Proteomic Identification of the Galectin-1-Involved Molecular Pathways in Urinary Bladder Urothelial Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Search for the De-Regulated Proteins in Sh-Gal(+120) T24 Cells by Two-Dimensional Gel Electrophoresis
2.2. Identification of the Differentially Expressed Proteins in Sh-Gal-1(+120) T24 Cells
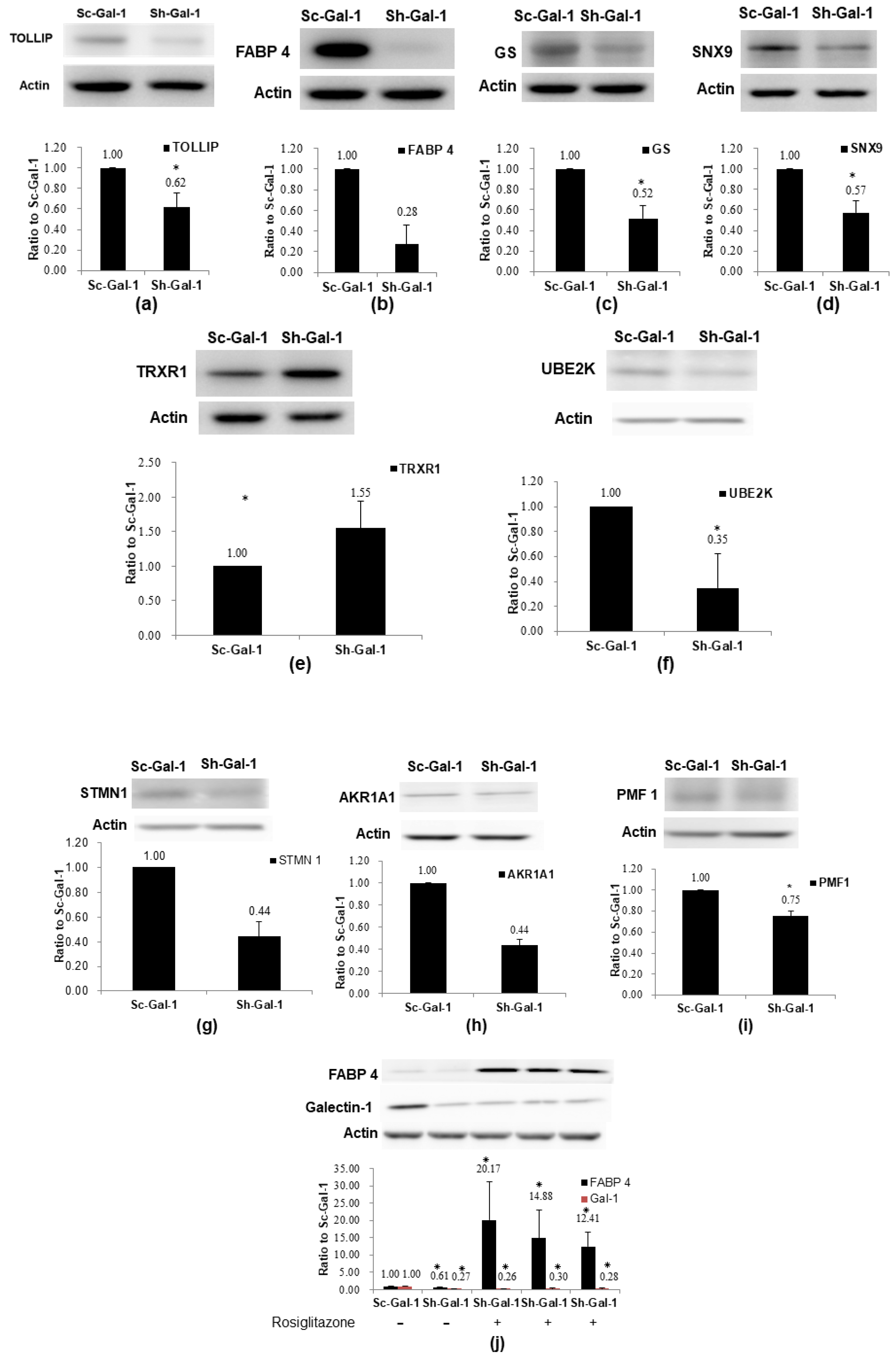
2.3. Confirmation of De-Regulated Proteins Found in Sh-Gal-1(+120) T24 Cells by Western Immunoblotting
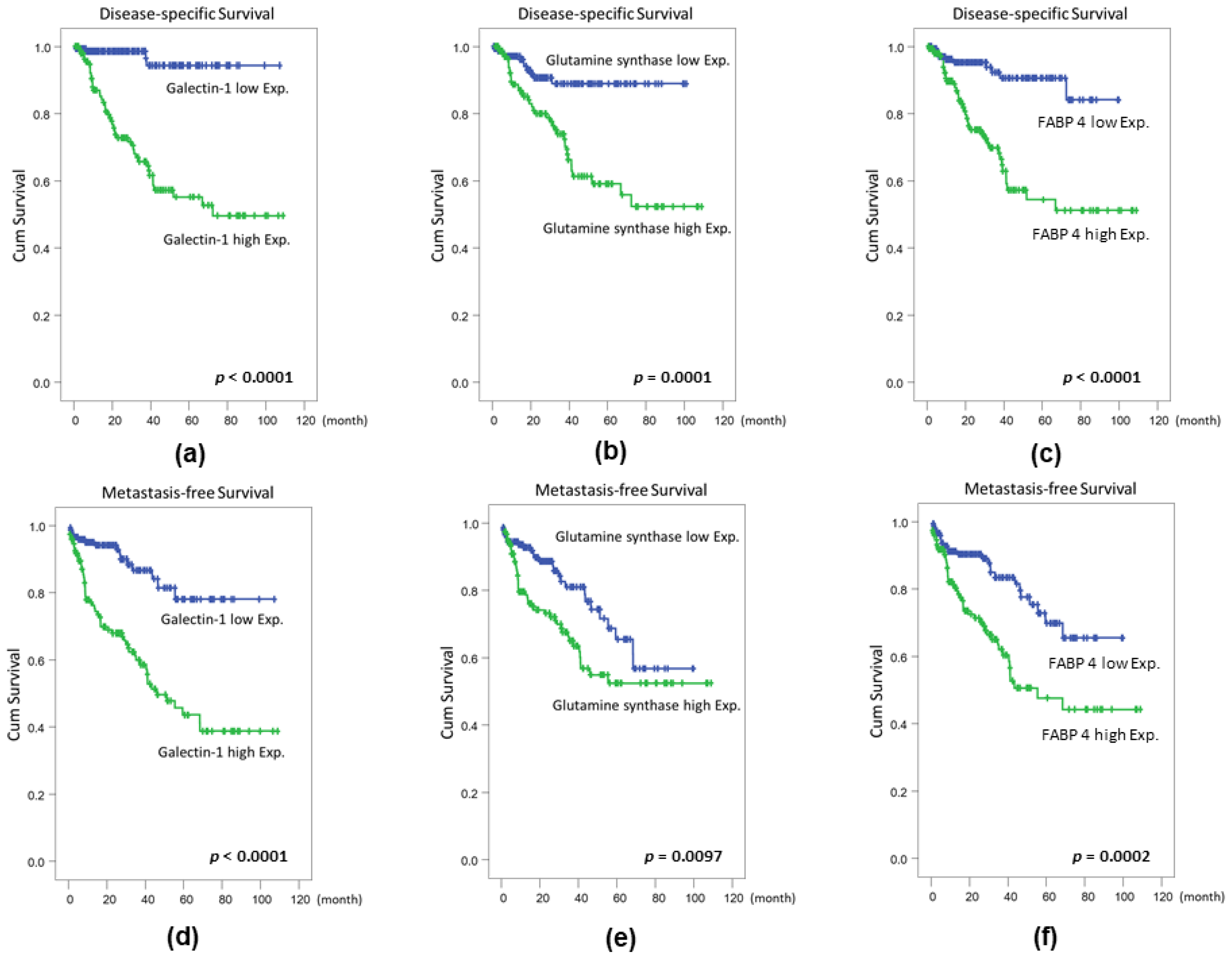
2.4. Confirmation of Proteomics Data Using Cohort Studies of GS, FABP 4, TOLLIP, and AKR1A1
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Construction of Galectin-1 Knockdown T24 Cells with Short-Hairpin RNA (shRNA)
4.3. Preparation of Protein Lysates for Two-Dimensional Gel Electrophoresis
4.4. Isoelectric Focusing (IEF) and SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.5. Lavapurple Staining, Image Analysis, and Statistical Analysis
4.6. In-Gel Digestion and Protein Identification Analysis Via Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
4.7. Western Immunoblotting
4.8. Case Selection
4.9. Immunohistochemistry (IHC) and Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| UBUC | Urinary bladder urothelial carcinoma |
| IHC | Immunohistochemistry |
| MMP | Matrix metalloproteinase protein |
| OSCC | Oral squamous cell carcinoma |
| 2-DE | Two-dimensional gel electrophoresis |
| LC-MS/MS | Liquid chromatography/tandem mass spectrometry |
| PPAR-γ | Peroxisome proliferator-activated receptor gamma |
| RNH1 | Ribonuclease/angiogenin inhibitor 1 |
| RCN1 | Reticulocalbin 1 |
| LGALS1 | Galectin 1 |
| TBCA | Tubulin specific chaperone A |
| UBE2K | Ubiquitin conjugating enzyme E2 K |
| PMF1 | Polyamine-modulated factor 1 |
| STMN1 | Stathmin 1 |
| TOLLIP | toll interacting protein |
| CWC15 | protein CWC15 homolog |
| SNX9 | sorting nexin 9 |
| DCPS | Scavenger mRNA-decapping enzyme |
| FABP 4 | fatty acid binding protein 4 |
