Invasion-Related Factors as Potential Diagnostic and Therapeutic Targets in Oral Squamous Cell Carcinoma—A Review
Abstract
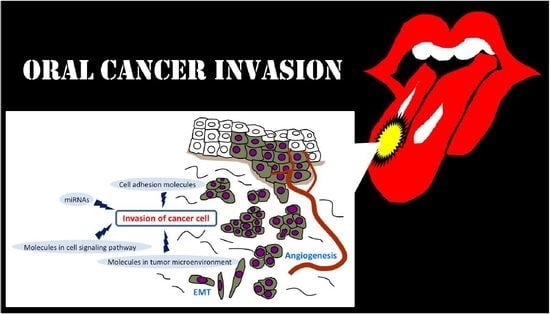
:1. Introduction
2. Invasion-Related Cell Adhesion Molecules
2.1. E-Cadherin
2.2. N-Cadherin
2.3. Claudin
2.4. DSG
3. Invasion-Related Molecules in Tumor Microenvironment (TME)
3.1. Matrix Metalloproteinases (MMPs)
3.2. Periostin
3.3. Hepatocyte Growth Factor (HGF)
3.4. Vascular Endothelial Growth Factor (VEGF)
3.5. Galanin (GAL)
4. Invasion-Related Molecules in Cell Signaling Pathway
4.1. Receptor Activator of Nuclear Factor-κB Ligand (RANKL/RANK)
4.2. Epidermal Growth Factor Receptor (EGFR)
4.3. Signal Transducer and Activator of Transcription (STAT)
4.4. Focal Adhesion Kinase (FAK)
4.5. EMT Related Signaling Pathways
5. Invasion-Related miRNAs
5.1. EMT-Related miRNAs
5.2. Invasion-Related Oncogenic miRNAs
5.3. Invasion-Related Tumor Suppressive miRNAs
6. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bryne, M.; Boysen, M.; Alfsen, C.G.; Abeler, V.M.; Sudbø, J.; Nesland, J.M.; Kristensen, G.B.; Piffko, J.; Bankfalvi, A. The invasive front of carcinomas: The most important area for tumor prognosis? Anticancer Res. 1998, 18, 4757–4764. [Google Scholar] [PubMed]
- Dissanayaka, W.L.; Pitiyage, G.; Kumarasiri, P.V.; Liyanage, R.L.; Dias, K.D.; Tilakaratne, W.M. Clinical and histopathologic parameters in survival of oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2012, 113, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Siriwardena, B.S.M.S.; Tilakaratne, A.; Amaratunga, E.A.P.D.; Udagama, M.N.G.P.K.; Ogawa, I.; Kudo, Y.; Takata, T.; Tilakaratne, W.M. Analysis of histopathological and immunohistochemical differences of oral squamous cell carcinoma in young and old patients in Sri Lanka. J. Oral Pathol. Med. 2007, 36, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Rajapakshe, R.M.; Pallegama, R.W.; Jayasooriya, P.R.; Siriwardena, B.S.; Attygalla, A.M.; Hewapathirana, S.; Weerasinghe, J.U.; Dias, D.K.; Tilakaratne, W.M. A retrospective analysis to determine factors contributing to the survival of patients with oral squamous cell carcinoma. Cancer Epidemiol. 2015, 39, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Sandu, K.; Nisa, L.; Monnier, P.; Simon, C.; Andrejevic-Blant, S.; Bron, L. Clinicobiological progression and prognosis of oral squamous cell carcinoma in relation to the tumor invasive front: Impact on prognosis. Acta Otolaryngol. 2014, 134, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.; Gil, Z.; Amit, M.; Yen, T.C.; Liao, C.T.; Chaturvedi, P.; Agarwal, J.P.; Kowalski, L.P.; Kreppel, M.; Cernea, C.R.; et al. International Consortium for Outcome Research (ICOR) in Head and Neck Cancer. Primary tumor staging for oral cancer and a proposed modification incorporating depth of invasion: An international multicenter retrospective study. JAMA Otolaryngol. Head Neck Surg. 2014, 140, 1138–1148. [Google Scholar] [PubMed]
- Wong, R.J.; Keel, S.B.; Glynn, R.J.; Varvares, M.A. Histological pattern of mandibular invasion by oral squamous cell carcinoma. Laryngoscope 2000, 110, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Cracchiolo, J.R.; Xu, B.; Migliacci, J.C.; Katabi, N.; Pfister, D.G.; Lee, N.Y.; Patel, S.G.; Ghossein, R.A.; Wong, R.J. Patterns of recurrence in oral tongue cancer with perineural invasion. Head Neck 2018. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Glentis, A.; Gurchenkov, V.; Matic Vignjevic, D. Assembly, heterogeneity, and breaching of the basement membranes. Cell Adhes. Migr. 2014, 8, 236–245. [Google Scholar] [CrossRef]
- Nakayama, S.; Sasaki, A.; Mese, H.; Alcalde, R.E.; Tsuji, T.; Matsumura, T. The E-cadherin gene is silenced by CpG methylation in human oral squamous cell carcinomas. Int. J. Cancer 2001, 93, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Kudo, Y.; Kitajima, S.; Ogawa, I.; Hiraoka, M.; Sargolzaei, S.; Keihaee, M.R.; Sato, S.; Miyauchi, M.; Takata, T. Invasion and metastasis of oral cancer cells require methylation of E-cadherin and/or degradation of membranous beta-catenin. Clin. Cancer Res. 2004, 10, 5455–5463. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, M.; Yoshida, M.; Tsunematsu, T.; Ogawa, I.; Sasahira, T.; Kuniyasu, H.; Imoto, I.; Abiko, Y.; Xu, D.; Fukunaga, S.; et al. MicroRNA-203 Suppresses Invasion and Epithelial-mesenchymal Transition Induction via Targeting NUAK1 and SNAI2 in Head and Neck Cancer. Oncotarget 2016, 7, 8223–8239. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.T.; Kudo, Y.; Yoshida, M.; Kamata, N.; Ogawa, I.; Takata, T. N-cadherin expression is involeved in malignant behaviour of head and neck cancer in relation to epithelial-mesenchymal transition. Histol. Histopathol. 2011, 26, 147–156. [Google Scholar] [PubMed]
