Comparative Transcriptome Analysis of Waterlogging-Sensitive and Waterlogging-Tolerant Chrysanthemum morifolium Cultivars under Waterlogging Stress and Reoxygenation Conditions
Abstract
:1. Introduction
2. Results
2.1. Morphological Changes and Ethylene Measurement in “Nannongxuefeng” and “Qinglu” Following Waterlogging Stress
2.2. Transcriptome Sequencing and Read Assembly
2.3. Gene Annotation and Functional Classification
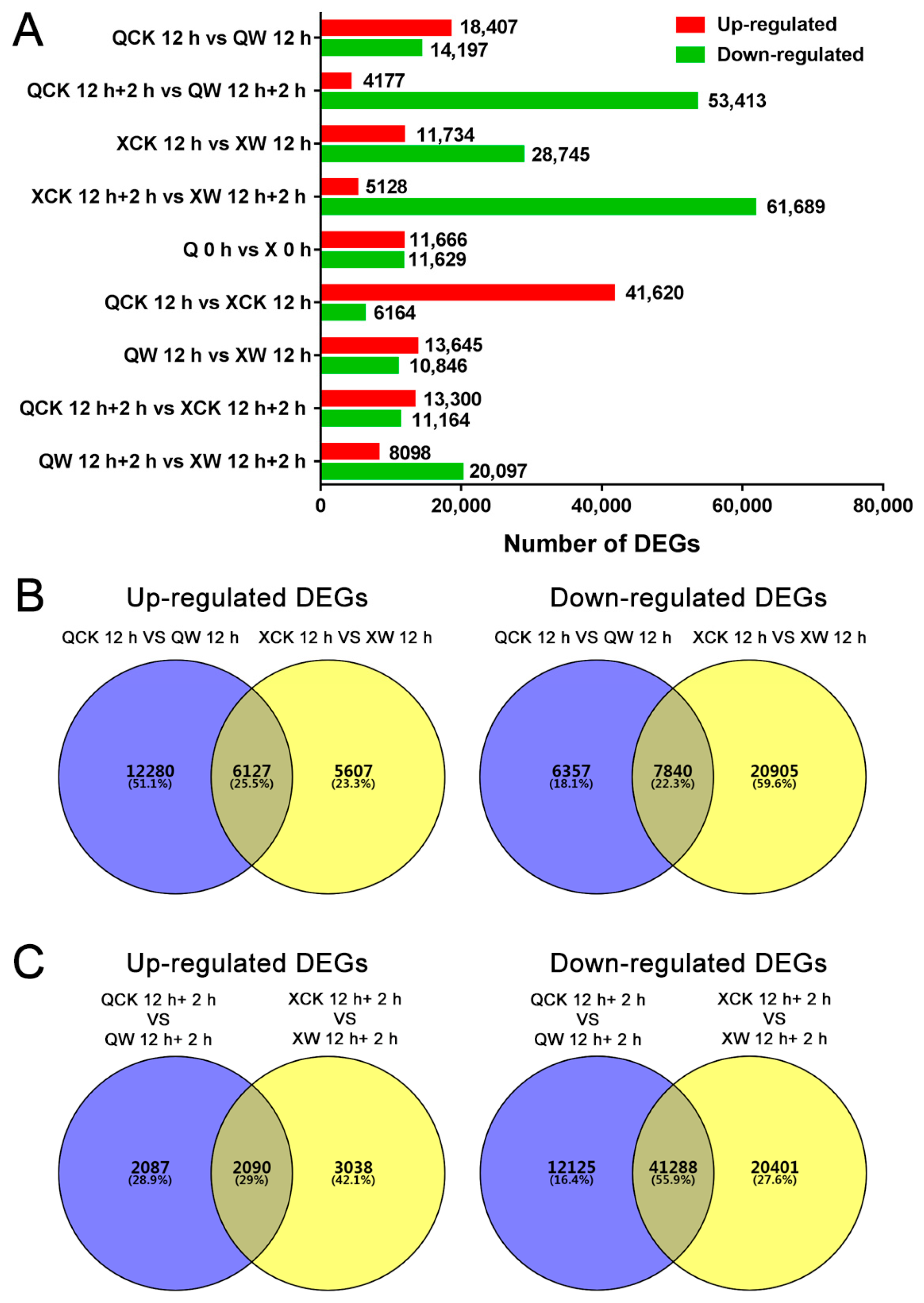
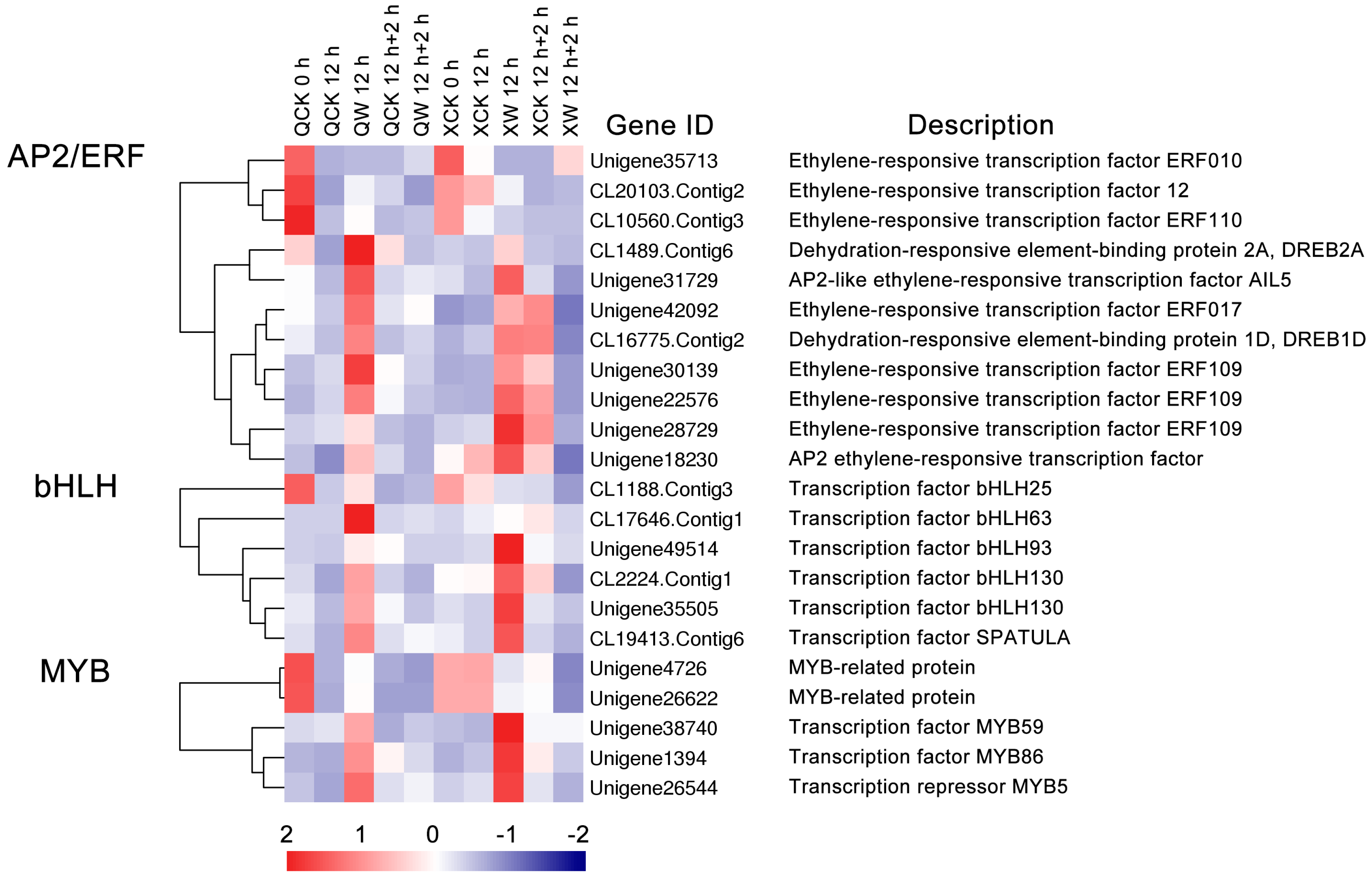
2.4. Identification of Transcription Factors (TFs) Involved in Waterlogging and Recovery in the Two Cultivars
2.5. Hormone Response and Biosynthesis
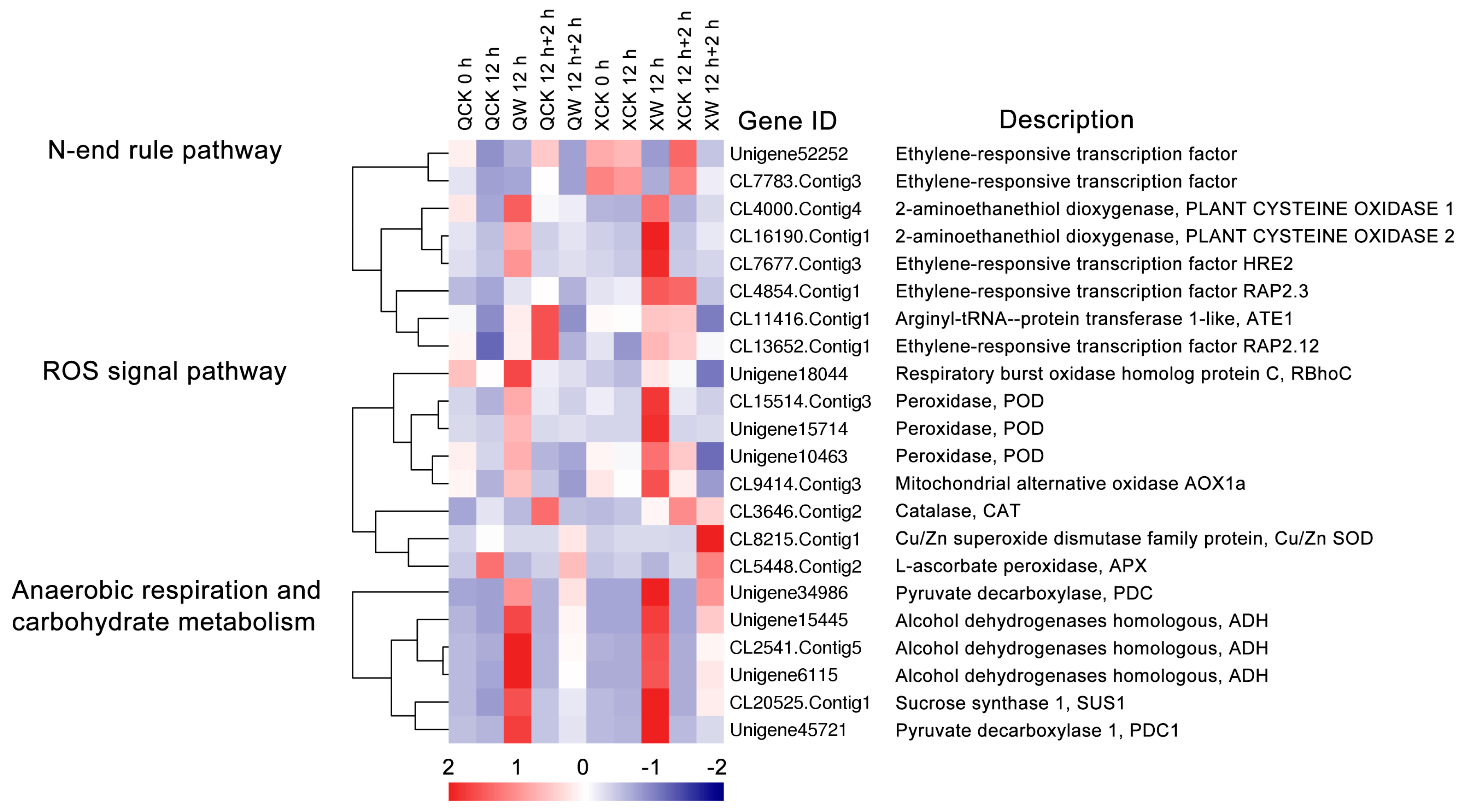
2.6. Other DEGs Involved in Waterlogging and Recovery in the Two Cultivars
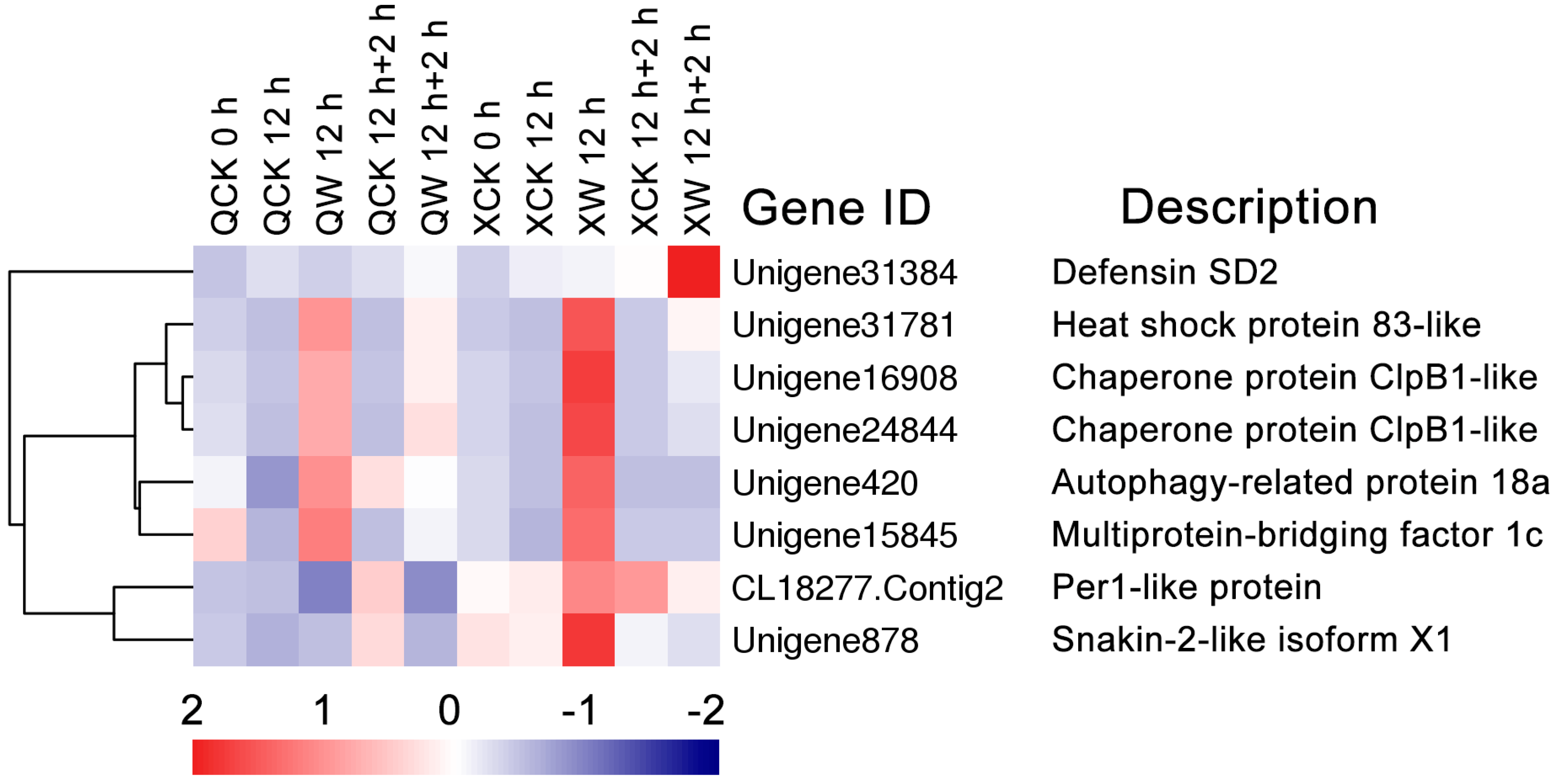
2.7. Differential Transcription of Other Novel Genes Under the Waterlogging and Recovery Conditions
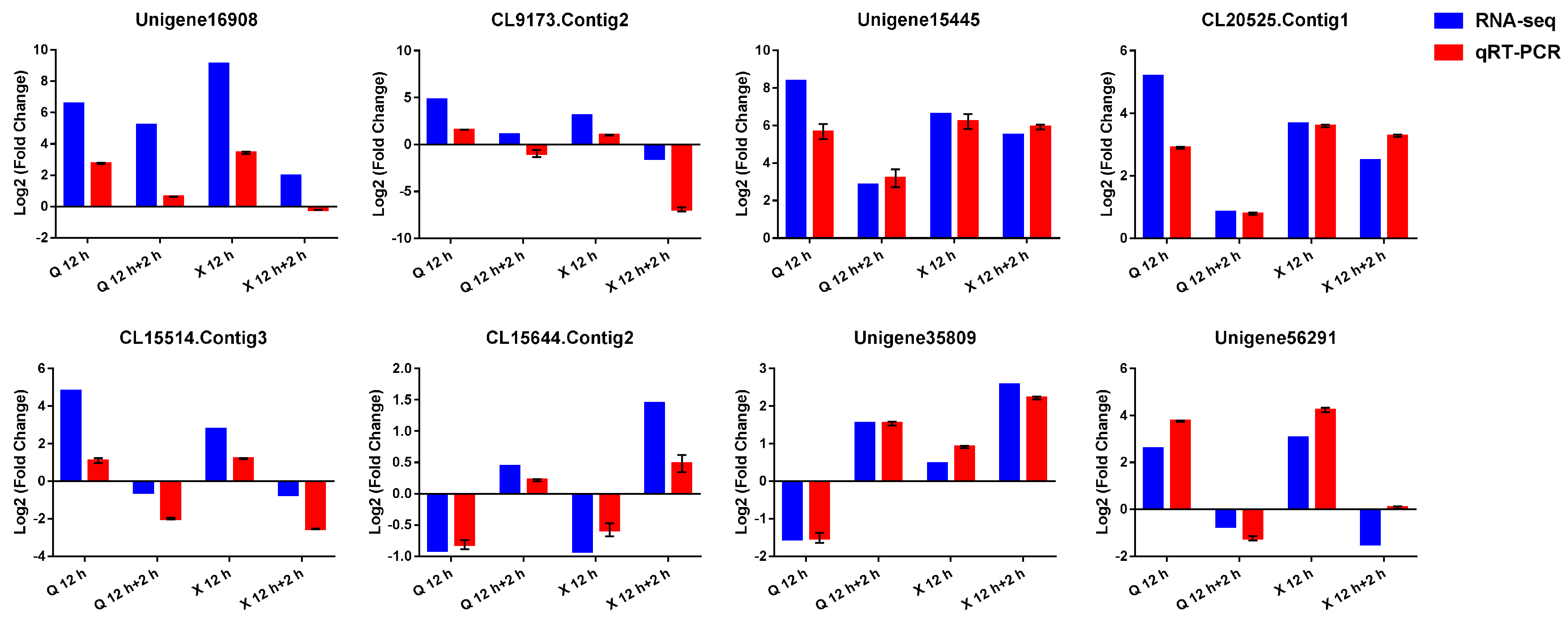
2.8. Verification of RNA-seq Data by Quantitative Real-Time PCR (qRT-PCR)
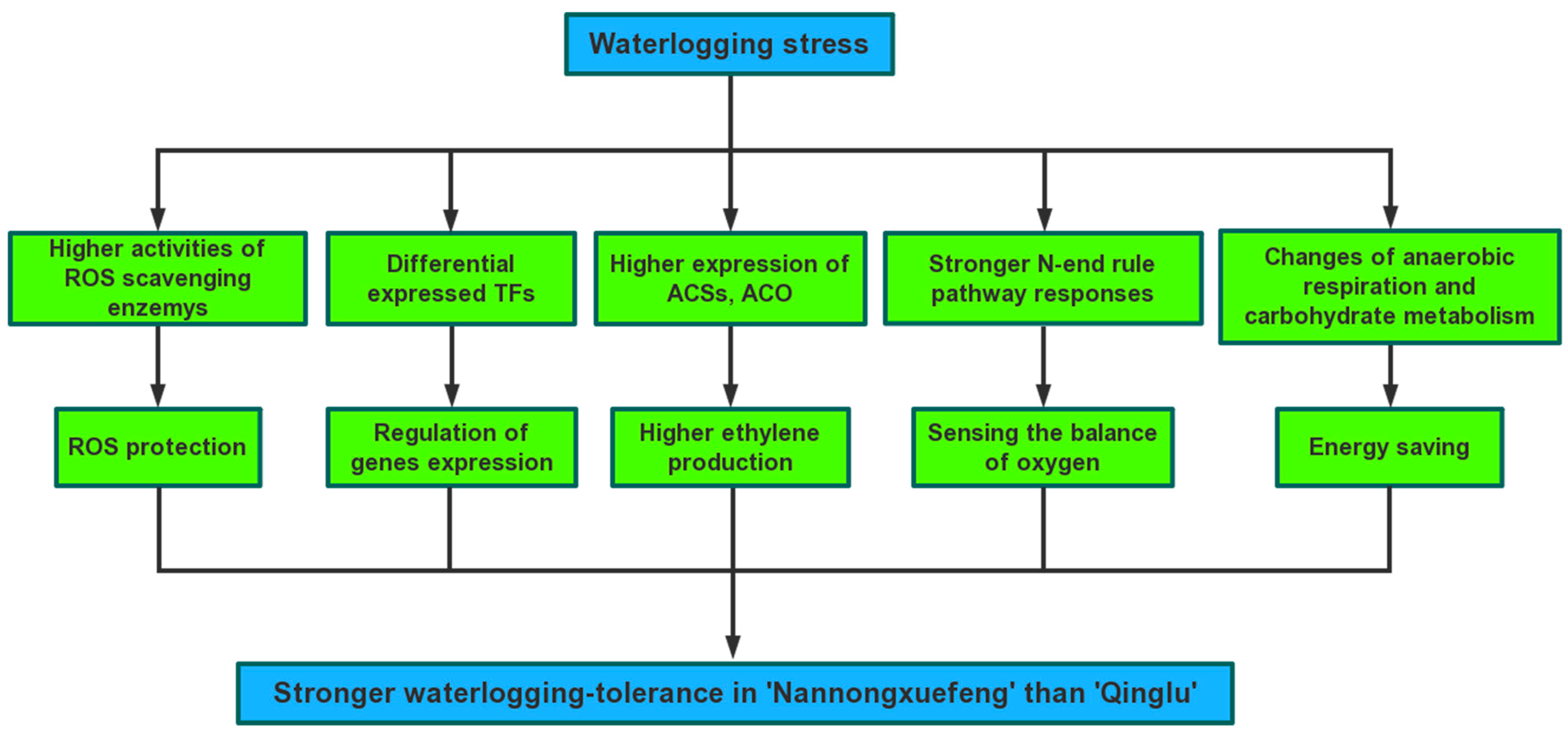
3. Discussion
3.1. Ethylene Results in Phenotypical Differences Between “Nannongxuefeng” and “Qinglu” after Waterlogging
3.2. Changes in the Expression of Transcription Factors under the Waterlogging and Reoxygenation Conditions
3.3. Waterlogging and Recovery Lead to Changes in Hormonal Responses and Biosynthesis-Related DEGs
3.4. ROS Signaling Pathway, Anaerobic Respiration and Carbohydrate Metabolism
3.5. Other Genes Involved in Waterlogging and Reoxygenation
4. Material and Methods
4.1. Plant Materials
4.2. Waterlogging Treatments
4.3. Ethylene Measurements
4.4. RNA Extraction, cDNA Library Construction and Sequencing
4.5. Transcriptome Data Processing and de novo Assembly
4.6. qRT-PCR Validation and Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Jackson, M.B.; Colmer, T.D. Response and adaptation by plants to flooding stress. Ann. Bot. 2005, 96, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Bailey-Serres, J.; Ashikari, M.; Atwell, B.J.; Colmer, T.D.; Fagerstedt, K.; Fukao, T.; Geigenberger, P.; Hebelstrup, K.H.; Hill, R.D.; et al. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Phytol. 2017, 214, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Drew, M. Plant injury and adaptation to oxygen deficiency in the root environment: A review. Plant Soil 1983, 75, 179–199. [Google Scholar] [CrossRef]
