Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light
Abstract
:1. Introduction
2. Results
2.1. Variation of CRK Genes in Arabidopsis sp.
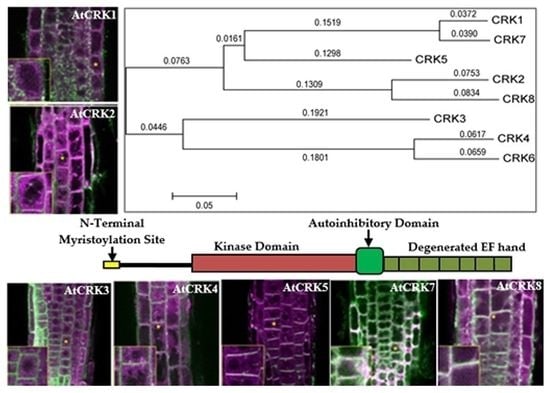
2.2. Intracellular Localization of AtCRK Proteins
2.3. Function of AtCRK Genes, Analysis of T-DNA Insertion Mutants
2.4. AtCRK1 Is Implicated in Light Sensitivity
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, Protoplast Transformation, Gravitropic Assays
4.2. Gene Cloning, Plant Transformation
4.3. Hairy Root Transformation of the 35S::cCRK1-GFP
4.4. Physiological Characterization of the Arabidopsis CRKs
4.4.1. PSII Photochemical Activity Measurements
4.4.2. Detection of 1O2 Production
4.4.3. Determination of Cell Death
4.5. Microsomal Membrane Preparation by Differential Centrifugation and Immunoblotting
4.6. RNA Isolation and Real Time Quantitative PCR
4.7. PIN2 Immunolocalization
4.8. Microscopy
4.9. Bioinformatics Analysis
4.10. Accession Numbers
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Xu, Y.P.; Munyampundu, J.P.; Liu, T.Y.; Cai, X.Z. Calcium dependent protein kinase (CDPK) and CDPK related kinase (CRK) gene families in tomato: Genome wide identification and functional analyses in disease resistance. Mol. Genet. Genom. 2016, 291, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Huang, W. Calcium-Dependent Protein Kinases in Phytohormon Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 2436. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sagar, S.; Biswas, D.K. Calcium Dependent Protein Kinase, a Versatile Player in Plant Stress Management and Development. Crit. Rev. Plant Sci. 2018. [Google Scholar] [CrossRef]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.; Sheen, J. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-Dependent Protein Kinases: Hubs in Plant Stress Signaling and Development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Cox, K.L., Jr.; He, P. Functions of Calcium-Dependent Protein Kinases in Plant Innate Immunity. Plants 2014, 3, 160–176. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Murata, Y. Microbe Associated Molecular Pattern Signaling in Guard Cells. Front. Plant Sci. 2016, 7, 583. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Furumoto, T.; Ogawa, N.; Hata, S.; Izui, K. Plant calcium dependent protein kinase-related kinases (CRKs) do not require calcium for their activities. FEBS Lett. 1996, 396, 147–151. [Google Scholar] [CrossRef]
- Rigó, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovács, H.; Páy, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, J.; Ranty, B.; Sanchez-Ballesta, M.T.; Li, Z.; Jones, B.; Jauneau, A.; Pech, J.C.; Latché, A.; Ranjeva, R.; Bouzayen, M. Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J. Exp. Bot. 2005, 56, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Rigó, G.; Ayadin, F.; Szabados, L.; Koncz, C.; Cséplő, Á. Suspension protoplasts as useful experimental tool to study localization of GFP-tagged proteins in Arabidopsis thaliana. Acta Biol. Szeged. 2008, 52, 59–61. [Google Scholar]
- Renna, L.; Stefano, G.; Majeran, W.; Micalella, C.; Meinnel, T.; Giglione, C.; Brandizzi, F. Golgi traffic integrity depends on N-Myristoyl transferase-1 in Arabidopsis. Plant Cell 2013, 25, 1756–1773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, B.F.; Liang, S.; Jones, R.L.; Lu, Y.T. Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem. J. 2002, 368, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, S.; Lu, Y.T. Characterization, physical location and expression of the genes encoding calcium/calmodulin dependent protein kinases in maize (Zea mays L.). Planta 2001, 21, 556–564. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, S.; Xie, Q.G.; Lu, Y.T. Characterizationof a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis. Biochem. J. 2004, 383, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Farmer, P.K.; Choi, J.H. Calcium and phospholipid activation of a recombinant calcium-dependent protein kinase (DcCPK1) from carrot (Daucus carota L.). Biochim. Biophys. Acta 1999, 1434, 6–17. [Google Scholar] [CrossRef]
- Tao, X.C.; Lu, Y.T. Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt. J. Plant Biol. 2013, 56, 306–314. [Google Scholar] [CrossRef]