- Nguyen, P.T.; Tsunematsu, T.; Yanagisawa, S.; Kudo, Y.; Miyauchi, M.; Kamata, N.; Takata, T. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br. J. Cancer 2013, 109, 2248–2258. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, P.P.; Bharadwaj, R.R.; Machado, J.; Macmillan, C.; Pintilie, M.; Sukhai, M.A.; Perez-Ordonez, B.; Gullane, P.; Irish, J.; Kamel-Reid, S. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 2008, 113, 3169–3180. [Google Scholar] [CrossRef] [PubMed]
- Oku, N.; Sasabe, E.; Ueta, E.; Yamamoto, T.; Osaki, T. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res. 2006, 66, 5251–5257. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, T.; Wang, Y.; Cao, L.; Nishioka, M.; Aguirre, R.L.; Ishikawa, A.; Geng, L.; Okada, N. Altered expression of desmocollin 3, desmoglein 3, and beta-catenin in oral squamous cell carcinoma: Correlation with lymph node metastasis and cell proliferation. Virchows Arch. 2007, 451, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Xi, L.; Seethala, R.R.; Chan, J.; Desai, S.; Hoch, B.; Gooding, W.; Godfrey, T.E. Intraoperative qRT-PCR for detection of lymph node metastasis in head and neck cancer. Clin. Cancer Res. 2011, 17, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, S.; Ishimaru, N.; Kudo, Y. Matrix Metalloproteinases: The Gene Expression Signatures of Head and Neck Cancer Progression. Cancers 2014, 6, 396–415. [Google Scholar] [CrossRef] [PubMed]
- Nyalendo, C.; Beaulieu, E.; Sartelet, H.; Michaud, M.; Fontaine, N.; Gingras, D.; Béliveau, R. Impaired tyrosine phosphorylation of membrane type 1-matrix metalloproteinase reduces tumor cell proliferation in three-dimensional matrices and abrogates tumor growth in mice. Carcinogenesis 2008, 29, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Siriwardena, B.S.; Kudo, Y.; Ogawa, I.; Kitagawa, M.; Kitajima, S.; Hatano, H.; Tilakaratne, W.M.; Miyauchi, M.; Takata, T. Periostin is frequently overexpressed and enhances invasion and angiogenesis in oral cancer. Br. J. Cancer 2006, 95, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Kudo, Y.; Siriwardena, B.S.M.S.; Hatano, H.; Ogawa, I.; Takata, T. Periostin: Novel diagnostic and therapeutic target for cancer. Histol. Histopathol. 2007, 22, 1167–1174. [Google Scholar] [PubMed]
- Deraz, E.M.; Kudo, Y.; Yoshida, M.; Obayashi, M.; Tsunematsu, T.; Tani, H.; Siriwardena, S.B.; Keikhaee, M.R.; Qi, G.; Iizuka, S.; et al. MMP-10/stromelysin-2 promotes invasion of head and neck cancer. PLoS ONE 2011, 6, e25438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, Y.; Iizuka, S.; Yoshida, M.; Nguyen, P.T.; Siriwardena, S.B.; Tsunematsu, T.; Ohbayashi, M.; Ando, T.; Hatakeyama, D.; Shibata, T.; et al. Periostin directly and indirectly promotes tumor lymphangiogenesis of head and neck cancer. PLoS ONE 2012, 7, e44488. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.C.; Chan, A.T. Novel therapeutic target for head and neck squamous cell carcinoma: HGF-MET signaling pathway. Anticancer Drugs 2011, 22, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Knowles, L.M.; Stabile, L.P.; Egloff, A.M.; Rothstein, M.E.; Thomas, S.M.; Gubish, C.T.; Lerner, E.C.; Seethala, R.R.; Suzuki, S.; Quesnelle, K.M.; et al. HGF and c-Met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin. Cancer Res. 2009, 15, 3740–3750. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Jagadeeswaran, R.; Faoro, L.; Janamanchi, V.; Nallasura, V.; El Dinali, M.; Yala, S.; Kanteti, R.; Cohen, E.E.; Lingen, M.W.; et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009, 69, 3021–3031. [Google Scholar] [CrossRef] [PubMed]
- Shintani, S.; Li, C.; Ishikawa, T.; Mihara, M.; Nakashiro, K.; Hamakawa, H. Expression of vascular endothelial growth factor A, B, C, and D in oral squamous cell carcinoma. Oral Oncol. 2004, 40, 13–20. [Google Scholar] [CrossRef]
- Vassilakopoulou, M.; Psyrri, A.; Argiris, A. Targeting angiogenesis in head and neck cancer. Oral Oncol. 2015, 51, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Scanlon, C.S.; Banerjee, R.; Inglehart, R.C.; Liu, M.; Russo, N.; Hariharan, A.; van Tubergen, E.A.; Corson, S.L.; Asangani, I.A.; Mistretta, C.M.; et al. Galanin modulates the neural niche to favour perineural invasion in head and neck cancer. Nat. Commun. 2015, 6, 6885. [Google Scholar] [CrossRef] [PubMed]
- Chuang, F.H.; Hsue, S.S.; Wu, C.W.; Chen, Y.K. Immunohistochemical expression of RANKL, RANK, and OPG in human oral squamous cell carcinoma. J. Oral Pathol. Med. 2009, 38, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Jimi, E.; Shin, M.; Furuta, H.; Tada, Y.; Kusukawa, J. The RANKL/RANK system as a therapeutic target for bone invasion by oral squamous cell carcinoma (Review). Int. J. Oncol. 2013, 42, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Sambandam, Y.; Ethiraj, P.; Hathway-Schrader, J.; Novince, C.; Panneerselvam, E.; Sundaram, K.; Reddy, S.V. Autoregulation of RANK ligand in oral squamous cell carcinoma tumor cells. J. Cell. Physiol. 2018, 233, 6125–6134. [Google Scholar] [CrossRef] [PubMed]