- Drew, M.C. Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Biol. 1997, 48, 223–250. [Google Scholar] [CrossRef] [PubMed]
- Kato-Noguchi, H.; Morokuma, M. Ethanolic fermentation and anoxia tolerance in four rice cultivars. J. Plant Physiol. 2007, 164, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Ismond, K.P.; Dolferus, R.; De Pauw, M.; Dennis, E.S.; Good, A.G. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 2003, 132, 1292–1302. [Google Scholar] [CrossRef] [PubMed]
- Vodnik, D.; Strajnar, P.; Jemc, S.; Macek, I. Respiratory potential of maize (Zea mays L.) roots exposed to hypoxia. Environ. Exp. Bot. 2009, 65, 107–110. [Google Scholar] [CrossRef]
- Lee, S.C.; Mustroph, A.; Sasidharan, R.; Vashisht, D.; Pedersen, O.; Oosumi, T.; Voesenek, L.A.; Bailey-Serres, J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011, 190, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Vaultier, M.N.; Rochat, C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 2004, 55, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, J.L.; Zhang, X.Q.; Wei, H.; Cui, L.J. Effects of heat acclimation pretreatment on changes of membrane lipid peroxidation, antioxidant metabolites, and ultrastructure of chloroplasts in two cool-season turfgrass species under heat stress. Environ. Exp. Bot. 2006, 56, 274–285. [Google Scholar] [CrossRef]
- Selote, D.S.; Bharti, S.; Khanna-Chopra, R. Drought acclimation reduces O2− accumulation and lipid peroxidation in wheat seedlings. Biochem. Biophys. Res. Commun. 2004, 314, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Fleury, C.; Mignotte, B.; Vayssiere, J.L. Mitochondrial reactive oxygen species in cell death signaling. Biochimie 2002, 84, 131–141. [Google Scholar] [CrossRef]
- Zhang, G.P.; Tanakamaru, K.; Abe, J.; Morita, S. Influence of waterlogging on some anti-oxidative enzymatic activities of two barley genotypes differing in anoxia tolerance. Acta. Physiol. Plant. 2007, 29, 171–176. [Google Scholar] [CrossRef]
