Integrative Bioinformatics and Functional Analyses of GEO, ENCODE, and TCGA Reveal FADD as a Direct Target of the Tumor Suppressor BRCA1
Abstract
:1. Introduction
2. Results
2.1. Identification of Potential BRCA1 Target Genes from ENCODE, GEO, and TCGA Data Analysis
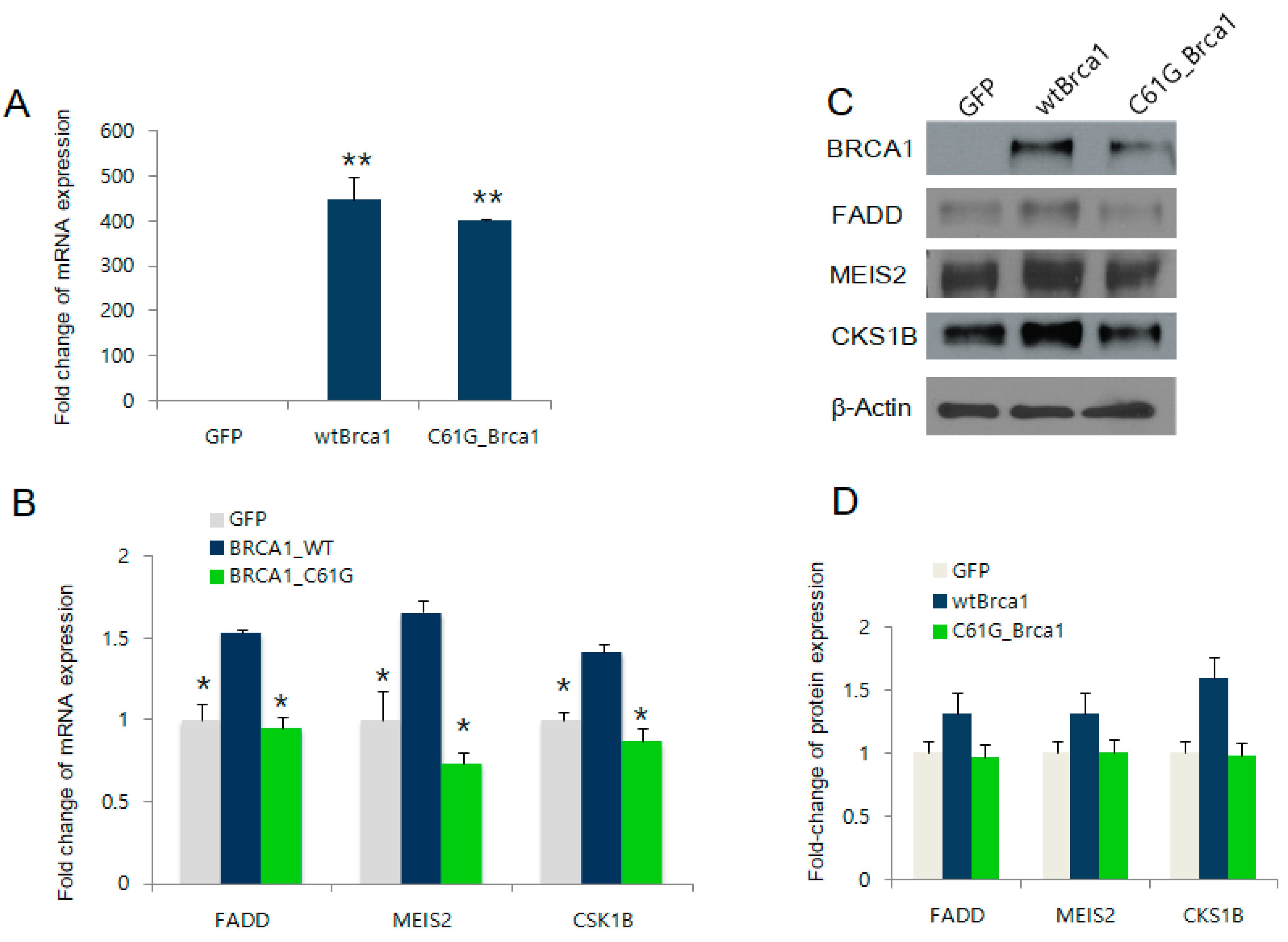
2.2. BRCA1 Overexpression Positively Regulates FADD, MEIS2, and CKS1B Expression
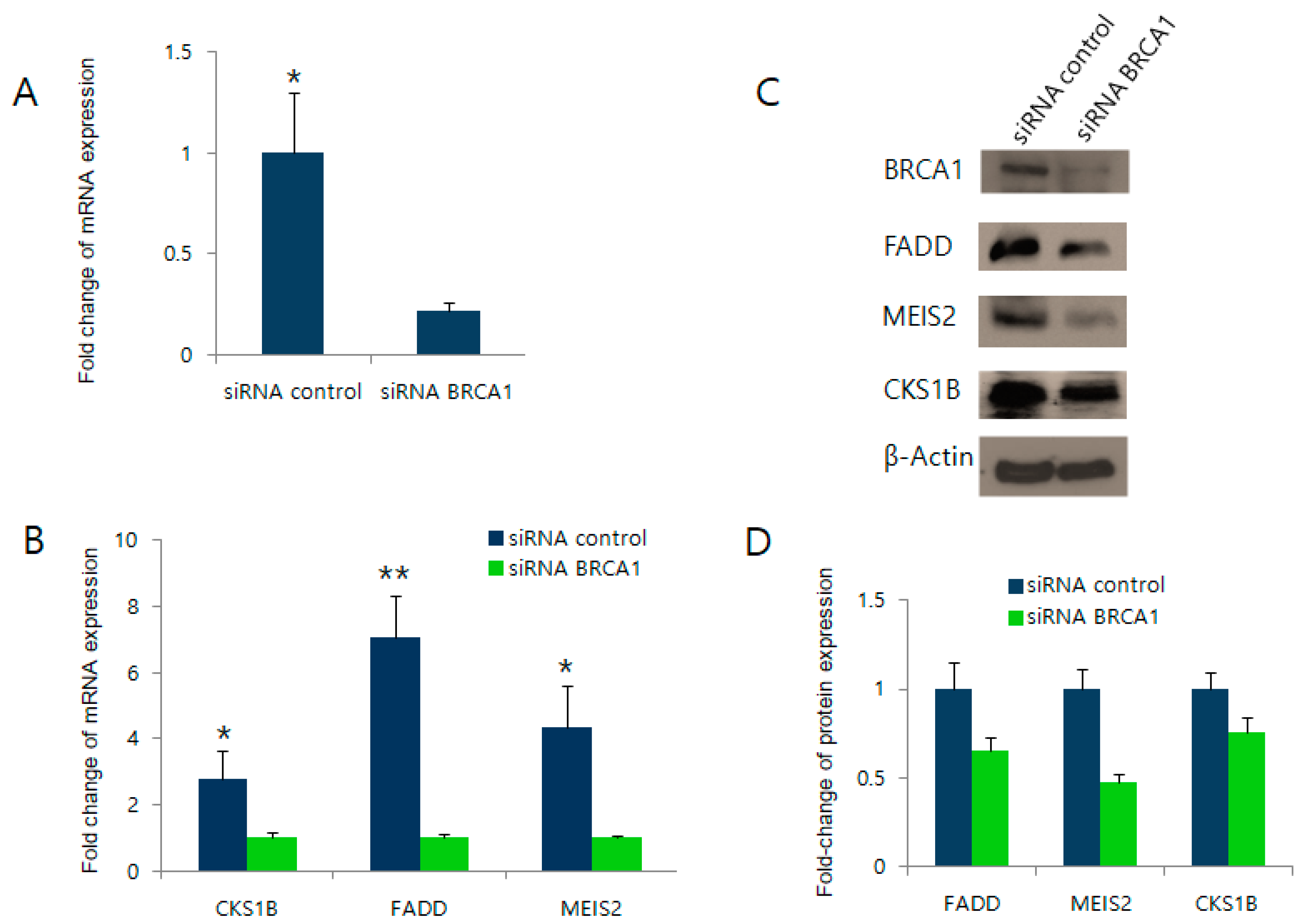
2.3. BRCA1 Knockdown Negatively Regulates FADD, MEIS2, and CKS1B Expression
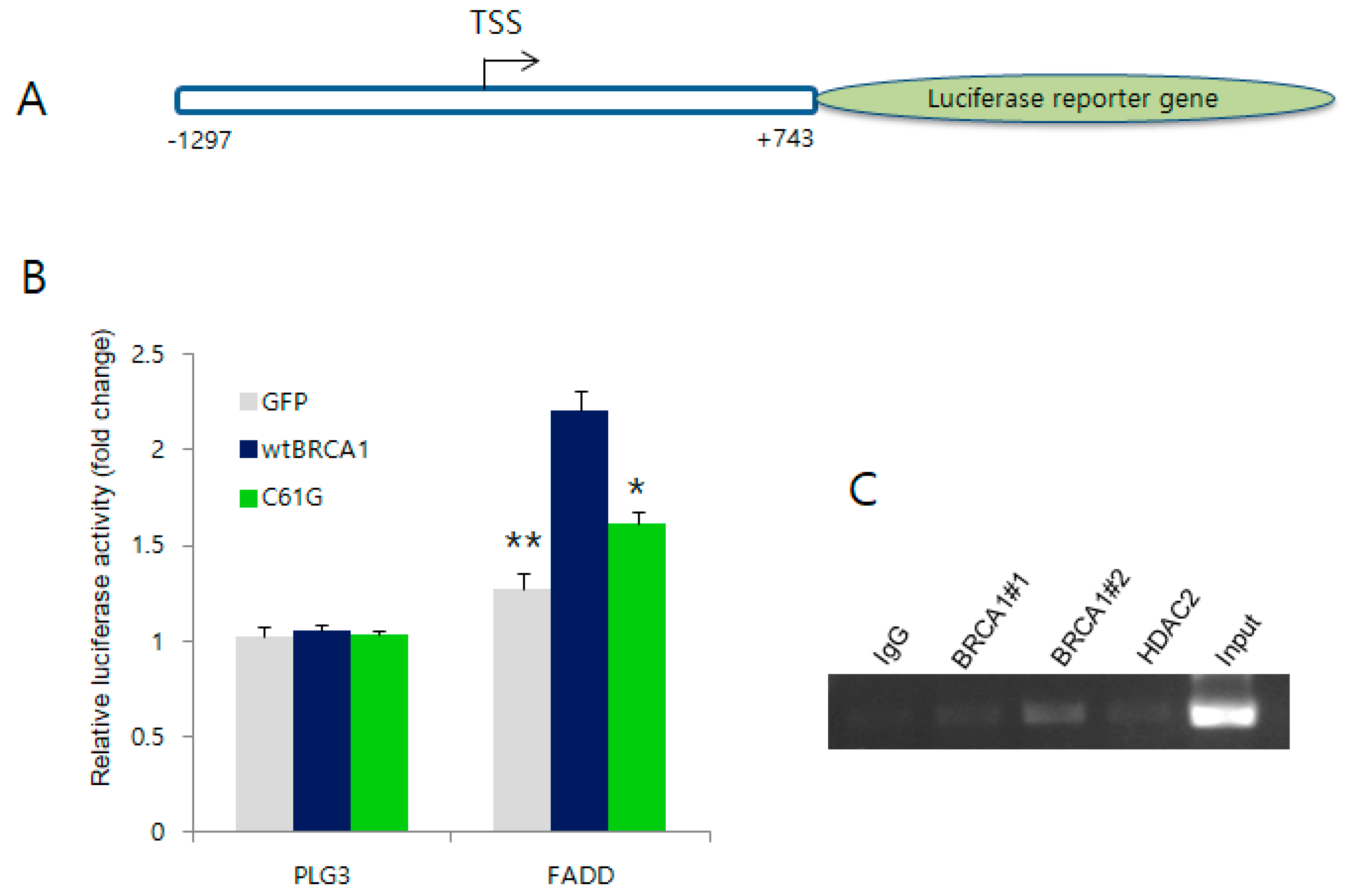
2.4. BRCA1 Activates the Promoter Region of FADD but Not the Other Target Genes
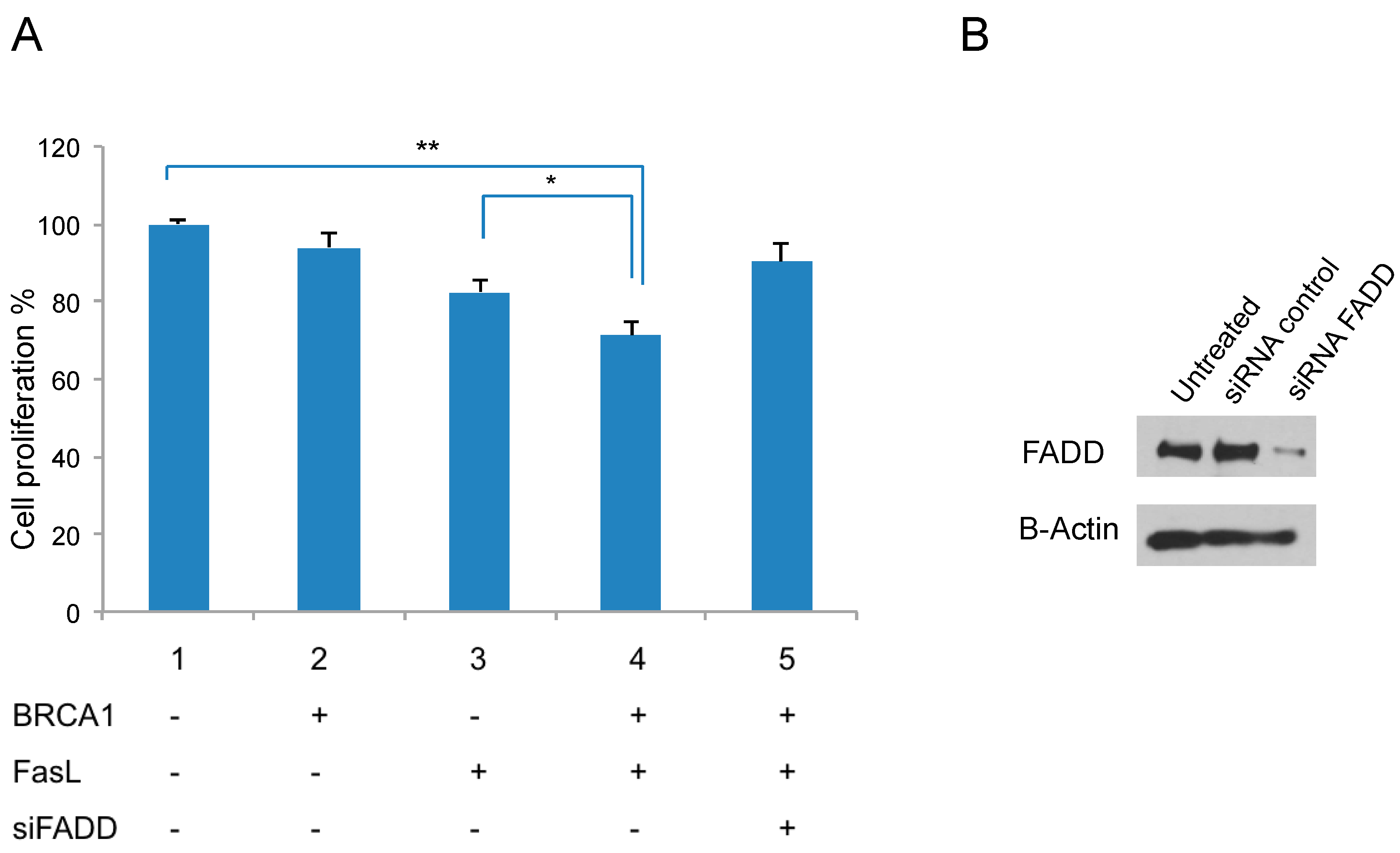
2.5. Fas Ligand (FasL) Signaling and BRCA1 Expression Reduce Proliferation
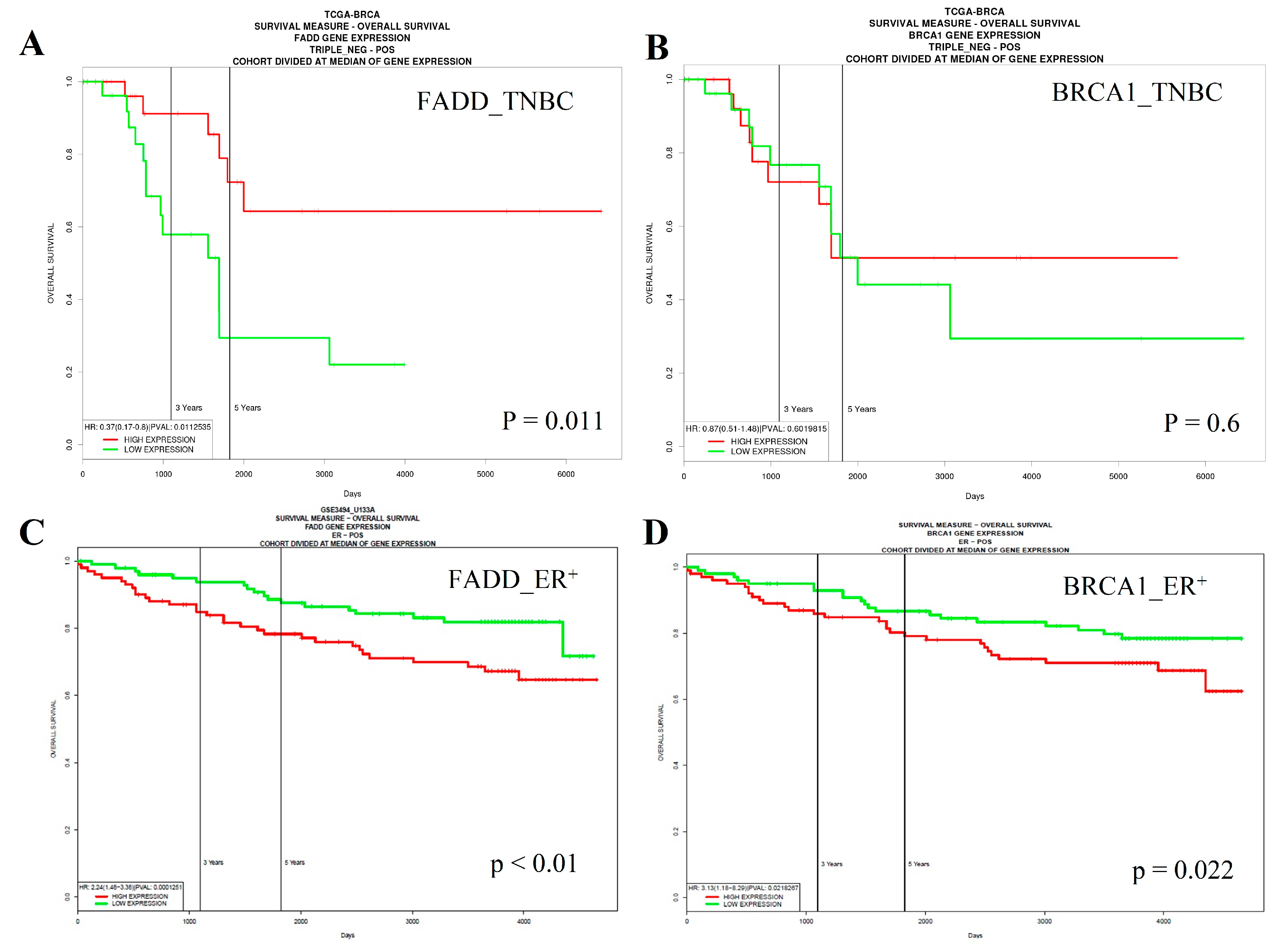
2.6. Low Expression of FADD is Correlated with Poor Survival of Triple-Negative Breast Cancer (TNBC) Patients, Whereas the Opposite Occurs in Estrogen Receptor (ER)-Positive Cases
3. Discussion
4. Materials and Methods
4.1. GEO, ENCODE, and TCGA Data Analysis
4.2. ChIP Assay
4.3. DNA Constructs
4.4. Transfection and Luciferase Assay
4.5. Knockdown Using siRNA
4.6. Real-Time PCR Analysis
4.7. Western Blot Analysis
4.8. AlamarBlue® Cell Proliferation Assay