- Juergens, R.A.; Bratman, S.V.; Tsao, M.S.; Laurie, S.A.; Sara Kuruvilla, M.; Razak, A.R.; Hansen, A.R. Biology and patterns of response to EGFR-inhibition in squamous cell cancers of the lung and head & neck. Cancer Treat. Rev. 2017, 54, 43–57. [Google Scholar] [PubMed]
- Wang, S.J.; Bourguignon, L.Y. Hyaluronan and the interaction between CD44 and epidermal growth factor receptor in oncogenic signaling and chemotherapy resistance in head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 2006, 132, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.B. Current challenges and clinical investigations of epidermal growth factor receptor (EGFR)- and ErbB family-targeted agents in the treatment of head and neck squamous cell carcinoma (HNSCC). Cancer Treat. Rev. 2014, 40, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Geiger, J.L.; Grandis, J.R.; Bauman, J.E. The STAT3 pathway as a therapeutic target in head and neck cancer: Barriers and innovations. Oral Oncol. 2016, 56, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, S.; Klijanienko, J.; Giaginis, C.; Alexandrou, P.; Patsouris, E.; Sastre-Garau, X. FAK and Src expression in mobile tongue squamous cell carcinoma: Associations with clinicopathological parameters and patients survival. J. Cancer Res. Clin. Oncol. 2012, 138, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, J.; Li, L.; Shao, S.; Wu, J.; Bian, L.; He, Y. Epithelial mesenchymal transition induced by the CXCL9/CXCR3 axis through AKT activation promotes invasion and metastasis in tongue squamous cell carcinoma. Oncol. Rep. 2018, 39, 1356–1368. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, M. Epithelial–mesenchymal transition is regulated at post-transcriptional levels by transforming growth factor-β signaling during tumor progression. Cancer Sci. 2015, 106, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Elhousiny, M.; Johnson, N.W.; Gao, J. Transforming growth factor-β1 treatment of oral cancer induces epithelial-mesenchymal transition and promotes bone invasion via enhanced activity of osteoclasts. Clin. Exp. Metastasis 2013, 30, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.L.; Xie, Y.G.; Li, Z.; Ma, J.H.; Xu, X. E-cadherin expression and prognosis of oral cancer: A meta-analysis. Tumor Biol. 2014, 35, 5533–5537. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shang, Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp. Cell Res. 2013, 319, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Berx, G.; Raspé, E.; Christofori, G.; Thiery, J.P.; Sleeman, J.P. Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clin. Exp. Metastasis 2007, 24, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates Epithelial-Mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Katayama, M.; Hirai, S.; Kamihagi, K.; Nakagawa, K.; Yasumoto, M.; Kato, I. Soluble E-cadherin fragments increased in circulation of cancer patients. Br. J. Cancer 1994, 69, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Hatta, K.; Nose, A.; Nagafuchi, A.; Takeichi, M. Cloning and expression of cDNA encoding a neural calcium-dependent cell adhesion molecule: Its identity in the cadherin gene family. J. Cell Biol. 1988, 106, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.; Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Suyama, K.; Shapiro, I.; Guttman, M.; Hazan, R.B. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2002, 2, 301–314. [Google Scholar] [CrossRef]
- Lal-Nag, M.; Morin, P.J. The claudins. Genome Biol. 2009, 10, 235. [Google Scholar] [CrossRef] [PubMed]
- Yuki, T.; Haratake, A.; Koishikawa, H.; Morita, K.; Miyachi, Y.; Inoue, S. Tight junction proteins in keratinocytes: Localization and contribution to barrier function. Exp. Dermatol. 2007, 16, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Berika, M.; Garrod, D. Desmosomal adhesion in vivo. Cell Commun. Adhes. 2014, 21, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Amagai, M.; Klaus-Kovtun, V.; Stanley, J.R. Autoantibodies against a novel epithelial cadherin in pemphigus vulgaris, a disease of cell adhesion. Cell 1991, 67, 869–877. [Google Scholar] [CrossRef]
- Oshiro, M.M.; Kim, C.J.; Wozniak, R.J.; Junk, D.J.; Muñoz-Rodríguez, J.L.; Burr, J.A.; Fitzgerald, M.; Pawar, S.C.; Cress, A.E.; Domann, F.E.; et al. Epigenetic silencing of DSC3 is a common event in human breast cancer. Breast Cancer Res. 2005, 7, R669–R680. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Parker, J.S.; Karaca, G.; Wu, J.; Funkhouser, W.K.; Moore, D.; Butterfoss, D.; Xiang, D.; Zanation, A.; Yin, X.; et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 2004, 5, 489–500. [Google Scholar] [CrossRef]
