Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI
Abstract
:1. Introduction
2. Results and Discussion
2.1. Ammonia Production
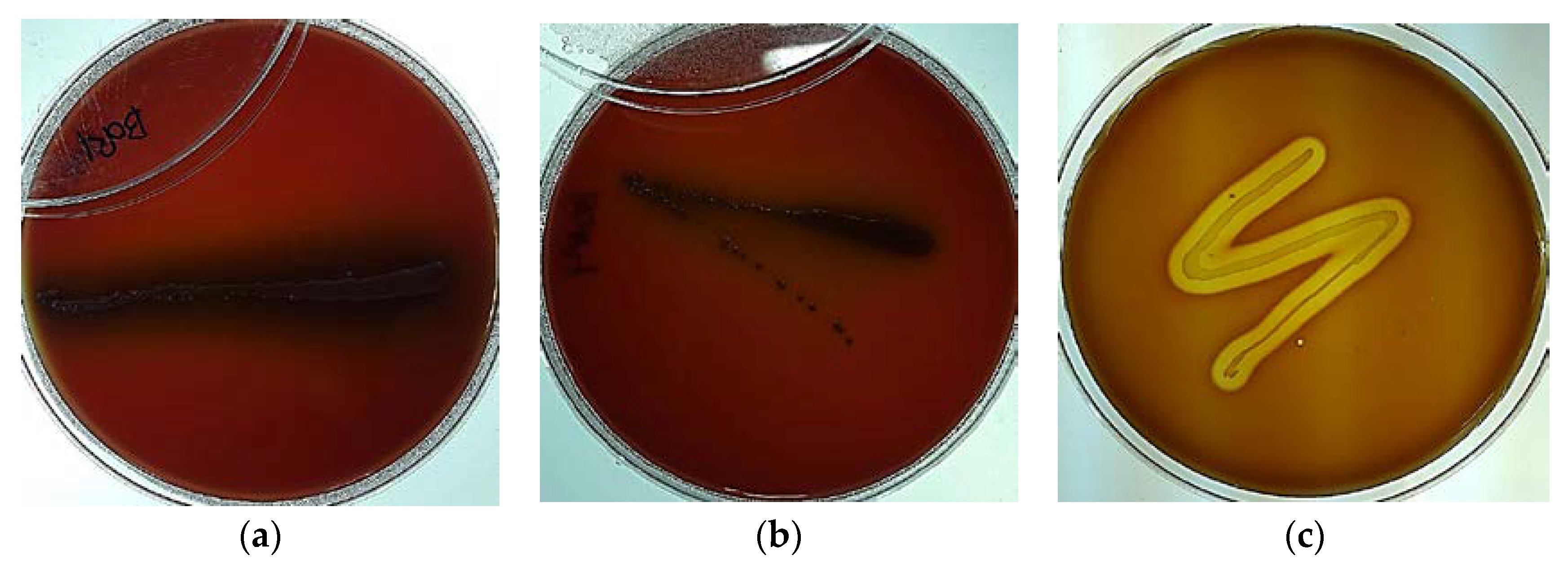
2.2. Hemolytic Property Test
2.3. Evaluation of Biogenic Amine Production
2.4. Antimicrobial Susceptibility and Antibiotic Resistance Transferability
2.4.1. Antibiotic Susceptibility
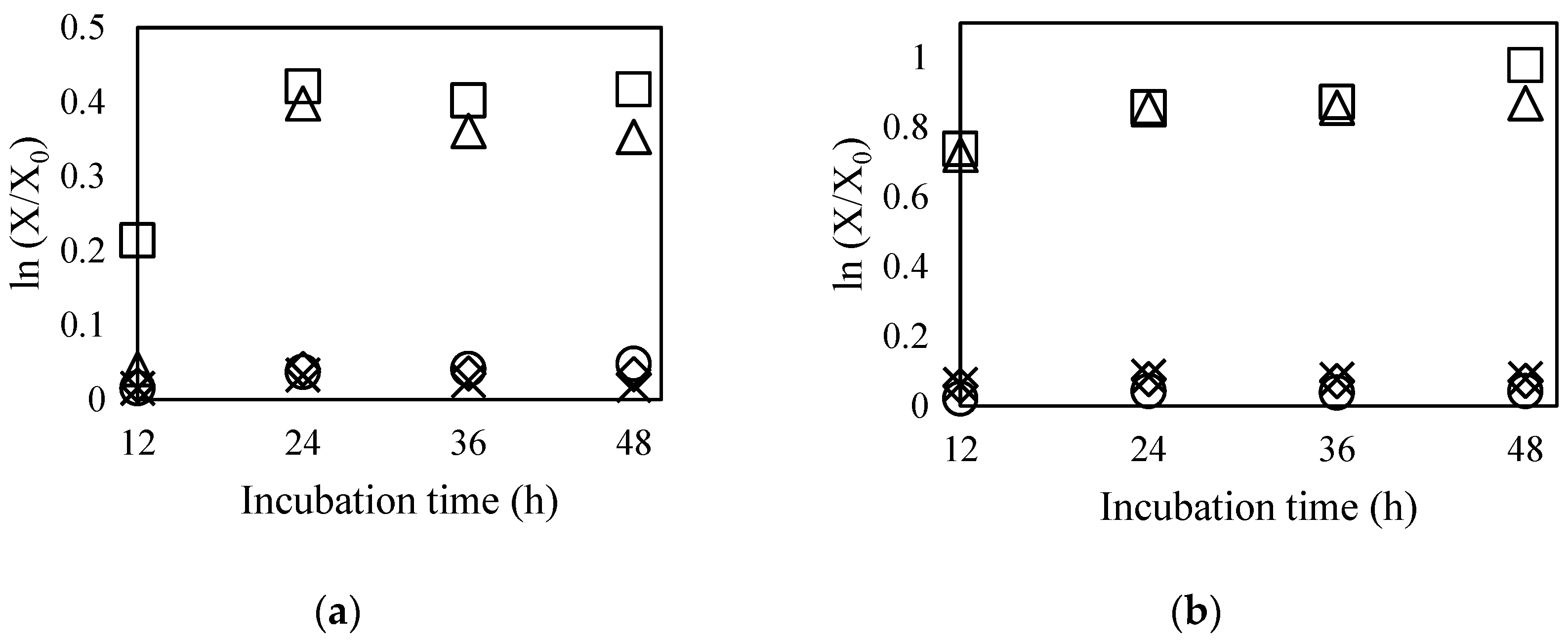
2.4.2. Antibiotic Resistance Transferability
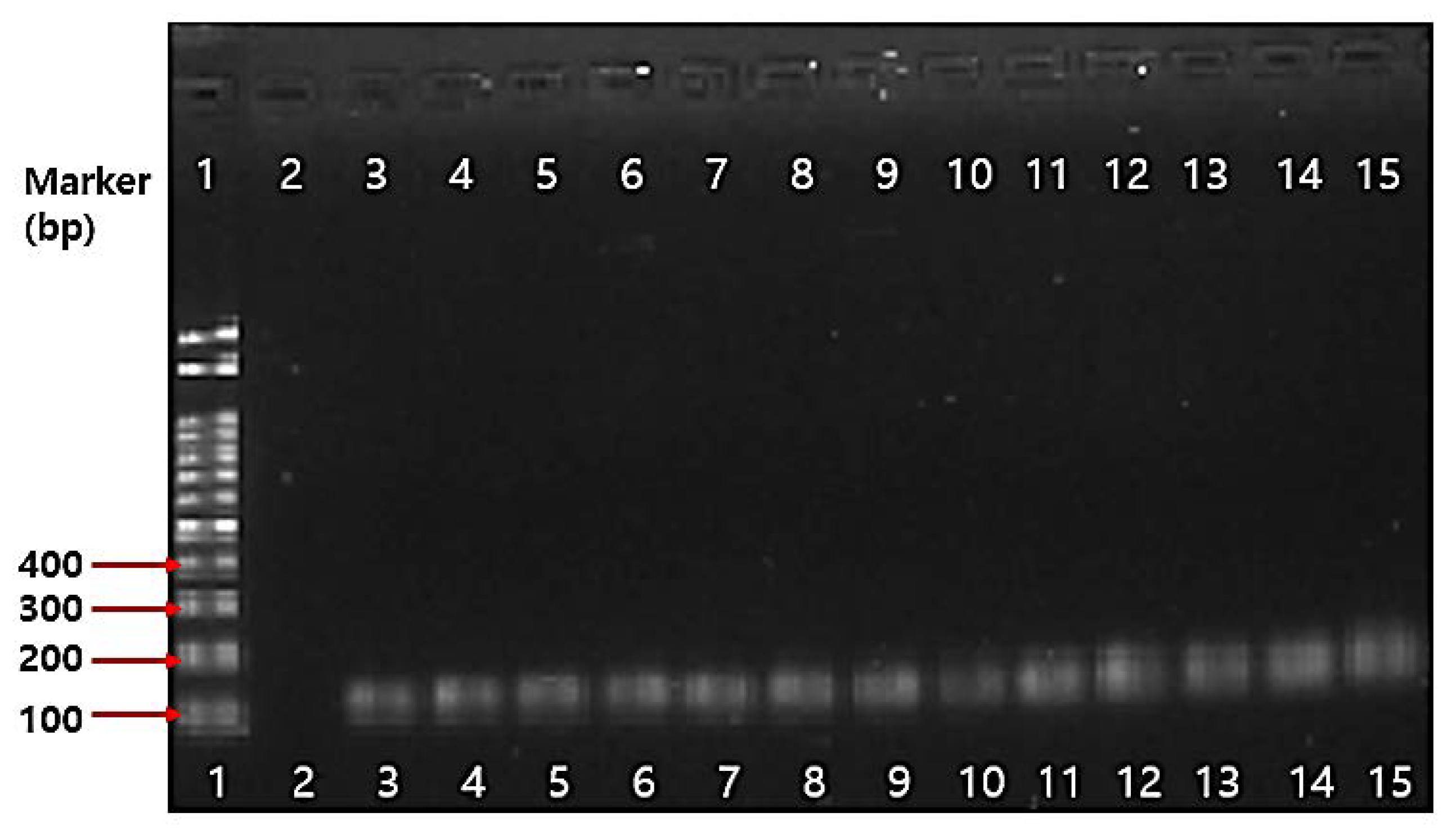
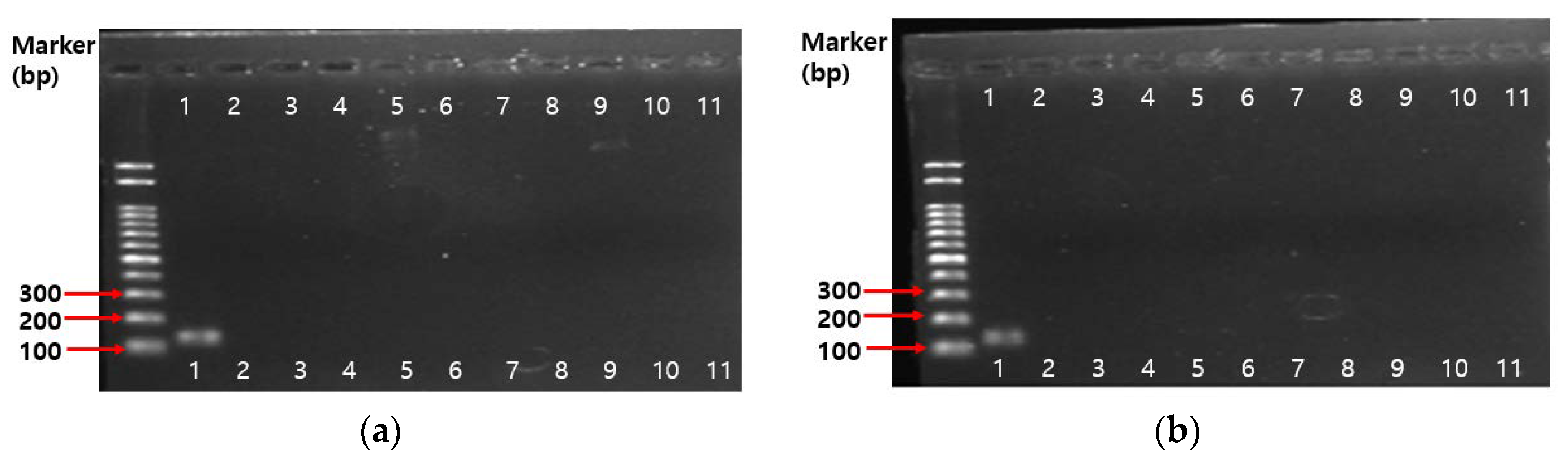
2.4.3. PCR Results on Antibiotic Resistance Genes
2.5. Mucin Degradation
2.6. Genetic Stability
2.7. Virulence Factors
3. Materials and Methods
3.1. Microorganisms
3.2. Ammonia Production Test
3.3. Hemolytic Test
3.4. Biogenic Amine Production Test
3.5. Antimicrobial Susceptibility and Antibiotic Resistance Transferability Test
3.5.1. Antimicrobial Agents
3.5.2. Antimicrobial Susceptibility Test
3.5.3. Antibiotic Resistance Transferability Test
3.5.4. PCR Assay on Antibiotic Resistance Genes
3.6. Mucin Degradation Test
3.7. Genetic Stability Test
3.8. Virulence Factors Researching
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lilly, D.M.; Stillwell, R.H. Probiotics: Growth-Promoting Factors Produced by Microorganisms. Science 1965, 147, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Probiotics: In Depth. Available online: https://nccih.nih.gov/health/probiotics/introduction.htm (accessed on 3 April 2018).