4.9. PROgeneV2 Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [PubMed]
- James, C.R.; Quinn, J.E.; Mullan, P.B.; Johnston, P.G.; Harkin, D.P. BRCA1, a potential predictive biomarker in the treatment of breast cancer. Oncologist 2007, 12, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Chen, J.; Ochs, R.L.; Keegan, K.; Hoekstra, M.; Feunteun, J.; Livingston, D.M. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 1997, 90, 425–435. [Google Scholar] [CrossRef]
- Cortez, D.; Wang, Y.; Qin, J.; Elledge, S.J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 1999, 286, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Anderson, S.F.; Chao, D.M.; Wei, W.; Ye, L.; Young, R.A.; Livingston, D.M.; Parvin, J.D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 5605–5610. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Wang, R.H.; Akagi, K.; Kim, K.A.; Martin, B.K.; Cavallone, L.; Breast, C.; Haines, D.C.; Basik, M.; Mai, P.; et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 2011, 17, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.N.; August, A.; Hanafusa, H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc. Natl. Acad. Sci. USA 1996, 93, 13595–13599. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.S.; Verma, I.M. Transcriptional activation by BRCA1. Nature 1996, 382, 678–679. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Pan, H.; Li, S.; Flesken-Nikitin, A.; Chen, P.L.; Boyer, T.G.; Lee, W.H. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol. Cell 2000, 6, 757–768. [Google Scholar] [CrossRef]
- Zhang, H.; Somasundaram, K.; Peng, Y.; Tian, H.; Zhang, H.; Bi, D.; Weber, B.L.; El-Deiry, W.S. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 1998, 16, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhao, H.; Fan, F.; Blanck, P.; Fan, W.; Colchagie, A.B.; Fornace, A.J., Jr.; Zhan, Q. BRCA1 activation of the GADD45 promoter. Oncogene 2000, 19, 4050–4057. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Jin, S.; Tong, T.; Zhao, H.; Fan, F.; Antinore, M.J.; Rajasekaran, B.; Wu, M.; Zhan, Q. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J. Biol. Chem. 2002, 277, 8061–8067. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Sharan, S.K. The role of epigenetic transcriptional regulation in BRCA1-mediated tumor suppression. Transcription 2013, 4, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. The ENCODE (ENCyclopedia