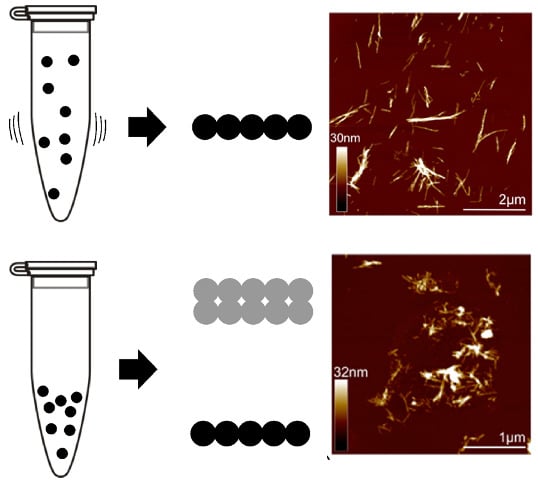
Insights into the Origin of Distinct Medin Fibril Morphologies Induced by Incubation Conditions and Seeding
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of Medin
4.2. Incubation of Medin
4.3. Thioflavin T Fluorescence
4.4. NMR
4.5. Intrinsic Fluorescence
4.6. Atomic Force Microscopy
4.7. Cell Viability
4.8. Circular Dichroism
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | Atomic Force Microscopy |
| CCK-8 | Cell Counting Kit-8 |
| CD | Circular Dichroism |
| GdmC | Guanidine hydrochloride |
| IF | Intrinsic Fluorescence |
| NMR | Nuclear Magnetic Resonance |
| ThT | Thioflavin T |
References
- Harper, J.D.; Lansbury, P.T. Models of amyloid seeding in Alzheimer‘s disease and scrapie: Mechanistic Truths and Physiological Consequences of the Time-Dependent Solubility of Amyloid Proteins. Ann. Rev. Biochem. 1997, 66, 385–407. [Google Scholar] [CrossRef] [PubMed]
- Faller, P.; Hureau, C.; Berthoumieu, O. Role of metal ions in the self-assembly of the Alzheimer’s amyloid-β peptide. Inorg. Chem. 2013, 52, 12193–12206. [Google Scholar] [CrossRef] [PubMed]
- McParland, V.J.; Kad, N.M.; Kalverda, A.P.; Brown, A.; Kirwin-Jones, P.; Hunter, M.G.; Sunde, M.; Radford, S.E. Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro. Biochemistry 2000, 39, 8735–8746. [Google Scholar] [CrossRef] [PubMed]
- Paravastu, A.K.; Leapman, R.D.; Yau, W.M.; Tycko, R. Molecular structural basis for polymorphism in Alzheimer‘s β-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 18349–18354. [Google Scholar] [CrossRef] [PubMed]
- Morel, B.; Varela, L.; Azuaga, A.I.; Conejero-Lara, F. Environmental conditions affect the kinetics of nucleation of amyloid fibrils and determine their morphology. Biophys. J. 2010, 99, 3801–3810. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.; Patil, S.M.; Gibson, J.; Nelson, C.E.; Alder, N.N.; Alexandrescu1, A.T. Mechanism of amylin fibrillization enhancement by heparin. J. Biol. Chem. 2011, 286, 22894–22904. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Fernandez, E.J.; Good, T.A. Role of aggregation conditions in structure, stability, and toxicity of intermediates in the Aβ fibril formation pathway. Protein Sci. 2007, 16, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, J.T.; Berger, E.P.; Lansbury, P.T., Jr. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer‘s disease. Biochemistry 1993, 32, 4693–4697. [Google Scholar] [CrossRef] [PubMed]
- Sasahara, K.; Yagi, H.; Sakai, M.; Naiki, H.; Goto, Y. Amyloid nucleation triggered by agitation of beta2-microglobulin under acidic and neutral pH conditions. Biochemistry 2008, 47, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.; Khurana, R.; Coats, A.; Frokjaer, S.; Brange, J.; Vyas, S.; Uversky, V.N.; Fink, A.L. Effect of environmental factors on the kinetics of insulin fibril formation: Elucidation of the molecular mechanism. Biochemistry 2001, 40, 6036–6046. [Google Scholar] [CrossRef] [PubMed]