- Food and Agriculture Organization-World Health Organization (FAO/WHO). Report on Joint FAO/WHO Guidelines for the Evaluation of Probiotics in Food. 2002. Available online: http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed on 3 April 2018).
- European Food and Feed Cultures Association (EFFCA). Definition of Microbial Food Culture (MFC); EFFCA: Brussels, Belgium, 2003; Available online: http://www.effca.org/content/food-culture (accessed on 3 April 2018).
- Akhter, N.; Wu, B.; Memon, A.M.; Mohsin, M. Probiotics and Prebiotics Associated with Aquaculture: A Review. Fish Shellfish Immunol. 2015, 45, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.C.; Quigley, E.M.; Lacy, B.E.; Lembo, A.J.; Saito, Y.A.; Schiller, L.R.; Soffer, E.E.; Spiegel, B.M.; Moayyedi, P. Efficacy of Prebiotics, Probiotics and Synbiotics in Irritable Bowel Syndrome and Chronic Idiopathic Constipation: Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2014, 109, 1547–1561. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, B. Bile Acid–Microbiota Crosstalk in Gastrointestinal Inflammation and Carcinogenesis: A Role for Bifidobacteria and Lactobacilli? Nat. Rev. Gastroenterol Hepatol. 2018, 15, 205. [Google Scholar] [CrossRef] [PubMed]
- Lukjancenko, O.; Ussery, D.W.; Wassenaar, T.M. Comparative genomics of Bifidobacterium, Lactobacillus and Related Probiotic Genera. Microb. Ecol. 2012, 63, 651–673. [Google Scholar] [CrossRef] [PubMed]
- Global Market Insights Inc. Probiotics Market Size to Exceed USD 64 Billion by 2023: Global Market Insights Inc. Available online: https://www.prnewswire.com/news-releases/probiotics-market-size-to-exceed-usd-64-billion-by-2023-global-market-insights-inc-578769201.html (accessed on 3 April 2018).
- EFSA. Update of the List of QPS-Recommended Biological Agents Intentionally Added to Food or Feed as Notified to EFSA 4: Suitability of Taxonomic Units Notified to EFSA until March 2016. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/4522 (accessed on 3 April 2018).
- Anadón, A.; Martínez-Larrañaga, M.R.; Ares, I.; Martínez, M.A. Nutraceuticals; Efficacy, Safety and Toxicity. Chapter 55—Probiotics: Safety and Toxicity Considerations; Hill-Parks, E., Ed.; Academic Press: London, UK, 2016; pp. 777–798. ISBN 978-0-12-802147-7. [Google Scholar]
- Huys, G.; Botteldoorn, N.; Delvigne, F.; de Vuyst, L.; Heyndrickx, M.; Pot, B.; Dubois, J.J.; Daube, G. Microbial Characterization of Probiotics-Advisory Report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol. Nutr. Food Res. 2013, 57, 1479–1504. [Google Scholar] [CrossRef] [PubMed]
- Shokryazdan, P.; Jahromi, M.F.; Liang, J.B.; Kalavathy, R.; Sieo, C.C.; Ho, Y.W. Safety Assessment of Two New Lactobacillus Strains as Probiotic for Human Using a Rat Model. PLoS ONE 2016, 11, e0159851. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Xu, H.; Aguilar, Z.P.; Peng, S.; Dong, S.; Wang, B.; Li, P.; Chen, T.; Xu, F.; Wei, H. Safety Assessment and Probiotic Evaluation of Enterococcus faecium Isolated from Sourdough. J. Food Sci. 2013, 78, M587–M593. [Google Scholar] [CrossRef] [PubMed]
- Endres, J.R.; Clewell, A.; Jade, K.A.; Farber, T.; Hauswirth, J.; Schauss, A.G. Safety Assessment of a Proprietary Preparation of a Novel Probiotic, Bacillus coagulans, as a Food Ingredient. Food Chem. Toxicol. 2009, 47, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Akkermans, L.M.A.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety Assessment of Probiotics for Human Use. Gut Microbes. 2010, 1, 164–185. [Google Scholar] [CrossRef] [PubMed]
- Salyers, A.A.; Gupta, A.; Wang, Y. Human Intestinal Bacteria as Reservoirs for Antibiotic Resistance Genes. Trends Microbiol. 2004, 12, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.O.A.; Dantas, G.; Church, G.M. Functional Characterization of the Antibiotic Resistance Reservoir in the Human. Science 2009, 325, 1128–1131. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-Throughput Sequencing Reveals the Incomplete, Short-Term, Recovery of the Infant Gut Microbiota Following Parenteral Antibiotic Treatment with Ampicillin and Gentamicin. Antimicrob. Agents. Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallani, M.; Young, D.; Scott, J.; Norin, E.; Amarri, S.; Adam, R.; Aguilera, M.; Khanna, S.; Gil, A.; Edwards, C.A.; et al. Intestinal microbiota of 6-week-old infants across Europe: Geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’Sullivan, O.; Fouhy, F.; Clarke, S.F.; O’Toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Imperial, I.C.V.J.; Ibana, J.A. Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Kwon, B.; Ku, S.; Ji, G.E. The Efficacy of Bifidobacterium longum BORI and Lactobacillus acidophilus AD031 Probiotic Treatment in Infants with Rotavirus Infection. Nutrients 2017, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.M.; Ji, G.E.; Cho, S.H.; Park, M.S.; Lee, H.J. Characterization of a Bifidobacterium longum BORI Dipeptidase Belonging to the U34 Family. Appl. Environ. Microbiol. 2007, 73, 5598–5606. [Google Scholar] [CrossRef] [PubMed]
- JI, G.E. Development of Bifidobacterium sp. BGN4 and BORI with Novel Probiotic Activity. Int. Meet. Microbiol. Soc. Korea 2005, 5, 81–84. [Google Scholar]
- Ku, S.; Park, M.S.; Ji, G.E.; You, H.J. Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism. Int. J. Mol. Sci. 2016, 17, 1544. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.; You, H.J.; Ji, G.E. Enhancement of Anti-Tumorigenic Polysaccharide Production, Adhesion, and Branch Formation of Bifidobacterium bifidum BGN4 by Phytic Acid. Food Sci. Biotechnol. 2009, 18, 749–754. [Google Scholar]
- Health Functional Food Code (HFFC). II.2.51. Probiotics. Ministry of Food and Drug Safety in Korea. 2010. Available online: http://www.mfds.go.kr/files/upload/eng/4.Health_Functioanl_Food_Code_(2010.09).pdf (accessed on 5 April 2018).
- Yu, D.S.; Jeong, H.; Lee, D.H.; Kwon, S.K.; Song, J.Y.; Kim, B.K.; Park, M.S.; Ji, G.E.; Oh, T.K.; Kim, J.F. Complete Genome Sequence of the Probiotic Bacterium Bifidobacterium bifidum strain BGN4. J. Bacteriol. 2012, 194, 4757–4758. [Google Scholar] [CrossRef] [PubMed]
- Igai, K.; Itakura, M.; Nishijima, S.; Tsurumaru, H.; Suda, W.; Tsutaya, T.; Tomitsuka, E.; Tadokoro, K.; Baba, J.; Odani, S.; et al. Nitrogen Fixation and nifH Diversity in Human Gut Microbiota. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Macfarlane, G.T. Formation of Phenolic and Indolic Compounds by Anaerobic Bacteria in the Human Large Intestine. Microbiol. Ecol. 1997, 33, 180–188. [Google Scholar] [CrossRef]
- Vince, A.J.; Burridge, S.M. Ammonia Production by Intestinal Bacteria: The Effects of Lactose, Lactulose and Glucose. J. Med. Microbiol. 1980, 13, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Najafzadeh, H.; Enayati, A.; Pashmforoush, M. Biogenic Amines Content of Canned Tuna Fish Marketed in Iran. Am.-Eurasian J. Toxicol. Sci. 2011, 3, 190–193. [Google Scholar] [CrossRef]
- Ku, S. Finding and Producing Probiotic Glycosylases for the Biocatalysis of Ginsenosides: A Mini Review. Molecules 2016, 21, 645. [Google Scholar] [CrossRef] [PubMed]
- Biji, K.B.; Ravishankar, C.N.; Venkateswarlu, R.; Mohan, C.O.; Srinivasa Gopal, T.K. Biogenic Amines in Seafood: A review. J. Food Sci. Technol. 2016, 53, 2210–2218. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.C.; van Dusseldorp, M.; Bottema, K.C.; Dubois, A.E. Intolerance to Dietary Biogenic Amines: A Review. Ann. Allergy Asthma Immunol. 2003, 91, 233–240. [Google Scholar] [CrossRef]
- Burdychová, R.; Komprda, T. Biogenic Amine-Forming Microbial Communities in Cheese. FEMS Microbiol. Lett. 2007, 276, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshani, W.M.D.; Rakshit, S.K. Screening Selected Strains of Probiotic Lactic Acid Bbacteria for Their Ability to Produce Biogenic Amines (Histamine and Tyramine). Int. J. Food. Sci. Technol. 2011, 46, 2062–2069. [Google Scholar] [CrossRef]
- Lorencová, E.; Buňková, L.; Matoulková, D.; Dráb, V.; Pleva, P.; Kubáň, V.; Buňká, F. Production of Biogenic Amines by Lactic Acid Bacteria and Bifidobacteria Isolated from Dairy Products and Beer. Int. J. Food. Sci. Technol. 2012, 47, 2086–2091. [Google Scholar] [CrossRef]
- Kalač, P.; Švecová, S.; Pelikánová, T. Levels of Biogenic Amines in Typical Vegetable Products. Food Chem. 2002, 77, 349–351. [Google Scholar] [CrossRef]
- Ten Brink, B.; Damink, C.; Joosten, H.M.L.J.; Huis in’t Veld, J.H.J. Occurrence and Formation of Biologically Active Amines in Foods. Int. J. Food Microbiol. 1990, 11, 73–84. [Google Scholar] [CrossRef]