of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Goswami, C.P.; Nakshatri, H. PROGgeneV2: Enhancements on the existing database. BMC Cancer 2014, 14, 970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.; Sharan, S.K. Epigenetic control of an oncogenic microRNA, miR-155, by BRCA1. Oncotarget 2012, 3, 5–6. [Google Scholar] [PubMed]
- Zheng, L.; Annab, L.A.; Afshari, C.A.; Lee, W.H.; Boyer, T.G. BRCA1 mediates ligand-independent transcriptional repression of the estrogen receptor. Proc. Natl. Acad. Sci. USA 2001, 98, 9587–9592. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Ma, Y.X.; Wang, C.; Yuan, R.Q.; Meng, Q.; Wang, J.A.; Erdos, M.; Goldberg, I.D.; Webb, P.; Kushner, P.J.; et al. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene 2001, 20, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Yarden, R.I.; Brody, L.C. BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 1999, 96, 4983–4988. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kim, C.; Yel, L.; Gollapudi, S. A role of fas-associated death domain (FADD) in increased apoptosis in aged humans. J. Clin. Immunol. 2004, 24, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Chinnaiyan, A.M.; O’Rourke, K.; Tewari, M.; Dixit, V.M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 1995, 81, 505–512. [Google Scholar] [CrossRef]