| TRXR1 | thioredoxin reductase 1 |
| AKR1A1 | Alcohol dehydrogenase (NADP+) |
| NANS | N-acetylneuraminate synthase (sialic acid synthase) |
| GS | glutamine synthetase |
| R. R. | relative risk |
References
- Stein, J.P.; Grossfeld, G.D.; Ginsberg, D.A.; Esrig, D.; Freeman, J.A.; Figueroa, A.J.; Skinner, D.G.; Cote, R.J. Prognostic markers in bladder cancer: A contemporary review of the literature. J. Urol. 1998, 160, 645–659. [Google Scholar] [CrossRef]
- Camby, I.; Le Mercier, M.; Lefranc, F.; Kiss, R. Galectin-1: A small protein with major functions. Glycobiology 2006, 16, 137R–157R. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, G.; Carpentieri, M.L.; Salvatore, P.; Cindolo, L.; Bruni, C.B.; Chiariotti, L. Cell-specific transcriptional regulation and reactivation of galectin-1 gene expression are controlled by DNA methylation of the promoter region. Mol. Cell. Biol. 1996, 16, 2736–2743. [Google Scholar] [CrossRef] [PubMed]
- Paz, A.; Haklai, R.; Elad-Sfadia, G.; Ballan, E.; Kloog, Y. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 2001, 20, 7486–7493. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, V.L.; Barkan, B.; Shoji, H.; Aries, I.M.; Mathieu, V.; Deltour, L.; Hackeng, T.M.; Kiss, R.; Kloog, Y.; Poirier, F.; et al. Tumor cells secrete galectin-1 to enhance endothelial cell activity. Cancer Res. 2010, 70, 6216. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.Y.; Jung, S.; Ryu, H.H.; Jeong, Y.I.; Jin, Y.H.; Jin, S.G.; Kim, I.Y.; Kang, S.S.; Kim, H.S. Role of galectin-1 in migration and invasion of human glioblastoma multiforme cell lines. J. Neurosurg. 2008, 109, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.H.; Ying, N.W.; Wu, M.H.; Chiang, W.F.; Hsu, C.L.; Wong, T.Y.; Jin, Y.T.; Hong, T.M.; Chen, Y.L. Galectin-1, a novel ligand of neuropilin-1, activates VEGFR-2 signaling and modulates the migration of vascular endothelial cells. Oncogene 2008, 27, 3746–3753. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.H.; Hong, T.M.; Cheng, H.W.; Pan, S.H.; Liang, Y.R.; Hong, H.C.; Chiang, W.F.; Wong, T.Y.; Shieh, D.B.; Shiau, A.L.; et al. Galectin-1-mediated tumor invasion and metastasis, up-regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Mol. Cancer Res. 2009, 7, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, N.; Alvarez, M.; Zwirner, N.W.; Toscano, M.A.; Ilarregui, J.M.; Bravo, A.; Mordoh, J.; Fainboim, L.; Podhajcer, O.L.; Rabinovich, G.A. Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection; a potential mechanism of tumor immune privilege. Cancer Cell 2004, 5, 241–251. [Google Scholar] [CrossRef]
- Dalotto-Moreno, T.; Croci, D.O.; Cerliani, J.P.; Martinez-Allo, V.C.; Dergan-Dylon, S.; Méndez-Huergo, S.P.; Stupirski, J.C.; Mazal, D.; Osinaga, E.; Toscano, M.A.; et al. Targeting galectin-1 overcomes breast cancer associated immunosuppression and prevents metastatic disease. Cancer Res. 2013, 73, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.F.; Li, C.F.; Chien, L.H.; Shen, K.H.; Huang, H.Y.; Su, C.C.; Liao, A.C. Galectin-1 dysregulation independently predicts disease specific survival in bladder urothelial carcinoma. J. Urol. 2015, 193, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Barrow, H.; Rhodes, J.M.; Yu, L.G. The role of galectins in colorectal cancer progression. Int. J. Cancer 2011, 129, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Szoke, T.; Kayser, K.; Baumhakel, J.D.; Trojan, I.; Furak, J.; Tiszlavicz, L.; Horvath, A.; Szluha, K.; Gabius, H.J.; Andre, S. Prognostic significance of endogenous adhesion/growth-regulatory lectins in lung cancer. Oncology 2005, 69, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saussez, S.; Camby, I.; Toubeau, G.; Kiss, R. Galectins as modulators of tumor progression in head and neck squamous cell carcinomas. Head Neck 2007, 29, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Chow, S.N.; Chen, R.J.; Chen, C.H.; Chang, T.C.; Chen, L.C.; Lee, W.J.; Shen, J.; Chow, L.P. Analysis of protein profiles in human epithelial ovarian cancer tissues by proteomic technology. Eur. J. Gynaecol. Oncol. 2010, 31, 55–62. [Google Scholar] [PubMed]