- Chen, Y.J.; Chang, J.T.; Lee, L.; Wang, H.M.; Liao, C.T.; Chiu, C.C.; Chen, P.J.; Cheng, A.J. DSG3 is overexpressed in head neck cancer and is a potential molecular target for inhibition of oncogenesis. Oncogene 2007, 26, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Waseem, A.; Cruz, I.N.; Szary, J.; Gunic, E.; Mannan, T.; Unadkat, M.; Yang, M.; Valderrama, F.; Toole, E.A.O.; et al. Desmoglein 3 promotes cancer cell migration and invasion by regulating activator protein 1 and protein kinase C-dependent-Ezrin activation. Oncogene 2014, 33, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Solassol, J.; Burcia, V.; Costes, V.; Lacombe, J.; Mange, A.; Barbotte, E.; de Verbizier, D.; Cartier, C.; Makeieff, M.; Crampette, L.; et al. Pemphigus vulgaris antigen mRNA quantification for the staging of sentinel lymph nodes in head and neck cancer. Br. J. Cancer 2010, 102, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Neilson, E.G. Epithelialmesenchymal transition and its implications for fibrosis. J. Clin. Investig. 2003, 112, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. Coming full circle-from endless complexity to simplicity and back again. Cell 2014, 157, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Lohmer, L.L.; Kelley, L.C.; Hagedorn, E.J.; Sherwood, D.R. Invadopodia and basement membrane invasion in vivo. Cell Adhes. Migr. 2014, 8, 246–255. [Google Scholar] [CrossRef]
- Basu, B.; Correa de Sampaio, P.; Mohammed, H.; Fogarasi, M.; Corrie, P.; Watkins, N.A.; Smethurst, P.A.; English, W.R.; Ouwehand, W.H.; Murphy, G. Inhibition of MT1-MMP activity using functional antibody fragments selected against its hemopexin domain. Int. J. Biochem. Cell Biol. 2012, 44, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.; Leyssen, A.; Van Aelst, I.; Fiten, P.; Piccard, H.; Hu, J.; Descamps, F.J.; Van den Steen, P.E.; Proost, P.; Van Damme, J.; et al. A monoclonal antibody inhibits gelatinase B/MMP-9 by selective binding to part of the catalytic domain and not to the fibronectin or zinc binding domains. Biochim. Biophys. Acta 2007, 1770, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Paemen, L.; Opdenakker, G.; Froyen, G. Cloning and expression in Escherichia coli of a human gelatinase B-inhibitory single-chain immunoglobulin variable fragment (scFv). FEBS Lett. 1997, 414, 562–566. [Google Scholar] [CrossRef]
- Kudo, Y.; Ogawa, I.; Kitajima, S.; Kitagawa, M.; Kawai, H.; Gaffney, P.M.; Miyauchi, M.; Takata, T. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res. 2006, 66, 6928–6935. [Google Scholar] [CrossRef] [PubMed]
- Kudo, Y.; Iizuka, S.; Yoshida, M.; Tsunematsu, T.; Kondo, T.; Subarnbhesaj, A.; Deraz, E.M.; Siriwardena, S.B.S.M.; Tahara, H.; Ogawa, I.; et al. Matrix metalloproteinase-13 directly and indirectly promotes tumor angiogenesis. J. Biol. Chem. 2012, 287, 38716–38728. [Google Scholar] [CrossRef] [PubMed]
- Conway, S.J.; Izuhara, K.; Kudo, Y.; Litvin, J.; Markwald, R.; Ouyang, G.; Arron, J.R.; Holweg, C.T.; Kudo, A. The role of periostin in tissue remodeling across health and disease. Cell. Mol. Life Sci. 2014, 71, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Ouyang, G.; Bai, X.; Huang, Z.; Ma, C.; Liu, M.; Shao, R.; Anderson, R.M.; Rich, J.N.; Wang, X.F. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell 2004, 5, 329–339. [Google Scholar] [CrossRef]
- Ruan, K.; Bao, S.; Ouyang, G. The multifaceted role of periostin in tumorigenesis. Cell. Mol. Life Sci. 2009, 66, 2219–2230. [Google Scholar] [CrossRef] [PubMed]
- Ghajar, C.M.; Peinado, H.; Mori, H.; Matei, I.R.; Evason, K.J.; Brazier, H.; Almeida, D.; Koller, A.; Hajjar, K.A.; Stainier, D.Y.; et al. The perivascular niche regulates breast tumor dormancy. Nat. Cell Biol. 2013, 15, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.; Wang, Z.; Huang, Y.; Liu, W.; Zhu, X.; Cai, Y.; Fang, X.; Lin, S.; Yuan, L.; et al. Periostin contributes to the acquisition of multipotent stem cell-like properties in human mammary epithelial cells and breast cancer cells. PLoS ONE 2013, 8, e72962. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Saxton, R.E.; Ramos, L.; Chang, D.D.; Karlan, B.Y.; Gasson, J.C.; Slamon, D.J. Neutralizing monoclonal antibody to periostin inhibits ovarian tumor growth and metastasis. Mol. Cancer Ther. 2001, 10, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, I.S.; Park, S.A.; Kim, Y.; Lee, J.E.; Noh, D.Y.; Kim, K.T.; Ryu, S.H.; Suh, P.G. Periostin-binding DNA aptamer inhibits breast cancer growth and metastasis. Mol. Ther. 2013, 21, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.Y.; Li, Q.; Lee, J.H.; Arango, M.E.; McDonnell, S.R.; Yamazaki, S.; Koudriakova, T.B.; Alton, G.; Cui, J.J.; Kung, P.P.; et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007, 67, 4408–4417. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Kandl, C.; Hamilton, C.D.; Shnayder, Y.; Tsue, T.T.; Kakarala, K.; Ledgerwood, L.; Sun, X.S.; Huang, H.J.; Girod, D.; et al. Mitigation of Tumor-Associated Fibroblast-Facilitated Head and Neck Cancer Progression with Anti-Hepatocyte Growth Factor Antibody Ficlatuzumab. JAMA Otolaryngol. Head Neck Surg. 2015, 141, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011, 473, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Body, J.J.; Greipp, P.; Coleman, R.E.; Facon, T.; Geurs, F.; Fermand, J.P.; Harousseau, J.L.; Lipton, A.; Mariette, X.; Williams, C.D.; et al. A phase I study of AMGN-0007, a recombinant osteoprotegerin construct, in patients with multiple myeloma or breast carcinoma related bone metastases. Cancer 2003, 97, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Roodman, G.D.; Dougall, W.C. RANK ligand as a therapeutic target for bone metastases and multiple myeloma. Cancer Treat. Rev. 2008, 34, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Finkel, K.A.; Warner, K.A.; Kerk, S.; Bradford, C.R.; McLean, S.A.; Prince, M.E.; Zhong, H.; Hurt, E.M.; Hollingsworth, R.E.; Wicha, M.S.; et al. IL-6 Inhibition with MEDI5117 Decreases the Fraction of Head and Neck Cancer Stem Cells and Prevents Tumor Recurrence. Neoplasia 2016, 18, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Argiris, A.; Harrington, K.J.; Tahara, M.; Schulten, J.; Chomette, P.; Ferreira Castro, A.; Licitra, L. Evidence-Based Treatment Options in Recurrent and/or Metastatic Squamous Cell Carcinoma of the Head and Neck. Front. Oncol. 2017, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Monkman, J.; Toh, A.K.L.; Nagaraj, S.H.; Thompson, E.W. Targeting epithelial-mesenchymal plasticity in cancer: Clinical and preclinical advances in therapy and monitoring. Biochem. J. 2017, 474, 3269–3306. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Grandis, J.R.; Sok, J.C. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol. Ther. 2004, 102, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, C.; Che Mat, N.F.; Gee, K. Interleukin-27 induces a STAT1/3- and NF-kappaB-dependent proinflammatory cytokine profile in human monocytes. J. Biol. Chem. 2010, 285, 24404–24411. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Zhang, X.; Paladino, D.; Sengupta, B.; Ahmad, S.; Holloway, R.W.; Ingersoll, S.B.; Turkson, J. Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene 2012, 31, 2309–2322. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.T. Focal adhesion kinase: The first ten years. J. Cell Sci. 2003, 116, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Sulzmaier, F.J.; Jean, C.; Schlaepfer, D.D. FAK in cancer: Mechanistic findings and clinical applications. Nat. Rev. Cancer 2014, 14, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wang, L.; Gao, W.; Meng, J.; Dai, B.; Wu, S.; Minna, J.; Roth, J.A.; Hofstetter, W.L.; Swisher, S.G.; et al. IGFBP2/FAK pathway is causally associated with dasatinib resistance in non-small cell lung cancer cells. Mol. Cancer Ther. 2013, 12, 2864–2873. [Google Scholar] [CrossRef] [PubMed]
- Infante, J.R.; Camidge, D.R.; Mileshkin, L.R.; Chen, E.X.; Hicks, R.J.; Rischin, D.; Fingert, H.; Pierce, K.J.; Xu, H.; Roberts, W.G.; et al. Safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF 00562271, an inhibitor of focal adhesion kinase, in advanced solid tumors. J. Clin. Oncol. 2012, 30, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; Böttinger, E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 2005, 24, 5764–5774. [Google Scholar] [CrossRef] [PubMed]
- Micalizzi, D.S.; Ford, H.L. Epithelial-mesenchymal transition in development and cancer. Future Oncol. 2009, 5, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Shirakihara, T.; Horiguchi, T.; Miyazawa, M.; Ehata, S.; Shibata, T.; Morita, I.; Miyazono, K.; Saitoh, M. TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J. 2011, 30, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, K.; Shirakihara, T.; Nakano, A.; Imamura, T.; Miyazono, K.; Saitoh, M. Role of Ras signaling in the induction of snail by transforming growth factor-β. J. Biol. Chem. 2009, 284, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Izuishi, K.; Sano, T.; Hossain, M.A.; Kimura, S.; Masaki, T.; Suzuki, Y. Modulating effect of the PI3- kinase inhibitor LY294002 on cisplatin in human pancreatic cancer cells. J. Exp. Clin. Cancer Res. 2008, 27, 76. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.L.; Lin, C.I.; Whang, E.E.; Carothers, A.M.; Moore, F.D., Jr.; Ruan, D.T. Sulindac reverses aberrant expression and localization of beta-catenin in papillary thyroid cancer cells with the BRAFV600E mutation. Thyroid 2010, 20, 615–622. [Google Scholar]