- Manca, G.; Porcu, A.; Ru, A.; Salaris, M.; Franco, M.A.; De Santis, E.P.L. Comparison of γ-Aminobutyric Acid and Biogenic Amine Content of Different Types of Ewe’s Milk Cheese Produced in Sardinia. Ital. J. Food Saf. 2015, 4, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Pollack, P.F.; Koldovskỳ, O.; Nishioka, K. Polyamines in Human and Rat Milk and in the Infant Formulas. Am. J. Clin. Nutr. 1992, 56, 371–375. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific Opinion on Risk Based Control of Biogenic Amine Formation in Fermented Foods. EFSA J. 2011, 9, 2393. [Google Scholar] [CrossRef] [Green Version]
- Borriello, S.P.; Hammes, W.P.; Holzapfel, W.; Marteau, P.; Schrezenmeir, J.; Vaara, M.; Valtonen, V. Safety of Probiotics That Contain Lactobacilli or Bifidobacteria. Clin. Infect. Dis. 2003, 36, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic Resistance in Non-Enterococcal Lactic Acid Bacteria and Bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Flórez, A.B.; Ammor, M.S.; Mayo, B.; van Hoek, A.H.A.M.; Aarts, H.J.M.; Huys, G. Antimicrobial Susceptibility Profiles of 32 Type Strains of Lactobacillus, Bifidobacterium, Lactococcus and Streptococcus spp. Int. J. Antimicrob. Agents 2008, 31, 484–486. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, R.; Yochevab, L.; Tserovskab, L.; Zhelezovab, G.; Stefanovaa, N.; Atanasovaa, A.; Dangulevaa, A.; Ivanovaa, G.; Karapetkova, N.; Rumya, N.; et al. Antimicrobial Activity and Antibiotic Susceptibility of Lactobacillus and Bifidobacterium spp. Intended for Use as Starter and Probiotic Cultures. Biotechnol. Biotechnol. Equip. 2015, 29, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Milani, C.; Mangifesta, M.; Ferrario, C.; Anzalone, R.; Viappiani, A.; van Sinderen, D.; et al. Prevalence of Antibiotic Resistance Genes among Human Gut-Derived Bifidobacteria. Appl. Environ. Microbiol. 2017, 83, e02894-16. [Google Scholar] [CrossRef] [PubMed]
- GRAS notice (GRN), No. 685 Generally Recognized as Safe (GRAS) Determination for the Use of Lactobacillus plantarum 299v in Conventional Foods. Available online: https://www.fda.gov/downloads/Food/IngredientsPackagingLabeling/GRAS/NoticeInventory/ucm544492.pdf (accessed on 20 November 2017).
- Mättö, J.; van Hoek, A.H.A.M.; Domig, K.J.; Saarela, M.; Floréz, A.B.; Brockmann, E.; Amtmann, E.; Mayo, B.; Aarts, H.J.M.; Danielsen, M. Susceptibility of Human and Probiotic Bifidobacterium spp. to Selected Antibiotics as Determined by the Etest Method. Int. Dairy J. 2007, 17, 1123–1131. [Google Scholar] [CrossRef]
- Lim, K.S.; Huh, C.S.; Baek, Y.J.A. Selective Enumeration Medium for Bifidobacteria in Fermented Dairy Products. J. Dairy Sci. 1995, 78, 2018–2112. [Google Scholar] [CrossRef]
- Food Code (2018-8). 7. General Test Methods: 4.4.1 Medium 25) TOS-MUP Medium; 4.9.2 Acid-Bacterial Streptococcus and Bifidus. Ministry of Food and Drug Safety in Korea. 2018. Available online: http://www.foodsafetykorea.go.kr/portal/safefoodlife/food/foodRvlv/foodRvlv.do (accessed on 5 April 2018).
- National Food Safety Standard. Microbiological Examination of Food—Examination of Lactic Acid Bacteria(GB4789.35-2016) 4. Medium and Reagents 4.2 Medium and Reagents; China Food and Drug Ministration: Beijing, China, 2016.
- Agency for Healthcare Research and Quality Advancing Excellence in Health Care (AHRQ). Safety of Probiotics to Reduce Risk and Prevent or Treat Disease; Southern California Evidence-Based Practice Center: Santa Monica, CA, USA, 2011. [Google Scholar]
- Fouhy, F.; Motherway, M.O.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C.; van Sinderen, D.; Cotter, P.D. In Silico Assig ned Resistance Genes Confer Bifidobacterium with Partial Resistance to Aminoglycosides but Not to β-Lactams. PLoS ONE 2013, 8, e82653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gueimonde, M.; Sánchez, B.; Reyes-Gavilán, C.G.; Margolles, A. Antibiotic Resistance in Probiotic Bacteria. Front. Microbiol. 2013, 18, 202. [Google Scholar] [CrossRef] [PubMed]
- UniProt. UniProtKB-O52836 (TETW_BUTFI). Available online: http://www.uniprot.org/uniprot/O52836 (accessed on 28 March 2018).