- Laderach, D.J.; Gentilini, L.D.; Giribaldi, L.; Delgado, V.C.; Nugnes, L.; Croci, D.O.; Al Nakouzi, N.; Sacca, P.; Casas, G.; Mazza, O.; et al. A unique galectin signature in Human prostate cancer progression suggests galectin-1 as a key target for treatment of advanced disease. Cancer Res. 2013, 73, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Rorive, S.; Belot, N.; Decaestecker, C.; Lefranc, F.; Gordower, L.; Micik, S.; Maurage, C.A.; Kaltner, H.; Ruchoux, M.M.; Danguy, A.; et al. Galectin-1 is highly expressed in human gliomas with relevance for modulation of invasion of tumor astrocytes into the brain parenchyma. Glia 2001, 33, 241–255. [Google Scholar] [CrossRef]
- Croci, D.O.; Salatino, M.; Rubinstein, N.; Cerliani, J.P.; Cavallin, L.E.; Leung, H.J.; Ouyang, J.; Ilarregui, J.M.; Toscano, M.A.; Domaica, C.I.; et al. Disrupting galectin-1 interactions with N-glycans suppresses hypoxia-driven angiogenesis and tumorigenesis in Kaposi’s sarcoma. J. Exp. Med. 2012, 209, 1985–2000. [Google Scholar] [CrossRef] [PubMed]
- D’Haene, N.; Maris, C.; Sandras, F.; Dehou, M.F.; Remmelink, M.; Decaestecker, C.; Salmon, I. The differential expression of galectin-1 and galectin-3 in normal lymphoid tissue and non-Hodgkins and Hodgkins lymphomas. Int. J. Immunopathol. Pharmacol. 2005, 18, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Puchades, M.; Nilsson, C.L.; Emmett, M.R.; Aldape, K.D.; Ji, Y.; Lang, F.F.; Liu, T.J.; Conrad, C.A. Proteomic investigation of glioblastoma cell lines treated with wild-type p53 and cytotoxic chemotherapy demonstrates an association between galectin-1 and p53 expression. J. Proteome Res. 2007, 6, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Camby, I.; Belot, N.; Lefranc, F.; Sadeghi, N.; de Launoit, Y.; Kaltner, H.; Musette, S.; Darro, F.; Danguy, A.; Salmon, I.; et al. Galectin-1 modulates human glioblastoma cell migration into the brain through modifications to the actin cytoskeleton and levels of expression of small GTPases. J. Neuropathol. Exp. Neurol. 