- Jang, Y.H.; Shin, H.S.; Sun Choi, H.; Ryu, E.S.; Jin Kim, M.; Ki Min, S.; Lee, J.H.; Kook Lee, H.; Kim, K.H.; Kang, D.H. Effects of dexamethasone on the TGFbeta1-induced epithelial-to-mesenchymal transition in human peritoneal mesothelial cells. Lab. Investig. 2013, 93, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Harrington, N.; Moraes, R.C.; Wu, M.F.; Hilsenbeck, S.G.; Lewis, M.T. Cyclopamine inhibition of human breast cancer cell growth independent of Smoothened (Smo). Breast Cancer Res. Treat. 2009, 115, 505–521. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.J.; He, C.L.; Sun, T.; Duan, X.J.; Sun, Y.; Xiong, S.J. hsa-miR-485-5p reverses epithelial to mesenchymal transition and promotes cisplatin-induced cell death by targeting PAK1 in oral tongue squamous cell carcinoma. Int. J. Mol. Med. 2017, 40, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Xun, W.; Wei, K.; Yang, Y.; Shen, H. MicroRNA-27a-3p regulates epithelial to mesenchymal transition via targeting YAP1 in oral squamous cell carcinoma cells. Oncol. Rep. 2016, 36, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Jiang, Y.P.; Chen, W.; Li, K.D.; Liu, X.; Gao, S.Y.; Feng, H.; Wang, S.S.; Jiang, J.; Ma, X.R.; et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget 2015, 6, 6797–6810. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Sun, Q.; Zhang, J.; Yu, J.; Chen, W.; Zhang, Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis 2013, 34, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Kawakita, A.; Yanamoto, S.; Yamada, S.; Naruse, T.; Takahashi, H.; Kawasaki, G.; Umeda, M. MicroRNA-21 promotes oral cancer invasion via the Wnt/β-catenin pathway by targeting DKK2. Pathol. Oncol. Res. 2014, 20, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, J.; Jiang, L.; Wang, A.; Shi, F.; Ye, H.; Zhou, X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genom. Proteom. 2009, 6, 131–139. [Google Scholar]
- Lu, L.; Xue, X.; Lan, J.; Gao, Y.; Xiong, Z.; Zhang, H.; Jiang, W.; Song, W.; Zhi, Q. MicroRNA-29a upregulates MMP2 in oral squamous cell carcinoma to promote cancer invasion and anti-apoptosis. Biomed. Pharmacother. 2014, 68, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Chang, J.T.; Liao, C.T.; Kang, C.J.; Huang, S.F.; Chen, I.H.; Huang, C.C.; Huang, Y.C.; Chen, W.H.; Tsai, C.Y.; et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol. Cancer 2014, 13, 218. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Tao, X.; Huang, F.; Wu, T.; Wang, J.; Jiang, X.; Kuang, Z.; Cheng, B. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int. J. Mol. Med. 2016, 37, 1274–1280. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hu, C.; Chi, J.; Li, J.; Peng, C.; Yun, X.; Li, D.; Yu, Y.; Li, Y.; Gao, M.; et al. miR-24 promotes the proliferation, migration and invasion in human tongue squamous cell carcinoma by targeting FBXW7. Oncol. Rep. 2016, 36, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.D.; Wu, H.; Wang, S.; Pang, P.; Jin, S.; Sun, C.F.; Liu, F.Y. MiR-1275 promotes cell migration, invasion and proliferation in squamous cell carcinoma of head and neck via up-regulating IGF-1R and CCR7. Gene 2018, 646, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sakha, S.; Muramatsu, T.; Ueda, K.; Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38750. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, L.; Wang, A.; Yu, J.; Shi, F.; Zhou, X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009, 286, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, J.; Cao, W.; Wang, X.; Xu, Q.; Yan, M.; Wu, X.; Chen, W. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int. J. Biochem. Cell Biol. 2013, 45, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Chan, S.H.; Jang, T.H.; Chang, J.W.; Ko, Y.C.; Yen, T.C.; Chiang, S.L.; Chiang, W.F.; Shieh, T.Y.; Liao, C.T.; et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014, 74, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Peng, W.; Ling, W.; Jiebing, H.; Zhuan, B. Reciprocal effects between microRNA-140-5p and ADAM10 suppress migration and invasion of human tongue cancer cells. Biochem. Biophys. Res. Commun. 2014, 448, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Pan, H.; Zhou, Y.; Zhou, J.; Fan, Y.; Qu, P. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol. Med. Rep. 2014, 9, 2491–2498. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.F.; Huang, Y.P.; Zheng, Y.F.; Lyu, M.Y.; Wei, S.B.; Meng, Z.; Gan, Y.H. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014, 50, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Shivananda, S.; Gopinath, K.S.; Kumar, A. MicroRNA-125a reduces proliferation and invasion of oral squamous cell carcinoma cells by targeting estrogen-related receptor α: Implications for cancer therapeutics. J. Biol. Chem. 2014, 289, 32276–32290. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.F.; Wei, S.B.; Mitchelson, K.; Gao, Y.; Zheng, Y.F.; Meng, Z.; Gan, Y.H.; Yu, G.Y. miR-34a inhibits migration and invasion of tongue squamous cell carcinoma via targeting MMP9 and MMP14. PLoS ONE 2014, 9, e108435. [Google Scholar] [CrossRef] [PubMed]