- Boldin, M.P.; Varfolomeev, E.E.; Pancer, Z.; Mett, I.L.; Camonis, J.H.; Wallach, D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 1995, 270, 7795–7798. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Chai, Y.L.; Shyam, E.; Reddy, P.; Rao, V.N. Induction of apoptosis by the tumor suppressor protein BRCA1. Oncogene 1996, 13, 1–7. [Google Scholar] [PubMed]
- Deans, A.J.; Khanna, K.K.; McNees, C.J.; Mercurio, C.; Heierhorst, J.; McArthur, G.A. Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1-deficient cancers. Cancer Res. 2006, 66, 8219–8226. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, S.; Zangari, M.; Xu, H.; Cao, T.M.; Xu, C.; Wu, Y.; Xiao, F.; Liu, Y.; Yang, Y.; et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget 2010, 1, 22–33. [Google Scholar] [PubMed]
- Chen, J.L.; Li, J.; Kiriluk, K.J.; Rosen, A.M.; Paner, G.P.; Antic, T.; Lussier, Y.A.; Vander Griend, D.J. Deregulation of a Hox protein regulatory network spanning prostate cancer initiation and progression. Clin. Cancer Res. 2012, 18, 4291–4302. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Haas, J.P.; Kim, M.; Sgagias, M.K.; Cowan, K.H. BRCA1-induced apoptosis involves inactivation of ERK1/2 activities. J. Biol. Chem. 2002, 277, 33422–33430. [Google Scholar] [CrossRef] [PubMed]