- De Groote, M.A.; Frank, D.N.; Dowell, E.; Glode, M.P.; Pace, N.R. Lactobacillus rhamnosus GG Bacteremia Associated with Probiotic Use in a Child with Short Gut Syndrome. Pediatr. Infect. Dis. J. 2005, 24, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Liong, M.T. Safety of Probiotics: Translocation and Infection. Nutr. Rev. 2008, 66, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Ruas-Madiedo, P.; Gueimonde, M.; Fernández-García, M.; de los Reyes-Gavilán, C.G.; Margolles, A. Mucin Degradation by Bifidobacterium Strains Isolated from the Human Intestinal Microbiota. Appl. Environ. Microbiol. 2008, 74, 1936–1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruseler-van Embden, J.G.; Liesholt, L.M.; Gosselink, M.J.; Marteau, P. Inability of Lactobacillus casei Strain GG, L. acidophilus and Bifidobacterium bifidum to Degrade Intestinal Mucus Glycoproteins. Scand. J. Gastroenterol. 1995, 30, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Abe, F.; Muto, M.; Yaeshima, T.; Iwatsuki, K.; Aihara, H.; Ohashi, Y.; Fujisawa, T. Safety Evaluation of Probiotic Bifidobacteria by Analysis of Mucin Degradation Activity and Translocation Ability. Anaerobe 2010, 16, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Chaney, A.L.; Marbach, E.P. Modified Reagents for Determination of Urea and Ammonia. Clin. Chem. 1962, 8, 130–132. [Google Scholar] [PubMed]
- Kim, N.Y.; Ji, G.E. Characterization of the Production of Biogenic Amines and Gamma-Aminobutyric Acid in the Soybean Pastes Fermented by Aspergillus oryzae and Lactobacillus brevis. J. Microbiol. Biotechnol. 2015, 25, 464–468. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization (ISO). Milk and Milk Products-Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB); ISO 10932:2010 (IDF 223:2010); ISO: Geneve, Switzerland, 2010. [Google Scholar]
- Tannock, G.W. Conjugal Transfer of Plasmid pAMßi in Lactobacillus reuteri and between Lactobacilli and Enterococcus faecalis. Appl. Environ. Microbiol. 1987, 53, 2693–2695. [Google Scholar] [PubMed]
- Guo, H.L.; Pan, L.; Li, L.N.; Lu, J.; Kwok, L.; Menghe, B.; Zhang, H.P.; Zhang, W.Y. Characterization of Antibiotic Resistance Genes from Lactobacillus Isolated from Traditional Dairy Products. J. Food Sci. 2017, 82, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Thapa, D.; Louis, P.; Losa, R.; Zweifel, B.; Wallace, R.J. Essential oils have different effects on human pathogenic and commensal bacteria in mixed faecal fermentations compared with pure cultures. Microbiology. 2015, 161, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Bezares, B.; Saenz, Y.; Poeta, P.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 2006, 111, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Ouoba, L.I.; Lei, V.; Jensen, L.B. Resistance of potential probiotic lactic acid bacteria and bifi-dobacteria of African and European origin to antimicrobials: Determination and transferability of the resistance genes to other bacteria. Int. J. Food Microbiol. 2008, 121, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Z.Y.; Dong, K.; Yuan, J.P.; Guo, X.K. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed Environ Sci. 2009, 22, 401–412. [Google Scholar] [CrossRef]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Silvestri, G.; Vignaroli, C.; Clementi, F. Isolation and molecular characterization of antibiotic-resistant lactic acid bacteria from poultry and swine meat products. J. Food. Prot. 2007, 70, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Gad, G.F.; Abdel-Hamid, A.M.; Farag, Z.S. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz. J. Microbiol. 2014, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Gerbaud, G.; Courvalin, P. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 1993, 131, 27–34. [Google Scholar] [CrossRef]
- Mellmann, A.; Andersen, P.S.; Bletz, S.; Friedrich, A.W.; Kohl, T.A.; Lilje, B.; Niemann, S.; Prior, K.; Rossen, J.W.; Harmsen, D. High Interlaboratory Reproducibility and Accuracy of Next-Generation-Sequencing-Based Bacterial Genotyping in a Ring Trial. J. Clin. Microbiol. 2017, 55, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Micobiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
| Strain | Ammonia (μg/mL) |
|---|---|
| Bifidobacterium bifidum BGN4 | negative |
| Bifidobacterium longum BORI | negative |
| Bifidobacterium breve ATCC 15701 | negative |
| Lactobacillus plantarum KFRI 708 | negative |
| Bacteroides fragilis ATCC 25285 | 14.7 ± 1.5 |
| Bacteroides thetaiotaomicron ATCC 29741 | 23.3 ± 3.0 |
| Clostridium perfringens ATCC 13124 | 23.5 ± 1.6 |
| Enterobacter cloacae ATCC 13047 | 161.0 ± 6.6 |
| Enterobacter faecalis ATCC 19433 | 12.9 ± 1.3 |