- Shiah, S.G.; Hsiao, J.R.; Chang, W.M.; Chen, Y.W.; Jin, Y.T.; Wong, T.Y.; Huang, J.S.; Tsai, S.T.; Hsu, Y.M.; Chou, S.T.; et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014, 74, 7560–7572. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Li, Y.; Yang, S.; Yang, H.; Tang, J.; Li, M. Micro-ribonucleic acid 143 (MiR-143) inhibits oral squamous cell carcinoma (OSCC) cell migration and invasion by downregulation of phospho-c-Met through targeting CD44 v3. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015, 120, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, L. MicroRNA-143 suppresses oral squamous cell carcinoma cell growth, invasion and glucose metabolism through targeting hexokinase 2. Biosci. Rep. 2017, 37, BSR20160404. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ren, Y.; Tang, H.; Wang, W.; He, Q.; Sun, J.; Zhou, X.; Wang, A. Deregulation of the miR-222-ABCG2 regulatory module in tongue squamous cell carcinoma contributes to chemoresistance and enhanced migratory/invasive potential. Oncotarget 2015, 6, 44538–44550. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumor Biol. 2016, 37, 4105–4113. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.Y.; You, J.J.; Yang, C.M.; Pan, H.W.; Chen, H.C.; Lee, J.H.; Lin, Y.S.; Liou, H.H.; Liu, P.F.; Chi, C.C.; et al. Aberrant DNA hypomethylation of miR-196b contributes to migration and invasion of oral cancer. Oncol. Lett. 2016, 11, 4013–4021. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, I.; Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Matsushita, R.; Kurozumi, A.; Kato, M.; Okato, A.; Okamoto, Y.; Seki, N. The tumor-suppressive microRNA-23b/27b cluster regulates the MET oncogene in oral squamous cell carcinoma. Int. J. Oncol. 2016, 49, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Kawakubo-Yasukochi, T.; Morioka, M.; Hazekawa, M.; Yasukochi, A.; Nishinakagawa, T.; Ono, K.; Kawano, S.; Nakamura, S.; Nakashima, M. miR-200c-3p spreads invasive capacity in human oral squamous cell carcinoma microenvironment. Mol. Carcinog. 2018, 57, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Hasegawa, S.; Uchida, F.; Terabe, T.; Ishibashi Kanno, N.; Kato, K.; Yamagata, K.; Sakai, S.; Kawashiri, S.; Sato, H.; et al. MicroRNA-205-5p suppresses the invasiveness of oral squamous cell carcinoma by inhibiting TIMP‑2 expression. Int. J. Oncol. 2018, 52, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Luo, Q.; Wang, H.; Zhang, H.; Chen, F. MicroRNA-22 suppresses cell proliferation, migration and invasion in oral squamous cell carcinoma by targeting NLRP3. J. Cell Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ren, Y.; Liu, R.; Ma, J.; Shi, Y.; Zhang, L.; Bu, R. miR-195-5p Suppresses the Proliferation, Migration, and Invasion of Oral Squamous Cell Carcinoma by Targeting TRIM14. Biomed. Res. Int. 2017, 2017, 7378148. [Google Scholar] [CrossRef] [PubMed]
- Ruan, P.; Tao, Z.; Tan, A. Low expression of miR-30a-5p induced the proliferation and invasion of oral cancer via promoting the expression of FAP. Biosci. Rep. 2018, 38, BSR20171027. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jin, J.; Ma, T.; Zhai, H. MiR-376c-3p regulates the proliferation, invasion, migration, cell cycle and apoptosis of human oral squamous cancer cells by suppressing HOXB7. Biomed. Pharmacother. 2017, 91, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Cheng, J.L.; Zhang, Y.; Bo, C.X.; Li, Y.L. MicroRNA‑375 inhibits oral squamous cell carcinoma cell migration and invasion by targeting platelet‑derived growth factor‑A. Mol. Med. Rep. 2017, 15, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Wang, C.; Zhuang, Z.; Hou, J.; Liu, X.; Wu, Y.; Liu, H.; Huang, H. Decreased miR-320a promotes invasion and metastasis of tumor budding cells in tongue squamous cell carcinoma. Oncotarget 2016, 7, 65744–65757. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liang, J.; Wang, Q.; Li, Z.; Du, Y.; Xu, X. MicroRNA-137 suppresses tongue squamous carcinoma cell proliferation, migration and invasion. Cell Prolif. 2016, 49, 628–635. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Aberrant Expression in OSCC | Specific Function in OSCC | References | |
|---|---|---|---|---|
| E-cadherin | Cell adhesion molecule | Downregulation hypermethylation | Acquisition of EMT phenotype including promoting invasion | [11,12,13] |
