.; Antoni, F.A. Dex-ras1 and serum- and glucocorticoid-inducible protein kinase 1: Regulation of expression by dexamethasone in HEK293 cells. Neurochem. Res. 2008, 33, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Guney, S.; Schuler, A.; Ott, A.; Hoschele, S.; Zugel, S.; Baloglu, E.; Bartsch, P.; Mairbaurl, H. Dexamethasone prevents transport inhibition by hypoxia in rat lung and alveolar epithelial cells by stimulating activity and expression of Na+-K+-ATPase and epithelial Na+ channels. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 293, L1332–L1338. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Jin, L.; Fujii, A.; Furihata, T.; Nagahara, Y.; Chiba, K.; Hosokawa, M. Dexamethasone-mediated transcriptional regulation of rat carboxylesterase 2 gene. Xenobiotica 2012, 42, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.; Garside, H.; Ray, D. RU486, the glucocorticoid receptor (GR) antagonist, recruits NCoR, but not SRC-1: Explaining type II antagonism. Endocr. Abstr. 2002, 3, 40. [Google Scholar]
- Harger, J.W.; Dinman, J.D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 2003, 9, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Ikeda, M.; Ohmiya, Y.; Nakajima, Y. A dual-color luciferase assay system reveals circadian resetting of cultured fibroblasts by co-cultured adrenal glands. PLoS ONE 2012, 7, e37093. [Google Scholar] [CrossRef] [PubMed]







| Genotype | Number of Geese | Number of Eggs | Lower Mean | Upper Mean | p Value |
|---|---|---|---|---|---|
| AA | 170 | 79.60 ± 11.08 a | 77.94 | 81.47 | 0.04 |
| TA | 73 | 77.61 ± 11.11 a,b | 74.79 | 80.28 | |
| TT | 13 | 73.00 ± 9.61 b | 69.62 | 76.23 | |
| Total | 256 | 78.73 ± 11.61 | 77.43 | 80.11 |
| Species | Nucleotide Accession Number | Amino Acid Accession Number | Nucleotide | Amino Acid |
|---|---|---|---|---|
| Platyrhynchos (Mallard) | XM_005027879.2 | XP_005027936 | 88.73% | 86.91% |
| Gallus gallus (chicken) | XM_418578.5 | XP_418578.4 | 87.67% | 85.80% |
| Meleagris gallopavo (turkey) | XM_003206999.2 | XP_003207047.2 | 86.68% | 84.01% |
| Columba livia (Rock pigeon) | XM_005504208.1 | XP_005504265.1 | 83.74% | 80.19% |
| Canis lupus familiaris (Dog) | XM_535151.5 | XP_535151.2 | 52.60% | 40.56% |
| Homo sapiens (Human) | NM_020848.2 | NP_065899.1 | 55.01% | 40.68% |
| Ovis aries (Sheep) | XM_004014272.3 | XP_004014321.1 | 51.04% | 35.64% |
| Bos taurus (Cattle) | NM_001082474.1 | NP_001075943.1 | 50.68% | 36.66% |
| Sus scrofa (Pig) | XM_003130706.5 | XP_003130754.3 | 52.06% | 36.67% |
| Name | Sequences (5′→3′) | Function | Size (bp) | Tm (°C) |
|---|---|---|---|---|
| S1 | GCTGACAGCTCATTTGATA | AS-PCR | 67 | 58 |
| S2 | GCTGACAGCTCATTTGATT | AS-PCR | ||
| AS | CAGGATCACGTCCTCAAC | AS-PCR | ||
| P1-F | ATGTTCAGTGTCGAGGACCTCC | Partial cDNA sequences | 1305 | 58 |
| P1-R | AACAGAACGCAGGTAGTCA | |||
| P2-F | GACCGCCTGCGAATAGTGT | Partial cDNA sequences | 1424 | 56 |
| P2-R | CGTTTCCAACCTCCCACC | |||
| P3-F | CTGAAGCCCGTAAGTCG | Partial cDNA sequences | 1300 | 56 |
| P3-R | CTATTTGAGCGTCATTACGTGGG | |||
| P4-F | CGAATTCCATGTTCAGTGTCGAGGACCTCC | KIAA1462 expression vector | 4029 | 62 |
| P4-R | CCCCGGGGCTATTTGAGCGTCATTACGTGGG | |||
| P5-F | AGCATGAGGTGCGTGGAGATG | q-PCR | 200 | 60 |
| P5-R | CTCCAAACCCGAGTCTTGAACG | |||
| caspase-3-F | CTGGTATTGAGGCAGACAGTGG | q-PCR | 158 | 50 |
| caspase-3-R | CAGCACCCTACACAGAGACTGAA | |||
| GAPDH-F | GCTGATGCTCCCATGTTCGTGAT | q-PCR | 86 | 60 |
| GAPDH-R | GTGGTGCAAGAGGCATTGCTGAC | |||
| GR-F | CTCTGGGTGTCATTACGGTGTT | q-PCR | 230 | 62 |
| GR-R | CATTAGCTTGCTGGATTCCTTT | |||
| KIAA1462-siRNA-1200 | GCCUGCGAAUAGUGUUGAATT | KIAA1462 knockdown experiments | ||
| UUCAACACUAUUCGCAGGCTT | ||||
| KIAA1462-siRNA-1847 | CCCAGAGCCCUGAUAAGAATT | KIAA1462 knockdown experiments | ||
| UUCUUAUCAGGGCUCUGGGTT | ||||
| KIAA1462-siRNA-2180 | CCAAACUGCUGUCUCCAAATT | KIAA1462 knockdown experiments | ||
| UUUGGAGACAGCAGUUUGGTT | ||||
| P6-F | TGGTTGATGTCCTGCGGG | Partial promoter sequences | 2440 | 59 |
| P6-R | CCACCCTTACAATAAAGCACA | |||
| P7-S1 | TCCCAGAATACAGAGCACTCC | AS-PCR | 265 | 58 |
| P7-S2 | TCCCAGAATACAGAGCACTCG | AS-PCR | ||
| P7-AS | CCACCCTTACAATAAAGCACATC | AS-PCR | ||
| P8-F | GGGGTACCCCGGGAATCATTGAGACACGACA | Dual luciferase reporter vector | 630 | 64 |
| P8-R | CCCTCGAGGGGCTGCTGAAGTGAAGGGTTT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Wei, W.; Jiang, Z.; He, D.; Li, Z.; Yu, S.; Wang, Q.; Liu, H.; Chen, J. A Functional Mutation in KIAA1462 Promoter Decreases Glucocorticoid Receptor Affinity and Affects Egg-Laying Performance in Yangzhou Geese. Int. J. Mol. Sci. 2018, 19, 1531. https://doi.org/10.3390/ijms19051531
Xia M, Wei W, Jiang Z, He D, Li Z, Yu S, Wang Q, Liu H, Chen J. A Functional Mutation in KIAA1462 Promoter Decreases Glucocorticoid Receptor Affinity and Affects Egg-Laying Performance in Yangzhou Geese. International Journal of Molecular Sciences. 2018; 19(5):1531. https://doi.org/10.3390/ijms19051531
Chicago/Turabian StyleXia, Mengyuan, Wei Wei, Zaohang Jiang, Dandan He, Zhen Li, Shigang Yu, Qiushi Wang, Honglin Liu, and Jie Chen. 2018. "A Functional Mutation in KIAA1462 Promoter Decreases Glucocorticoid Receptor Affinity and Affects Egg-Laying Performance in Yangzhou Geese" International Journal of Molecular Sciences 19, no. 5: 1531. https://doi.org/10.3390/ijms19051531