- Li, C.Y.; Jiang, D.; Wollenweber, B.; Li, Y.; Dai, T.B.; Cao, W.X. Waterlogging pretreatment during vegetative growth improves tolerance to waterlogging after anthesis in wheat. Plant Sci. 2011, 180, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Srivastava, J.P. Antioxidative defense system in pigeonpea roots under waterlogging stress. Acta. Physiol. Plant. 2012, 34, 515–522. [Google Scholar] [CrossRef]
- Li, H.W.; Cai, J.; Liu, F.L.; Jiang, D.; Dai, T.B.; Cao, W.X. Generation and scavenging of reactive oxygen species in wheat flag leaves under combined shading and waterlogging stress. Funct. Plant Biol. 2012, 39, 71–81. [Google Scholar] [CrossRef]
- Sasidharan, R.; Voesenek, L.A. Ethylene-mediated acclimations to flooding stress. Plant Physiol. 2015, 169, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Rieu, I.; Cristescu, S.M.; Harren, F.J.M.; Huibers, W.; Voesenek, L.A.C.J.; Mariani, C.; Vriezen, W.H. RP-ACS1, a flooding-induced 1-aminocyclopropane-1-carboxylate synthase gene of Rumex palustris, is involved in rhythmic ethylene production. J. Exp. Bot. 2005, 56, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Visser, E.J.W.; Bogemann, G.M.; Blom, C.W.P.M.; Voesenek, L.A.C.J. Ethylene accumulation in waterlogged Rumex plants promotes formation of adventitious roots. J. Exp. Bot. 1996, 47, 403–410. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Mahmood, T. Functional role of DREB and ERF transcription factors: Regulating stress-responsive network in plants. Acta. Physiol. Plant. 2015, 37, 178. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.; Bailey-Serres, J. Plant biology: Genetics of high-rise rice. Nature 2009, 460, 959–960. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [PubMed]
- Niroula, R.K.; Pucciariello, C.; Ho, V.T.; Novi, G.; Fukao, T.; Perata, P. SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. Plant J. 2012, 72, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Colmer, T.D. Plant tolerance of flooding stress - recent advances. Plant Cell Environ. 2014, 37, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Septiningsih, E.M.; Pamplona, A.M.; Sanchez, D.L.; Neeraja, C.N.; Vergara, G.V.; Heuer, S.; Ismail, A.M.; Mackill, D.J. Development of submergence-tolerant rice cultivars: The Sub1 locus and beyond. Ann. Bot. 2009, 103, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Pena-Castro, J.M.; van Zanten, M.; Lee, S.C.; Patel, M.R.; Voesenek, L.A.J.C.; Fukao, T.; Bailey-Serres, J. Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J. 2011, 67, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Papdi, C.; Perez-Salamo, I.; Joseph, M.P.; Giuntoli, B.; Bogre, L.; Koncz, C.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the Ethylene Response Factor-VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Hess, N.; Klode, M.; Anders, M.; Sauter, M. The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiol. Plant. 2011, 143, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Howell, K.A.; Carroll, A.; Ivanova, A.; Millar, A.H.; Whelan, J. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 2009, 151, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Branco-Price, C.; Kaiser, K.A.; Jang, C.J.H.; Larive, C.K.; Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008, 56, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Dai, Y.; Xie, L.; Yu, L.; Zhou, Y.; Lai, Y.; Yang, Y.; Xu, L.; Chen, Q.; Xiao, S. Jasmonate regulates plant responses to postsubmergence reoxygenation through transcriptional activation of antioxidant synthesis. Plant Physiol. 2017, 173, 1864–1880. [Google Scholar] [CrossRef] [PubMed]
- Teixeira da Silva, J.A.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum biotechnology: Quo vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Yin, D.; Chen, S.; Chen, F.; Guan, Z.; Fang, W. Morphological and physiological responses of two chrysanthemum cultivars differing in their tolerance to waterlogging. Environ. Exp. Bot. 2009, 67, 87–93. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Li, P.; Guan, Z.; Fang, W.; Chen, F. Genetic variation and association mapping of waterlogging tolerance in chrysanthemum. Planta 2016, 244, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Ni, D.; Song, L.; Zhang, Z. Isolation of an alcohol dehydrogenase cDNA from and characterization of its expression in chrysanthemum under waterlogging. Plant Sci. 2013, 212, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.C.J.; Jackson, M.B.; Toebes, A.H.W.; Huibers, W.; Vriezen, W.H.; Colmer, T.D. De-submergence induced ethylene production in Rumex palustris: Regulation and ecophysiological significance. Plant J. 2003, 33, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lu, X.