| N-cadherin | Cell adhesion molecule | Upregulation | Promote invasiveness via activating FGFR1 signaling pathway | [14,15] |
| Claudin-1 | Cell adhesion molecule | Upregulation | Promoting invasion via structural and functional alterations of tight junctions | [16,17] |
| Desmoglein-3 | Cell adhesion molecule | Downregulation Upregulation | Involved in desmosomal intercellular junction | [18,19] |
| MT1-MMP (MMP-14) | Matrix metalloprotease | Upregulation | Promoting invasion via degradation of ECM (Collagens I, II, and III; gelatins; aggrecan; fibronectin; laminin, fibrin) | [20,21] |
| MMP-2 | Matrix metalloprotease | Upregulation | Promoting invasion via degradation of ECM (gelatins; VII, X and, XI; fibronectin; laminin; elastin; aggrecan) | [20] |
| MMP-9 | Matrix metalloprotease | Upregulation | Promoting invasion via degradation of ECM (gelatins; collagens III, IV, and, V; aggrecan; elastin; entactin; vitronectin; N-telopeptide of collagen I) | [20] |
| Periostin | Component of ECM | Upregulation | Promoting angiogenesis, lymphangiogenesis, migration, and invasion | [22,23,24,25] |
| HGF | Growth factor | Upregulation | Promoting EMT induction via HGF/c-Met signaling | [26,27] |
| c-Met | Receptor | Upregulation | Promoting EMT, proliferation, and angiogenesis induction via HGF/c-Met signaling | [26,27,28] |
| VEGF | Growth factor | Upregulation | Angiogenesis | [29,30] |
| GAL | Neuropeptide | Downregulation | Perineural invasion | [31] |
| RANKL | Membrane protein | Upregulation | Bone invasion via induction of osteoclastogenesis | [32,33,34] |
| EGFR | Receptor | Upregulation | Activating P13K and Akt pathways | [35,36,37] |
| STAT3 | Activator of transduction | Signal activation | Activating gene transcription involved in the essential components of invasion and metastasis | [38] |
| FAK | Mediator of signal transduction | Upregulation | Promoting invasion as a mediator of integrin and growth factors signaling | [39] |
| CXCL9 | Chemokine | Upregulation | EMT induction and cytoskeleton rearrangement via activation of Akt signaling pathway | [40] |
| CXCR3 | Chemokine receptor | Upregulation | EMT induction and cytoskeleton rearrangement via activation of Akt signaling pathway | [40] |
| TGF-β | Growth factor | Signal activation | EMT induction | [41,42] |
| Function | miRNA | Target Gene Etc. | References |
|---|---|---|---|
| EMT-related miRNAs | miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, miR-429) | ZEB1/ZEB2 | [13] |
| miR-203 | SNAI2/NUAK1 | [13] | |
| miR-485-5p | PAK1 | [109] | |
| miR-27a-3p | YAP1 | [110] | |
| miR-101 | EZH2 | [111] | |
| miR-153 | SNAI1/ZEB2 | [112] | |
| Oncogenic miRNAs | miR-21 | DKK2 | [113,114] |
| miR-29a | upregulating MMP2 | [115] | |
| miR-196 | NME4 | [116] | |
| miR-155 | BCL6 | [117] | |
| miR-24 | FBXW7 | [118] | |
| miR-1275 | upregulating IGF-1R/CCR7 | [119] | |
| miR-342-3p | included in exosome | [120] | |
| miR-1246 | included in exosome | [120] | |
| Tumor suppressive miRNAs | miR-222 | MP1/SOD2 | [114] |
| miR-138 | - | [121] | |
| miR-363 | podoplanin | [122] | |
| miR-491-5p | GIT1 | [123] | |
| miR-140-5p | - | [124] | |
| miR-133b | - | [125] | |
| miR-29b | SP1 | [126] | |
| miR-125a | ESRRA | [127] | |
| miR-34a | MMP9/MMP14 | [128] | |
| miR-329 | Wnt-7b | [129] | |
| miR-410 | Wnt-7b | [129] | |
| miR-143 | CD44v3/hrxokinase 2 | [130,131] | |
| miR-222 | ABCG2 | [132] | |
| miR-188 | SIX1 | [133] | |
| miR-196b | - | [134] | |
| miR-23b | MET | [135] | |
| miR-27b | MET | [135] | |
| miR-200c-3p | CHD9/WRN | [136] | |
| miR-205-5p | TIMP-2 | [137] | |
| miR-22 | NLRP3 | [138] | |
| miR-195-5p | TRIM14 | [139] | |
| miR-30a-5p | FAP | [140] | |
| miR-376c-3p | HOXB7 | [141] | |
| miR-375 | PDGF-A | [142] | |
| miR-320a | - | [143] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siriwardena, S.B.S.M.; Tsunematsu, T.; Qi, G.; Ishimaru, N.; Kudo, Y. Invasion-Related Factors as Potential Diagnostic and Therapeutic Targets in Oral Squamous Cell Carcinoma—A Review. Int. J. Mol. Sci. 2018, 19, 1462. https://doi.org/10.3390/ijms19051462
Siriwardena SBSM, Tsunematsu T, Qi G, Ishimaru N, Kudo Y. Invasion-Related Factors as Potential Diagnostic and Therapeutic Targets in Oral Squamous Cell Carcinoma—A Review. International Journal of Molecular Sciences. 2018; 19(5):1462. https://doi.org/10.3390/ijms19051462
Chicago/Turabian StyleSiriwardena, Samadarani B. S. M., Takaaki Tsunematsu, Guangying Qi, Naozumi Ishimaru, and Yasusei Kudo. 2018. "Invasion-Related Factors as Potential Diagnostic and Therapeutic Targets in Oral Squamous Cell Carcinoma—A Review" International Journal of Molecular Sciences 19, no. 5: 1462. https://doi.org/10.3390/ijms19051462