- Benezra, M.; Chevallier, N.; Morrison, D.J.; MacLachlan, T.K.; El-Deiry, W.S.; Licht, J.D. BRCA1 augments transcription by the NF-kappaB transcription factor by binding to the Rel domain of the p65/RelA subunit. J. Biol. Chem. 2003, 278, 26333–26341. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.E.; Kennedy, R.D.; Mullan, P.B.; Gilmore, P.M.; Carty, M.; Johnston, P.G.; Harkin, D.P. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003, 63, 6221–6228. [Google Scholar] [CrossRef]
- Thangaraju, M.; Kaufmann, S.H.; Couch, F.J. BRCA1 facilitates stress-induced apoptosis in breast and ovarian cancer cell lines. J. Biol. Chem. 2000, 275, 33487–33496. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Pierredon, S.; Wuillemin, C.; Delie, F.; Petignat, P. Acellular fraction of ovarian cancer ascites induce apoptosis by activating JNK and inducing BRCA1, Fas and FasL expression in ovarian cancer cells. Oncoscience 2014, 1, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Reddy, R.B.; Bhat, A.R.; James, B.L.; Govindan, S.V.; Mathew, R.; Ravindra, D.R.; Hedne, N.; Illiayaraja, J.; Kekatpure, V.; Khora, S.S.; et