- Tiiman, A.; Noormägi, A.; Friedemann, M.; Krishtal, J.; Palumaa, P.; Tõugu, V. Effect of agitation on the peptide fibrillization: Alzheimer‘s amyloid-beta peptide 1-42 but not amylin and insulin fibrils can grow under quiescent conditions. J. Pept. Sci. 2013, 19, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Petkova, A.T.; Leapman, R.D.; Guo, Z.; Yau, W.-M.; Mattson, M.P.; Tycko, R. Self-propagating, molecular-level polymorphism in Alzheimer’s ß-amyloid fibrils. Science 2005, 307, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Come, J.H.; Fraser, P.E.; Lansbury, P.T. A kinetic model for amyloid formation in the prion diseases: Importance of seeding. Proc. Natl. Acad. Sci. USA 1993, 90, 5959–5963. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Westermark, G.T. Seeding and Cross-seeding in Amyloid Diseases, in Proteopathic Seeds and Neurodegenerative Diseases; Jucker, M., Christen, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 47–60. [Google Scholar]
- Knowles, T.P.J.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nano 2011, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid—From bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Mostaert, A.S.; Higgins, M.J.; Fukuma, T.; Rindi, F.; Jarvis, S.P. Nanoscale mechanical characterisation of amyloid fibrils discovered in a natural adhesive. J. Biol. Phys. 2006, 32, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006, 4, e6. [Google Scholar] [CrossRef] [PubMed]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Brender, J.R.; Hartman, K.; Ramamoorthy, A.; Marsh, E.N.G. Alternative Pathways of Human Islet Amyloid Polypeptide Aggregation Distinguished by 19F Nuclear Magnetic Resonance-Detected Kinetics of Monomer Consumption. Biochemistry 2012, 51, 8154–8162. [Google Scholar] [CrossRef] [PubMed]
- Dusa, A.; Kaylor, J.; Edridge, S.; Bodner, N.; Hong, D.P.; Fink, A.L. Characterization of oligomers during alpha-synuclein aggregation using intrinsic tryptophan fluorescence. Biochemistry 2006, 45, 2752–2760. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, A.; Borowik, T.; Gröbner, G.; Sauer-Eriksson, A.E. Negatively charged phospholipid membranes induce amyloid formation of medin via an alpha-helical intermediate. J. Mol. Biol. 2007, 374, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.A.; Madine, J.; Middleton, D.A. Comparisons with amyloid-beta reveal an aspartate residue that stabilizes fibrils of the aortic amyloid peptide medin. J. Biol. Chem. 2015, 290, 7791–7803. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.A.; Phelan, M.M.; Wilkinson, M.C.; Migrino, R.Q.; Truran, S.; Franco, D.A.; Liu, L.N.; Longmore, C.J.; Madine, J. Oxidative stress alters the morphology and toxicity of aortic medial amyloid. Biophys. J. 2015, 109, 2363–2370. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.A.; Rigden, D.J.; Phelan, M.M.; Madine, J. Probing medin monomer structure and its amyloid nucleation using (13)C-direct detection NMR in combination with structural bioinformatics. Sci. Rep. 2017, 7, 45224. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Geyer, M.; Winter, R. NMR spectroscopic investigation of early events in IAPP amyloid fibril formation. Chembiochem 2009, 10, 1769–1772. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Martinez, M.T.; Fontes, P.; Zomosa-Signoret, V.; Arnaud, J.D.; Hingant, E.; Pujo-Menjouet, L.; Liautard, J.P. Dynamics of polymerization shed light on the mechanisms that lead to multiple amyloid structures of the prion protein. Biochim. Biophys. Acta 2011, 1814, 1305–1317. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.F.; Bird, S.; Shaw, M.; Jean, L.; Vaux, D.J. Combined effects of agitation, macromolecular crowding, and interfaces on amyloidogenesis. J. Biol. Chem. 2012, 287, 38006–38019. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, D.E.; Hamilton-Brown, P.; Asimakis, P.; Ducker, W.; Bertolini, J. Shear flow promotes amyloid-β fibrilization. Protein Eng. Des. Sel. 2009, 22, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Morinaga, A.; Hasegawa, K.; Nomura, R.; Ookoshi, T.; Ozawa, D.; Goto, Y.; Yamada, M.; Naiki, H. Critical role of interfaces and agitation on the nucleation of Abeta amyloid fibrils at low concentrations of Abeta monomers. Biochim. Biophys. Acta 2010, 1804, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Waudby, C.A.; Devlin, G.L.; Cohen, S.I.A.; Aguzzi, A.; Vendruscolo, M.; Terentjev, E.M.; Welland, M.E.; Dobson, C.M. An analytical solution to the kinetics of breakable filament assembly. Science 2009, 326, 1533–1537. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J. Evidence for bulk flow of brain interstitial fluid: Significance for physiology and pathology. Neurochem. Int. 2004, 45, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Stein, K.C.; True, H.L. Prion strains and amyloid polymorphism influence phenotypic variation. PLoS Path. 2014, 10, e1004328. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.; Malmström, S.; Westermark, P. Signs of cross-seeding: Aortic medin amyloid as a trigger for protein AA deposition. Amyloid 2011, 18, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Domigan, L.J.; Healy, J.P.; Meade, S.J.; Blaikie, R.J.; Gerrard, J.A. Controlling the dimensions of amyloid fibrils: Toward homogenous components for bionanotechnology. Biopolymers 2012, 97, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.A.; Wilkinson, M.C.; Gibson, R.P.; Middleton, D.A. Expression and purification of the aortic amyloid polypeptide medin. Protein Expr. Purif. 2014, 98, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Bioinf. 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.A.; Phelan, M.M.; Madine, J. 1H, 15N and 13C assignment of the amyloidogenic protein medin using fast-pulsing NMR techniques. Biomol. NMR Assign. 2015. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-N.; Duquesne, K.; Oesterhelt, F.; Sturgis, J.N.; Scheuring, S. Forces guiding assembly of light-harvesting complex 2 in native membranes. Proc. Natl. Acad. Sci. USA 2011, 108, 9455–9459. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]








| Secondary Structure | Time (h) | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 8 | 16 | 24 | 32 | 40 | 48 | |
| Helix | 18 | 23 | 0 | 0 | 0 | 0 | 0 |
| Sheet | 8 | 7 | 58 | 59 | 60 | 63 | 67 |
| Turn | 27 | 27 | 20 | 19 | 18 | 19 | 7 |
| Unordered | 47 | 42 | 22 | 21 | 22 | 18 | 25 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davies, H.A.; Lee, C.F.; Miller, L.; Liu, L.-N.; Madine, J. Insights into the Origin of Distinct Medin Fibril Morphologies Induced by Incubation Conditions and Seeding. Int. J. Mol. Sci. 2018, 19, 1357. https://doi.org/10.3390/ijms19051357
Davies HA, Lee CF, Miller L, Liu L-N, Madine J. Insights into the Origin of Distinct Medin Fibril Morphologies Induced by Incubation Conditions and Seeding. International Journal of Molecular Sciences. 2018; 19(5):1357. https://doi.org/10.3390/ijms19051357
Chicago/Turabian StyleDavies, Hannah A., Chiu Fan Lee, Leanne Miller, Lu-Ning Liu, and Jillian Madine. 2018. "Insights into the Origin of Distinct Medin Fibril Morphologies Induced by Incubation Conditions and Seeding" International Journal of Molecular Sciences 19, no. 5: 1357. https://doi.org/10.3390/ijms19051357