- Xiao, X.H.; Yang, M.; Sui, J.L.; Qi, J.Y.; Fang, Y.J.; Hu, S.N.; Tang, C.R. The calcium-dependent protein kinase (CDPK) and CDPK-related kinase gene families in Hevea brasiliensis-comparison with five other plant species in structure, evolution and expression. FEBS Open Bio 2017, 7, 4–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Gao, F.; Li, G.L.; Han, J.L.; Liu, D.L.; Sun, D.Y.; Zhou, R.G. The calmodulin-binding protein kinase 3 is a part of heat-shock signal trasduction in Arabidopsis thaliana. Plant J. 2008, 55, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Li, R.J.; Hua, W.; Lu, Y.T. Arabidopsis cytosolic glutamine synthetase AtGLN1; 1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem. Biophys. Res. Commun. 2006, 342, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, K.; Takemori, N.; Seki, M.; Shinozaki, K.; Sawasaki, T. Members of the Plant CRK Superfamily Are Capable of Trans- and Autophosphorylation of Tyrosine Residues. J. Biol. Chem. 2015, 290, 16665–16677. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provant, N.J. An ‘‘Electronic Fluorescent Pictograph’’ Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, E718. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Zádníková, P.; Petrášek, J.; Marhavý, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Alabadí, D.; Blázquez, M.A. Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 2013, 4, 441. [Google Scholar] [CrossRef] [PubMed]
- Zádnikova, P.; Wabnik, K.; Abuzeineh, A.; Gallemi, M.; Van Der Straeten, D.; Smith, R.S.; Inze, D.; Friml, J.; Prusinkiewicz, P.; Benkova, E. A model of differential growth-guided apical hook formation in plants. Plant Cell 2016, 28, 2464–2477. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Frimml, J. PIN-dependent auxin transport: Action, regulation and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Zádnikova, P.; Smet, D.; Zhu, Q.; Van der Straeten, D.; Benková, E. Strategies of seedlings to overcome their sessile nature: Auxin in mobility control. Front. Plant Sci. 2015, 6, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, A.; Guan, C.; Galweiler, L.; Tanzler, P.; Huijser, P.; Marchant, A.; Parry, G.; Bennett, M.; Wisman, E.; Palme, K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998, 17, 6903–6911. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Lee, S.Y.; Chung, I.K.; Lee, C.H.; Nam, H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 1996, 30, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Klughammer, C.; Schreiber, U. Complementary PS II quantum yields calculated from simple fluorescence parameters measured by PAM fluorometry and the Saturation Pulse method. PAM Appl. 2018, 1, 27–35. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and responding to excess light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Krieger-Liszkay, A. Singlet oxygen production in photosynthesis. J. Exp. Bot. 2005, 56, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Van Breusegem, F.; Mueller, M.J. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramel, F.; Ksas, B.; Akkari, E.; Mialoundama, A.S.; Monnet, F.; Krieger-Liszkay, A.; Ravanat, J.L.; Mueller, M.J.; Bouvier, F.; Havaux, M. Light-Induced Acclimation of the Arabidopsis chlorina1 Mutant to Singlet Oxygen. Plant Cell 2013, 25, 1445–1462. [Google Scholar] [CrossRef] [PubMed]
- Flors, C.; Fryer, M.J.; Waring, J.; Reeder, B.; Bechtold, U.; Mullineaux, P.; Nonell, S.; Wilson, M.T.; Baker, N.R. Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 2006, 57, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Hideg, E. A comparative study of fluorescent singlet oxygen probes in plant leaves. Cent. Eur. J. Biol. 2008, 3, 273–284. [Google Scholar] [CrossRef]
- Rehman, A.U.; Szabó, M.; Deak, Z.; Sass, L.; Larkum, L.A.; Ralph, P.; Vass, I. Symbiodinium sp. cells produce light-induced intra and extracellular singlet oxygen, which mediates photodamage of the photosynthetic apparatus and has the potential to interact with the animal host in coral symbiosis. New Phytol. 2016, 212, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Dammann, C.; Ichida, A.; Hong, B.; Romanowsky, S.M.; Hrabak, E.M.; Harmon, A.C.; Pickard, B.G.; Harper, J.F. Subcellular Targeting of Nine Calcium-Dependent Protein Kinase Isoforms from Arabidopsis. Plant Physiol. 2003, 132, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.I.; Rigó, G.; Andrási, N.; Tietz, O.; Palme, K.; Szabados, L.; Cséplő, A. Striving Towards Abiotic Stresses: Role of the Plant CDPK Super Family Members. In Adaptation in Agriculture, Forestry and Water Resources Economy; Springer: Dordrecht, The Netherlands, 2018; in press. [Google Scholar]