| Strains | Cadaverine (µg/mL) | Histamine (µg/mL) | Putrescine (µg/mL) | Tyramine (µg/mL) |
|---|---|---|---|---|
| B. bifidum BGN4 | N/D 1 | N/D 1 | 24.23 | N/D 1 |
| B. longum BORI | N/D 1 | N/D 1 | 16.58 | N/D 1 |
| Antibiotics | EFSA Cut-Off of Bifidobacterium spp. | B. longum ATCC 15707 | B. longum BB536 | B. longum KCCM 91563 | B. longum BORI | B. infantis ATCC 15697 | B. lactis BB-12 | B. bifidum BGN4 | B. bifidum KCTC 3440 | B. adolescentis ATCC 15703 | B. breve M-16V | E. faecalis ATCC 29212 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillin G | 0.25 | 0.125 | 0.5 | 1 | 0.125 | 0.125 | 0.063 | 0.063 | 0.25 | 0.25 | 0.5 | |
| Carbenicillin disodium salt | 2 | 2 | 4 | 8 | 0.5 | 2 | 0.5 | 0.5 | 4 | 4 | 8 | |
| Methicillin | 8 | 4 | 16 | 16 | 1 | 2 | 1 | 0.5 | 8 | 8 | 16 | |
| Ampicillin sodium salt | 2 | 0.5 | 0.25 | 1 | 0.5 | 0.125 | 0.125 | 0.063 | 0.063 | 0.25 | 0.25 | 0.25 |
| Dicloxacillin sodium salt hydrate | 4 | 4 | 8 | 8 | 0.5 | 4 | 0.5 | 1 | 256 | 8 | 4 | |
| Gentamicin sulfate | 64 | 32 | 64 | 32 | 32 | 16 | 128 | 128 | 256 | 128 | 128 | 256 |
| Streptomycin sulfate salt | 128 | 32 | 128 | 64 | 64 | >256 | 128 | 64 | 32 | 128 | 256 | >256 |
| Kanamycin sulfate | N/R 1 | 512 | 1024 | 1024 | 512 | 32 | 1024 | 1024 | 1024 | 1024 | 1024 | 256 |
| Neomycin sulfate | 1024 | 512 | 512 | 512 | 64 | 512 | 1024 | 512 | 512 | 1024 | 1024 | |
| Cephalothin sodium salt | 8 | 4 | 16 | 32 | 4 | 8 | 4 | 2 | 16 | 16 | 16 | |
| Tetracycline | 8 | 1 | 1 | 1 | 64 | 2 | 16 | 1 | 1 | 8 | 16 | 32 |
| Polymyxin B sulfate salt | 256 | 32 | 256 | 256 | 128 | 256 | 512 | 512 | 512 | 1024 | >1024 | |
| Erythromycin | 1 | 0.125 | 0.5 | 0.5 | 0.5 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 8 |
| Metronidazole | 8 | 8 | >256 | >256 | 8 | 4 | 4 | 64 | >256 | 8 | >256 | |
| Vancomycin hydrochloride | 2 | 0.5 | <0.25 | <0.25 | 1 | 0.5 | 0.5 | 1 | 2 | 0.5 | 0.5 | 2 |
| Chloramphenicol | 4 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 8 |
| Rifampicin | <0.125 | <0.125 | <0.125 | 0.25 | <0.125 | 2 | 0.5 | 0.25 | 0.5 | 1 | 0.5 | |
| Clindamycin hydrochloride | 1 | <0.032 | 0.063 | 0.063 | 0.125 | 0.25 | <0.032 | 0.063 | 0.063 | <0.032 | <0.032 | >16 |
| Phosphomycin disodium salt | 128 | 256 | 256 | 256 | 16 | 64 | 128 | 256 | 64 | 32 | 32 | |
| Mupirocin | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | |
| Trimethoprim–Sulfamethoxazole | 128 | 256 | 128 | 256 | 256 | 1 | 128 | 64 | 1 | 2 | 32 |
| Antibiotics | AGBG1 (Aerobic) | AGBG1 + BORI | BORI (Anaerobic) | |
|---|---|---|---|---|
| Aerobic | Anaerobic | |||
| None 1 | 4.38 × 108 | 3.38 × 108 | 2.27 × 108 | 4.56 × 108 |
| T8 2 | 0 | 0 | 4.44 × 106 | 7.11 × 107 |
| Antibiotics | ATCC 4356 (Aerobic) | ATCC 4356 + BORI | BORI (Anaerobic) | |
| Aerobic | Anaerobic | |||
| None 1 | 3.65 × 108 | 1.67 × 108 | 2.34 × 108 | 3.14 × 108 |
| G64 3 | 0 | 0 | 2.78 × 106 | 1.46 × 108 |
| Antibiotics | ATCC 4356 (Aerobic) | ATCC 4356 + BGN4 | BGN4 (Anaerobic) | |
| Aerobic | Anaerobic | |||
| None 1 | 3.65 × 108 | 3.29 × 108 | 2.54 × 108 | 3.86 × 108 |
| G64 3 | 0 | 0 | 4.64 × 106 | 1.43 × 108 |
| Strain/Sample | B. bifidum BGN4-1/13075.BBGN41.1 1 | B. bifidum BGN4-2/13075.BBGN42.1 2 | B. bifidum BGN4-3/13075.BBGN43.1 3 |
|---|---|---|---|
| B. bifidum BGN4-1/13075.BBGN41.1 1 | 100 | 99.9997 | 99.9996 |
| B. bifidum BGN4-2/13075.BBGN42.1 2 | 99.9997 | 100 | 99.9998 |
| B. bifidum BGN4-3/13075.BBGN43.1 3 | 99.9996 | 99.9998 | 100 |
| Strains | Origin | Medium | Method |
|---|---|---|---|
| Bifidobacterium bifidum BGN4 | BIFIDO Co., Ltd. (Hongcheon, Korea) | BHI 1, Blood agar 2, whole milk 3, LSM-Cys 4, MRS 5–8 | 3.2., 3.3., 3.4., 3.5.2., 3.5.3., 3.5.4., 3.6., 3.7 |
| Bifidobacterium longum BORI | BIFIDO Co., Ltd. (Hongcheon, Korea) | BHI 1, Blood agar 2, MRS 3,5–7, LSM-Cys 4 | 3.2., 3.3., 3.4., 3.5.2., 3.5.4., 3.6 |
| Bacteroides fragilis ATCC 25285 | American Type Culture Collection (Manassas, VA, USA) | BHI 1 | 3.2 |
| Bacteroides thetaiotaomicron ATCC 29741 | American Type Culture Collection (Manassas,VA, USA) | BHI 1 | 3.2 |
| Bifidobacterium adolescentis ATCC 15703 | American Type Culture Collection (Manassas,VA, USA) | LSM-Cys 4, MRS 6 | 3.5.2., 3.5.4 |
| Bifidobacterium animalis ATCC 25527 | American Type Culture Collection (Manassas,VA, USA) | MRS 6 | 3.5.4 |
| Bifidobacterium animalis subsp. lactis AD011 | BIFIDO Co., Ltd. (Hongcheon, Korea) | MRS 6 | 3.5.4 |
| Bifidobacterium animalis subsp. lactis AS60 | BIFIDO Co., Ltd. (Hongcheon, Korea) | MRS 6 | 3.5.4 |
| Bifidobacterium animalis subsp. lactis BB-12 | Isolated from a pharmaceutical product, USA | LSM-Cys 4, MRS 6 | 3.5.2., 3.5.4 |
| Bifidobacterium bifidum KCTC 3440 | Korean Collection for Type Cultures, (Jeongeup, Korea) | LSM-Cys 4, MRS 6 | 3.5.2., 3.5.4 |