2002, 61, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Chiang, W.F.; Liu, S.Y.; Fang, L.Y.; Lin, C.N.; Wu, M.H.; Chen, Y.C.; Chen, Y.L.; Jin, Y.T. Overexpression of galectin-1 at the tumor invasion front is associated with poor prognosis in early-stage oral squamous cell carcinoma. Oral Oncol. 2008, 44, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Blaževitš, O.; Mideksa, Y.G.; Šolman, M.; Ligabue, A.; Ariotti, N.; Nakhaeizadeh, H.; Fansa, E.K.; Papageorgiou, A.C.; Wittinghofer, A.; Ahmadian, M.R.; et al. Galectin-1 dimers can scaffold Raf-effectors to increase H-ras nanoclustering. Sci. Rep. 2016, 6, 24165. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.H.; Li, C.F.; Chien, L.H.; Huang, C.H.; Su, C.C.; Liao, A.C.; Wu, T.F. Role of galectin-1 in urinary bladder urothelial carcinoma cell invasion through the JNK pathway. Cancer Sci. 2016, 107, 1390–1398. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.A. Glutamine and cancer. J. Nutr. 2001, 131 (Suppl. S9), 2539S–2542S. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Cassimeris, L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002, 14, 18–24. [Google Scholar] [CrossRef]
- Tian, G.; Cowan, N.J. Tubulin-specific chaperones: Components of a molecular machine that assembles the α/β heterodimer. Methods Cell. Biol. 2013, 115, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Rubin, C.I.; Atweh, G.F. The role of stathmin in the regulation of the cell cycle. J. Cell. Biochem. 2004, 93, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; McCormack, S.A.; Viar, M.J.; Wang, H.; Tzen, C.Y.; Scott, R.E.; Johnson, L.R. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am. J. Physiol. 1993, 265 Pt 1, G331–G338. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Sarcinelli, C.; Sheng, J.; Fusco, S.; Formiggini, F.; Netti, P.; Yu, W.; D’Alessio, G.; Hu, G.F. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J. Cell Sci. 2013, 126 Pt 18, 4308–4319. [Google Scholar] [CrossRef] [PubMed]