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol. Plant 2015, 8, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Foley, R.C.; Onate-Sanchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Zhang, Z.G.; He, X.J.; Zhou, H.L.; Wen, Y.X.; Dai, J.X.; Zhang, J.S.; Chen, S.Y. A rice transcription factor OsbHLH1 is involved in cold stress response. Theor. Appl. Genet. 2003, 107, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Mustroph, A. Plant oxygen sensing is mediated by the N-end rule pathway: A milestone in plant anaerobiosis. Plant Cell 2011, 23, 4173–4183. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Kim, D.S.; Park, J.H.; Kim, D.Y.; Seo, Y.W. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 2007, 129, 375–385. [Google Scholar] [CrossRef]
- Giuntoli, B.; Lee, S.C.; Licausi, F.; Kosmacz, M.; Oosumi, T.; van Dongen, J.T.; Bailey-Serres, J.; Perata, P. A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 2014, 12, e1001950. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Sirikhachornkit, A.; Pimjan, R.; Sonthirod, C.; Sangsrakru, D.; Yoocha, T.; Tangphatsornruang, S.; Srinives, P. Elucidation of the molecular responses to waterlogging in jatropha roots by transcriptome profiling. Front. Plant Sci. 2014, 5, 658. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.A.; Llewellyn, D.J.; Dennis, E.S.; Wilson, I.W. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, M.; Ji, J.; Xu, Q.; Qi, X.; Chen, X. Comparative RNA-seq based transcriptome profiling of waterlogging response in cucumber hypocotyls reveals novel insights into the de novo adventitious root primordia initiation. BMC Plant Biol. 2017, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Visser, E.J.W.; Cohen, J.D.; Barendse, G.W.M.; Blom, C.W.P.M.; Voesenek, L.A.C.J. An ethylene-mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palustris Sm. Plant Physiol. 1996, 112, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.R.; Udvardi, M.K.; Kazan, K. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Caarls, L.; Van der Does, D.; Hickman, R.; Jansen, W.; Van Verk, M.C.; Proietti, S.; Lorenzo, O.; Solano, R.; Pieterse, C.M.J.; Van Wees, S.C.M. Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. 2017, 58, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.G.; Wang, M.M.; Gong, Z.Y.; Fang, F.; Sun, N.J.; Li, X.; Grierson, D.; Yin, X.R.; Chen, K.S. Involvement of DkTGA1 transcription factor in anaerobic response leading to persimmon fruit postharvest de-astringency. PLoS ONE 2016, 11, e0155916. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Liu, J.; Dai, T.; Jing, Q.; Cao, W.; Jiang, D. Alterations in photosynthesis and antioxidant enzyme activity in winter wheat subjected to post-anthesis waterlogging. Photosynthetica 2008, 46, 21–27. [Google Scholar] [CrossRef]
- Blokhina, O.; Fagerstedt, K.V. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol. Bioch. 2010, 48, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Amor, Y.; Babiychuk, E.; Inze, D.; Levine, A. The involvement of poly (ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett. 1998, 440, 1–7. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Jaruga, P.; Birincioglu, M.; Rodriguez, H. Free radical-induced damage to DNA: Mechanisms and measurement. Free Radic. Biol. Med. 2002, 32, 1102–1115. [Google Scholar] [CrossRef]
- Roldan-Arjona, T.; Ariza, R.R. Repair and tolerance of oxidative DNA damage in plants. Mutat. Res-Rev. Mutat. 2009, 681, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Vanderauwera, S.; Suzuki, N.; Miller, G.; van de Cotte, B.; Morsa, S.; Ravanat, J.L.; Hegie, A.; Triantaphylides, C.; Shulaev, V.; Van Montagu, M.C.E.; et al. Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1711–1716. [Google Scholar] [CrossRef] [PubMed]
- Subbaiah, C.C.; Sachs, M.M. Molecular and cellular adaptations of maize to flooding stress. Ann. Bot. 2003, 91, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Biemelt, S.; Keetman, U.; Albrecht, G. Re-aeration following hypoxia or anoxia leads to activation of the antioxidative defense system in roots of wheat seedlings. Plant Physiol. 1998, 116, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Rawyler, A.; Arpagaus, S.; Braendle, R. Impact of oxygen stress and energy availability on membrane stability of plant cells. Ann. Bot. 2002, 90, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lyu, D.; Jia, L.; He, J.; Qin, S. Physiological and de novo transcriptome analysis of the fermentation mechanism of Cerasus sachalinensis roots in response to short-term waterlogging. BMC Genom. 