al. Meta-Analyses of Microarray Datasets Identifies ANO1 and FADD as Prognostic Markers of Head and Neck Cancer. PLoS ONE 2016, 11, e0147409. [Google Scholar] [CrossRef] [PubMed]
- Tanic, M.; Zajac, M.; Gomez-Lopez, G.; Benitez, J.; Martinez-Delgado, B. Integration of BRCA1-mediated miRNA and mRNA profiles reveals microRNA regulation of TRAF2 and NFκB pathway. Breast Cancer Res. Treat. 2012, 134, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Rhee, J.K.; Yoo, H.J.; Lee, H.J.; Lee, E.J.; Lee, J.W.; Yu, J.H.; Son, B.H.; Gong, G.; Kim, S.B.; et al. Bioinformatic and metabolomic analysis reveals miR-155 regulates thiamine level in breast cancer. Cancer Lett. 2015, 357, 488–497. [Google Scholar] [CrossRef] [PubMed]
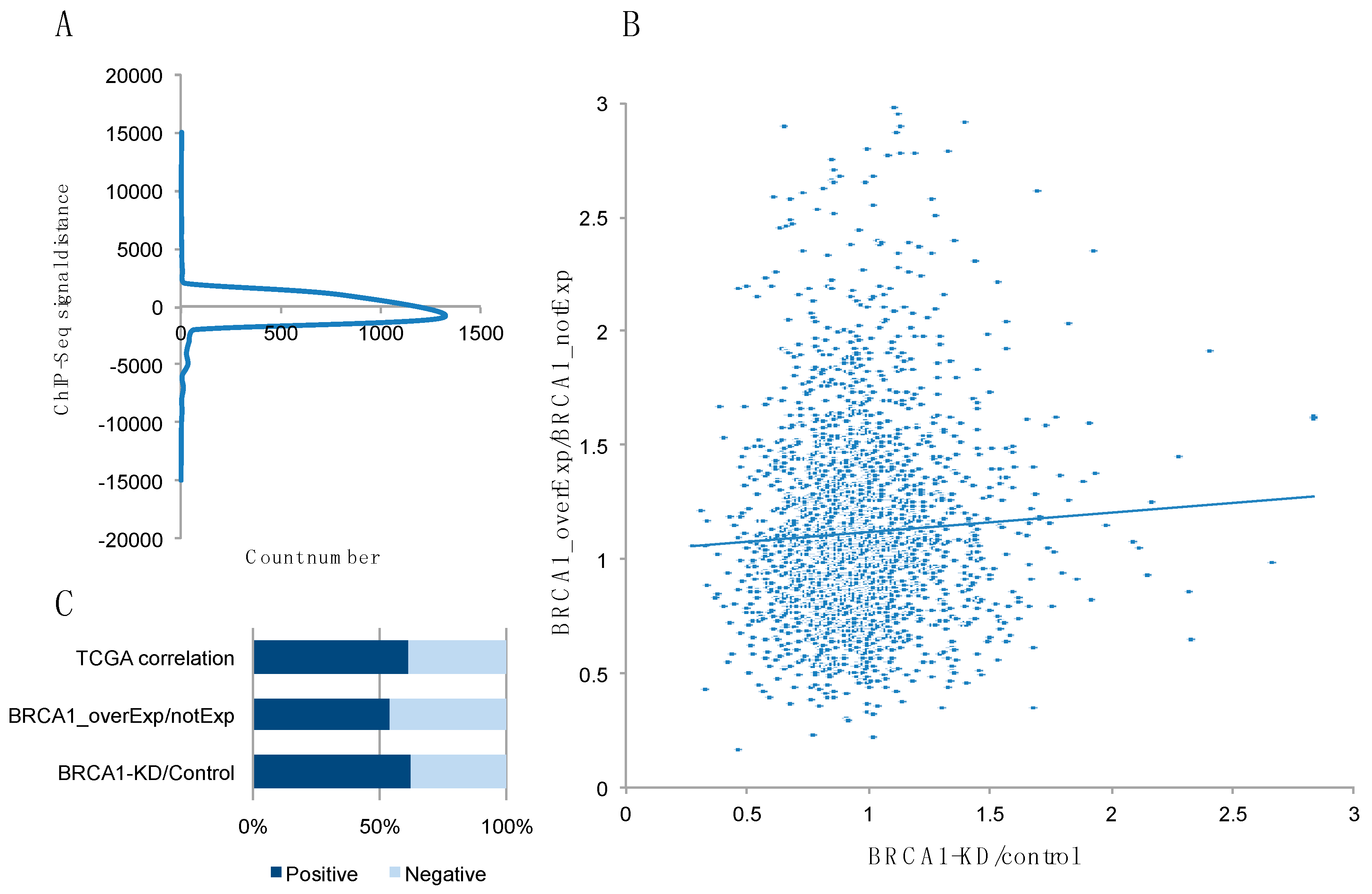
| Gene Name | Function | ENCODE Data (CHIP seq) a | BRCA1 KD/Ctr b | BRCA1 overExp/Ctr c | TCGA Correlation d |
|---|---|---|---|---|---|
| CKS1B | binding to the catalytic subunit of the cyclin-dependent kinases and is essential for their biological function | −319 | 0.66 | 1.90 | 0.29 |
| FADD | an adaptor molecule that interacts with various cell surface receptors and mediates cell apoptotic signals | +152 | 1.06 | 2.10 | 0.24 |