| Bifidobacterium breve ATCC 15701 | American Type Culture Collection (Manassas,VA, USA) | BHI 1 | 3.2 |
| Bifidobacterium breve M-16V | Isolated from a pharmaceutical product, USA | LSM-Cys 4, MRS 6 | 3.5.2., 3.5.4 |
| Bifidobacterium infantis ATCC 15697 | American Type Culture Collection (Manassas, VA, USA) | LSM-Cys 4, MRS 6 | 3.5.2., 3.5.4 |
| Bifidobacterium longum ATCC 15707 | American Type Culture Collection (Manassas,VA, USA) | LSM-Cys 4 | 3.5.2 |
| Bifidobacterium longum BB536 | Isolated from a pharmaceutical product, USA | LSM-Cys 4 | 3.5.2 |
| Bifidobacterium longum KCCM 91563 | Korean Culture Center of Microorganisms (Seoul, Korea) | LSM-Cys 4, MRS 6 | 3.5.2., 3.5.4 |
| Bifidobacterium longum RD47 | BIFIDO Co., Ltd. (Hongcheon, Korea) | MRS 6 | 3.5.4 |
| Bifidobacterium thermophilum KCCM 12097 | Korean Culture Center of Microorganisms (Seoul, Korea) | MRS 6 | 3.5.4 |
| Clostridium perfringens ATCC 13124 | American Type Culture Collection (Manassas,VA, USA) | BHI 1 | 3.2 |
| Enterococcus faecalis ATCC 29212 | American Type Culture Collection (Manassas,VA, USA) | LSM-Cys 4 | 3.5.2 |
| Enterobacter cloacae subsp. cloaca ATCC 13047 | American Type Culture Collection (Manassas,VA, USA) | BHI 1 | 3.2 |
| Enterobacter faecalis ATCC 19433 | American Type Culture Collection (Manassas,VA, USA) | BHI 1 | 3.2 |
| Lactobacillus acidophilus ATCC 4356 | American Type Culture Collection (Manassas,VA, USA) | MRS 5 | 3.5.3 |
| Lactobacillus fermentum AGBG1 | BIFIDO Co., Ltd. (Hongcheon, Korea) | MRS 5 | 3.5.3 |
| Lactobacillus plantarum KFRI 708 | Korea Food Research Institute (Wanju, Korea) | BHI 1 | 3.2 |
| Listeria ivanovii subsp. ivanovii ATCC 19119 | American Type Culture Collection (Manassas,VA, USA) | Blood Agar 2 | 3.3 |
| Parameters | Conditions | ||
|---|---|---|---|
| HPLC | Thermo Dionex Ultimate 3000 HPLC (Thermo Fisher Scientific, St Peters, MO, USA) | ||
| Column | VDSpher C-18 column (4.6 × 250 mm, 5 µm) (VDS optilab Chromatographietechnik GmbH, Berlin, Germany) | ||
| Mobile solvent | Time (min) | Distilled Water (%) | Acetonitrile (%) |
| 0 | 40 | 60 | |
| 1 | 40 | 60 | |
| 20 | 0 | 100 | |
| 25 | 0 | 100 | |
| 26 | 40 | 60 | |
| 30 | 40 | 60 | |
| Flow rate | 0.8 mL | ||
| Column temperature | 30 °C | ||
| Injection volume | 20 µL | ||
| Detector | UV 250 nm | ||
| Donor Strains Recipient Strains | B. bifidum BGN4 | B. longum BORI |
|---|---|---|
| L. fermentum AGBG1 | N/A 1 | BORI + AGBG1 |
| L. acidophilus ATCC 4356 | BGN4 + ATCC 4356 | BORI + ATCC 4356 |
| No. | Primer Name | Oligo Sequence | TM (°C) | Product Size | Reference | |
|---|---|---|---|---|---|---|
| 1 | Bifidobacterium genus-specific primers | - | F: 5′-TCGCGTCYGGTGTGAAAG-3′ R: 5′-GGTGTTCTTCCCGATATCTACA-3′ | 55 | 128 bp | [72] |
| 2 | Gentamicin | aaac(6)-aph(2) | F: 5′-CCAAGAGCAATAAGGGCATA-3′ R: 5′-CACTATCATAACCACTACCG-3′ | 60 | 220 bp | [73] |
| 3 | Kanamycin | AphA3 | F: 5′-GCCGATGTGGATTGCGAAAA-3′ R: 5′-GCTTGATCCCCAGTAAGTCA-3′ | 52 | 292 bp | [74] |
| 4 | Streptomycin | aadE | F: 5′-ATGGAATTATTCCCACCTGA-3′ R: 5′-TCAAAACCCCTATTAAAGCC-3′ | 50 | 565 bp | [74] |
| 5 | Trimethoprim | dfrA | F: 5′-AAAAGGGGCAGAGCATG-3′ R: 5′-AGAAAATGGCGTAATCGGTA-3′ | 50 | 474 bp | [75] |
| 6 | Tetracycline(K) | tet(K) | F: 5′-TTAGGTGAAGGGTTAGGTCC-3′ R: 5′-GCAAACTCATTCCAGAAGCA-3′ | 55 | 169 bp | [76] |
| 7 | Tetracycline(L) | tet(L) | F: 5′-GTTGCGCGCTATATTCCAAA-3′ R: 5′-TTAAGCAAACTCATTCCAGC-3′ | 55 | ||
| 8 | Tetracycline(M) | tet(M) | F: 5′-GTTAAATAGTGTTCTTGGAG-3′ R: 5′-CTAAGATATGGCTCTAACAA-3′ | 55 | 401 bp | [77] |
| 9 | Tetracycline(O) | tet(O) | F: 5′-GATGGCATACAGGCACAGAC-3′ R: 5′-CAATATCACCAGAGCAGGCT-3′ | 55 | ||
| 10 | Tetracycline(S) | tet(S) | F: 5′-TGGAACGCCAGAGAGGTATT-3′ R: 5′-ACATAGACAAGCCGTTGACC-3′ | 55 | 1923 bp | [78] |
| 11 | Kanamycin | aaaD | F: 5′-TGCGTTTTGACACATCCAC-3′ R: 5′-GGTGTTTATGGCTCTCTTGG-3′ | 55 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.J.; Ku, S.; Kim, S.Y.; Lee, H.H.; Jin, H.; Kang, S.; Li, R.; Johnston, T.V.; Park, M.S.; Ji, G.E. Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 2018, 19, 1422. https://doi.org/10.3390/ijms19051422
Kim MJ, Ku S, Kim SY, Lee HH, Jin H, Kang S, Li R, Johnston TV, Park MS, Ji GE. Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. International Journal of Molecular Sciences. 2018; 19(5):1422. https://doi.org/10.3390/ijms19051422
Chicago/Turabian StyleKim, Min Jeong, Seockmo Ku, Sun Young Kim, Hyun Ha Lee, Hui Jin, Sini Kang, Rui Li, Tony V. Johnston, Myeong Soo Park, and Geun Eog Ji. 2018. "Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI" International Journal of Molecular Sciences 19, no. 5: 1422. https://doi.org/10.3390/ijms19051422