2017, 18, 649. [Google Scholar] [CrossRef] [PubMed]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Woeste, K.E.; Theologis, A.; Kieber, J.J. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 1998, 95, 4766–4771. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Sun, J.; Chen, S.; Gao, J.; Dong, B.; Liu, Y.; Xia, X.; Wang, Y.; Liao, Y.; Teng, N.; et al. A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance. BMC Genom. 2014, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]


| Treatment | Ethylene Production (pl·fw−1·h−1) | |||||
|---|---|---|---|---|---|---|
| Time (Hours) | ||||||
| 0 h | 3 h | 6 h | 12 h | 24 h | 12 h + 2 h | |
| ”Qinglu” | ||||||
| Control | 1.49 ± 0.22 a | 1.28 ± 0.12 c | 1.74 ± 0.36 c | 1.14 ± 0.05 c | 1.48 ± 0.45 c | 1.43 ± 0.08 b |
| Waterlogging | 1.49 ± 0.22 a | 2.56 ± 0.10 b | 2.75 ± 0.11 b | 2.3 ± 0.18 b | 4.53 ± 0.23 b | 1.90 ± 0.57 b |
| Waterlogging + ACC | 1.49 ± 0.22 a | 27.67 ± 2.86 a | 39.30 ± 2.39 a | 23.16 ± 1.57 a | 73.78 ± 1.72 a | 55.89 ± 3.38 a |
| Waterlogging + AVG | 1.49 ± 0.22 a | 0.46 ± 0.02 d | 0.37 ± 0.01 d | 0.31 ± 0.02 d | 1.24 ± 0.15 c | 0.88 ± 0.05 c |
| “Nannongxuefeng” | ||||||
| Control | 2.35 ± 0.32 a | 2.19 ± 0.19 b | 3.33 ± 0.60 c | 2.48 ± 0.31 b | 2.40 ± 0.17 d | 2.19 ± 0.12 c |
| Waterlogging | 2.35 ± 0.32 a | 2.93 ± 0.04 b | 4.35 ± 0.26 b | 3.03 ± 0.45 b | 17.03 ± 2.37 b | 5.98 ± 0.28 b |
| Waterlogging + ACC | 2.35 ± 0.32 a | 42.93 ± 0.59 a | 51.85 ± 4.43 a | 32.83 ± 4.94 a | 74.22 ± 0.93 a | 62.58 ± 4.24 a |
| Waterlogging + AVG | 2.35 ± 0.32 a | 0.79 ± 0.07 c | 1.24 ± 0.14 d | 0.54 ± 0.07 c | 3.61 ± 0.61 c | 1.17 ± 0.07 d |
| Sample | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Total Clean Bases (Gb) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Clean Reads Ratio (%) |
|---|---|---|---|---|---|---|
| Q 0 h | 67.41 | 67.01 | 6.70 | 97.31 | 93.36 | 99.41 |
| QCK 12 h | 68.13 | 65.06 | 6.51 | 97.55 | 94.19 | 95.48 |
| QW 12 h | 65.86 | 65.43 | 6.54 | 98.02 | 95.02 | 99.34 |
| QCK 12 h + 2 h | 65.86 | 65.43 | 6.54 | 97.68 | 94.25 | 99.35 |
| QW 12 h + 2 h | 67.39 | 65.97 | 6.60 | 97.67 | 94.14 | 97.9 |