- Yemets, A.; Sheremet, Y.; Vissenberg, K.; Van Orden, J.; Verbelen, J.P.; Blume, Y.B. Effects of tyrosine kinase and phosphatase inhibitorson microtubules in Arabidopsis root cells. Cell Biol. Int. 2008, 32, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Sheremet, Y.A.; Yemets, A.I.; Azmi, A.; Vissenberg, K.; Verbelen, J.P.; Blume, Y.B. Effects of tyrosine kinase and phosphatase inhibitors on mitosis progression in synchronized tobacco BY-2 cells. Tsitol. Genet. 2012, 46, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Hrabak, E.M. The myristoylated amino-terminus of an Arabidopsis calcium dependent protein kinase mediates plasma membrane localization. Plant Mol. Biol. 2013, 82, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, L.; Tang, Y.; Tang, R.; Jing, Y.; Zhang, C.; Zhang, B.; Li, X.; Cui, Y.; Zhang, C.; Shi, J.; et al. Arabidopsis choline transporter-like 1 (CTL1) regulates secretory trafficking of auxin transporters to control seedling growth. PLoS Biol. 2017, 15, e2004310. [Google Scholar] [CrossRef] [PubMed]
- Retzer, K.; Lacek, J.; Skokan, R.; del Genio, C.I.; Vosolsobe, S.; Lanková, M.; Malínská, K.; Konstantinova, N.; Zažímalová, E.; Napier, R.M.; et al. Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED2 Distribution. Int. J. Mol. Sci. 2017, 18, 2274. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, R.; Uzilday, B.; Sekmen, H.A.; Turkan, I. The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in Arabidopsis. Ann. Bot. 2015, 116, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Howell, S.H. The Unfolded Protein ResponseSupports Plant Development and Defense as well as Responses to Abiotic Stress. Front. Plant Sci. 2017, 8, 344. [Google Scholar] [CrossRef] [PubMed]
- Vass, I.; Cser, K. Janus-faced charge recombination in photosystem II photoinhibition. Trends Plant Sci. 2009, 14, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M.; Dall’Osto, L.; Bassi, R. Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 2007, 145, 1506–1520. [Google Scholar] [CrossRef] [PubMed]
- Op den Camp, R.G.L.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- SIGnAl. Available online: http://signal.salk.edu/ (accessed on 27 March 2014).
- Ríos, G.; Lossow, A.; Hertel, B.; Breuer, F.; Schaefer, S.; Broich, M.; Kleinow, T.; Jásik, J.; Winter, J.; Ferrando, A.; et al. Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 2012, 32, 243–253. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Faragó, D.; Sass, L.; Valkai, I.; Andrási, N.; Szabados, L. PlantSize Offers an Affordable, Non-destructive Method to Measure Plant Size and Color in Vitro. Front. Plant Sci. 2018, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Koncz, C.; Martini, N.; Szabados, L.; Hrouda, M.; Bachmair, A.; Schell, J. Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual; Gelvin, S., Schilperoort, B., Eds.; Springer: Dordrecht, The Netherlands, 1994; pp. 1–22. ISBN 978-94-011-7654-5. [Google Scholar]
- Bechtold, N.; Ellis, J.; Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 1993, 316, 1194–1199. [Google Scholar]
- Stock: CD3-967. Available online: https://www.arabidopsis.org/servlet/TairObject?id=3001623306&type=stock (accessed on 10 December 2017).
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S.; et al. Hairy Root Transformation Using Agrobacterium rhizogenes as a Tool for Exploring Cell Type-Specific Gene Expression and Function Using Tomato as a Model. Plant Physiol. 2014, 166, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Genty, B.; Briantais, J.M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 1988, 990, 87–92. [Google Scholar] [CrossRef]
- Shumbe, L.; Chevalier, A.; Legeret, B.; Taconnat, L.; Monnet, F.; Havaux, M. Cell Death in Arabidopsis under High-Light Stress Is Controlled by OXI1 Kinase. Plant Physiol. 2016, 170, 1757–1771. [Google Scholar] [PubMed]
- Abas, L.; Luschnig, C. Maximum yields of microsomal-type membranes from small amount of plant material without requiring ultracentrifugation. Anal. Biochem. 2010, 401, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Primer3Plus. Available online: http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi (accessed on 20 October 2017).
- MEGA5.05 software. Available online: http://www.megasoftware.net/software (accessed on 10 May 2013).








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. https://doi.org/10.3390/ijms19051282
Baba AI, Rigó G, Ayaydin F, Rehman AU, Andrási N, Zsigmond L, Valkai I, Urbancsok J, Vass I, Pasternak T, et al. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. International Journal of Molecular Sciences. 2018; 19(5):1282. https://doi.org/10.3390/ijms19051282
Chicago/Turabian StyleBaba, Abu Imran, Gábor Rigó, Ferhan Ayaydin, Ateeq Ur Rehman, Norbert Andrási, Laura Zsigmond, Ildikó Valkai, János Urbancsok, Imre Vass, Taras Pasternak, and et al. 2018. "Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light" International Journal of Molecular Sciences 19, no. 5: 1282. https://doi.org/10.3390/ijms19051282