| MEIS2 | binding to HOX or PBX proteins to form dimers, or to a DNA-bound dimer of these proteins and is thought to play a role in stabilization of the homeoprotein-DNA complex for transcriptional regulation | +521,171 | 0.77 | 1.01 | −0.32 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.-D.; Lee, D.G.; Kim, S.; Kang, K.; Rhee, J.-k.; Chang, S. Integrative Bioinformatics and Functional Analyses of GEO, ENCODE, and TCGA Reveal FADD as a Direct Target of the Tumor Suppressor BRCA1. Int. J. Mol. Sci. 2018, 19, 1458. https://doi.org/10.3390/ijms19051458
Nguyen D-D, Lee DG, Kim S, Kang K, Rhee J-k, Chang S. Integrative Bioinformatics and Functional Analyses of GEO, ENCODE, and TCGA Reveal FADD as a Direct Target of the Tumor Suppressor BRCA1. International Journal of Molecular Sciences. 2018; 19(5):1458. https://doi.org/10.3390/ijms19051458
Chicago/Turabian StyleNguyen, Dinh-Duc, Dong Gyu Lee, Sinae Kim, Keunsoo Kang, Je-keun Rhee, and Suhwan Chang. 2018. "Integrative Bioinformatics and Functional Analyses of GEO, ENCODE, and TCGA Reveal FADD as a Direct Target of the Tumor Suppressor BRCA1" International Journal of Molecular Sciences 19, no. 5: 1458. https://doi.org/10.3390/ijms19051458