| X 0 h | 65.86 | 65.48 | 6.55 | 97.88 | 94.72 | 99.42 |
| XCK 12 h | 65.86 | 65.48 | 6.55 | 97.93 | 94.84 | 99.42 |
| XW 12 h | 65.86 | 65.02 | 6.50 | 98.09 | 95.17 | 98.72 |
| XCK 12 h + 2 h | 65.86 | 65.52 | 6.55 | 98.06 | 95.1 | 99.48 |
| XW 12 h + 2 h | 68.13 | 67.03 | 6.70 | 97.69 | 94.18 | 98.38 |
| Sample | Total Number | Total Length | Mean Length | N50 | N70 | N90 | GC (%) |
|---|---|---|---|---|---|---|---|
| Q 0 h | 154,416 | 91,195,520 | 590 | 924 | 490 | 239 | 40.91 |
| QCK 12 h | 76,075 | 39,322,976 | 516 | 738 | 389 | 220 | 41.74 |
| QW 12 h | 134,241 | 81,313,892 | 605 | 950 | 511 | 246 | 39.97 |
| QCK 12 h + 2 h | 149,040 | 90,100,401 | 604 | 956 | 511 | 244 | 40.36 |
| QW 12 h + 2 h | 70,777 | 31,838,872 | 449 | 567 | 327 | 209 | 42.13 |
| X 0 h | 149,070 | 91,615,155 | 614 | 983 | 522 | 245 | 40.02 |
| XCK 12 h | 151,868 | 94,221,259 | 620 | 1002 | 530 | 247 | 39.96 |
| XW 12 h | 148,994 | 87,558,806 | 587 | 898 | 489 | 241 | 40.18 |
| XCK 12 h + 2 h | 147,947 | 89,776,249 | 606 | 955 | 517 | 246 | 40.21 |
| XW 12 h + 2 h | 50,049 | 21,171,969 | 423 | 510 | 320 | 209 | 42.44 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, N.; Li, C.; Yan, Y.; Cao, W.; Song, A.; Wang, H.; Chen, S.; Jiang, J.; Chen, F. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Waterlogging-Tolerant Chrysanthemum morifolium Cultivars under Waterlogging Stress and Reoxygenation Conditions. Int. J. Mol. Sci. 2018, 19, 1455. https://doi.org/10.3390/ijms19051455
Zhao N, Li C, Yan Y, Cao W, Song A, Wang H, Chen S, Jiang J, Chen F. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Waterlogging-Tolerant Chrysanthemum morifolium Cultivars under Waterlogging Stress and Reoxygenation Conditions. International Journal of Molecular Sciences. 2018; 19(5):1455. https://doi.org/10.3390/ijms19051455
Chicago/Turabian StyleZhao, Nan, Chuanwei Li, Yajun Yan, Wen Cao, Aiping Song, Haibin Wang, Sumei Chen, Jiafu Jiang, and Fadi Chen. 2018. "Comparative Transcriptome Analysis of Waterlogging-Sensitive and Waterlogging-Tolerant Chrysanthemum morifolium Cultivars under Waterlogging Stress and Reoxygenation Conditions" International Journal of Molecular Sciences 19, no. 5: 1455. https://doi.org/10.3390/ijms19051455