- Büll, C.; Stoel, M.A.; den Brok, M.H.; Adema, G.J. Sialic acids sweeten a tumor’s life. Cancer Res. 2014, 74, 3199–31204. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zou, P.; Zhao, Z.; Weng, Q.; Chen, X.; Ying, S.; Ye, Q.; Wang, Z.; Ji, J.; Liang, G. Selective killing of gastric cancer cells by a small molecule via targeting TrxR1 and ROS-mediated ER stress activation. Oncotarget 2016, 7, 16593–16609. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xu, Y.; Chen, L.; Fang, Q.; Song, S.; Chen, J.; Teng, J. RCN1 suppresses ER stress-induced apoptosis via calcium homeostasis and PERK-CHOP signaling. Oncogenesis 2017, 6, e304. [Google Scholar] [CrossRef] [PubMed]
- Bendris, N.; Williams, K.C.; Reis, C.R.; Welf, E.S.; Chen, P.H.; Lemmers, B.; Hahne, M.; Leong, H.S.; Schmid, S.L. SNX9 promotes metastasis by enhancing cancer cell invasion via differential regulation of RhoGTPases. Mol. Biol. Cell. 2016, 27, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, L.; Luo, X.; Zhang, Y.; Ding, Q.; Jiang, X.; Wang, X.; Pan, Y.; Chen, Y. Tollip, an intracellular trafficking protein, is a novel modulator of the transforming growth factor-β signaling pathway. J. Biol. Chem. 2012, 287, 39653–39663. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, G.; Lascombe, I.; Roche, E.; Plissonnier, M.L.; Clairotte, A.; Bittard, H.; Fauconnet, S. A-FABP, a candidate progression marker of human transitional cell carcinoma of the bladder, is differentially regulated by PPAR in urothelial cancer cells. Int. J. Cancer 2009, 124, 1820–1828. [Google Scholar] [CrossRef] [PubMed]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–14503. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.J.; Herr, A.; Wissmann, C.; Stoehr, R.; Rosenthal, A.; Zaak, D.; Simon, R.; Knuechel, R.; Pilarsky, C.; Hartmann, A. Gene expression profiling of progressive papillary noninvasive carcinomas of the urinary bladder. Clin. Cancer Res. 2005, 11, 4415–4429. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Lang, Z.W.; Wang, H.G.; Wang, T.L.; Wang, B.E.; Liu, S.Q. Glutamine synthetase as an early marker for hepatocellular carcinoma based on proteomic analysis of resected small hepatocellular carcinomas. Hepatobiliary Pancreat. Dis. Int. 2001, 9, 296–305. [Google Scholar]
- Dal Bello, B.; Rosa, L.; Campanini, N.; Tinelli, C.; Torello Viera, F.; D’Ambrosio, G.; Rossi, S.; Silini, E.M. Glutamine synthetase immunostaining correlates with pathologic features of hepatocellular carcinoma and better survival after radiofrequency thermal ablation. Clin. Cancer Res. 2010, 16, 2157–2166. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Gelmann, E.P. The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 2005, 23, 4776–4789. [Google Scholar] [CrossRef] [PubMed]
- Belletti, B.; Baldassarre, G. Stathmin: A protein with many tasks. New biomarker and potential target in cancer. Expert Opin. Ther. Targets 2011, 15, 1249–1266. [Google Scholar] [CrossRef] [PubMed]
- Aleman, A.; Cebrian, V.; Alvarez, M.; Lopez, V.; Orenes, E.; Lopez-Serra, L.; Algaba, F.; Bellmunt, J.; López-Beltrán, A.; Gonzalez-Peramato, P.; et al. Identification of PMF1 methylation in association with bladder cancer progression. Clin. Cancer Res. 2008, 14, 8236–8243. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.F.; Hsu, L.T.; Tsang, B.X.; Huang, L.C.; Shih, W.Y.; Chen, L.Y. Clarification of the molecular pathway of Taiwan local pomegranate fruit juice underlying the inhibition of urinary bladder urothelial carcinoma cell by proteomics strategy. BMC Complement. Altern. Med. 2016, 16, 96. [Google Scholar] [CrossRef] [PubMed]
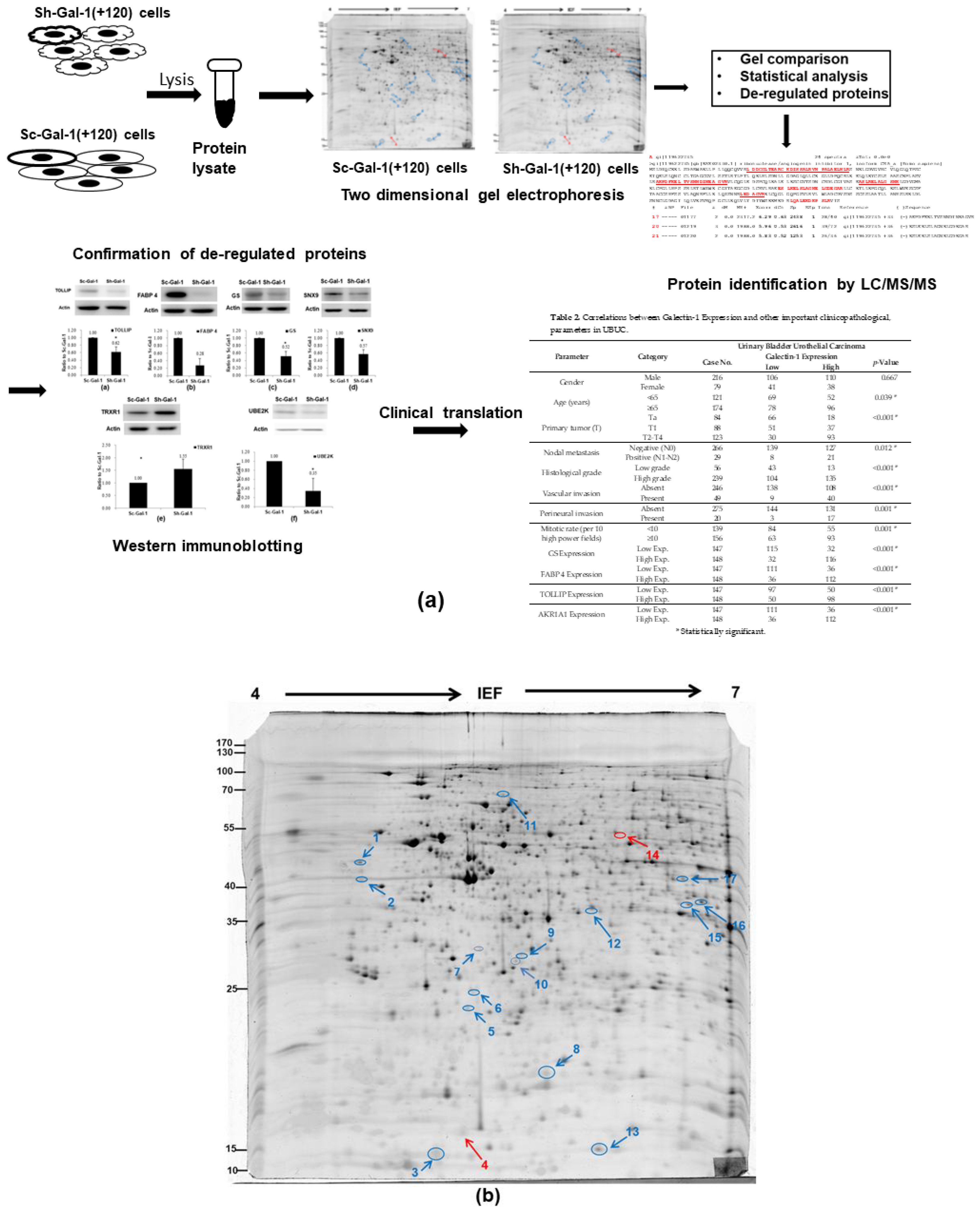

| Spot | Identity | Accession Number | Theo. b PI/Mr. | Exper. c PI/Mr. | Matched d Peptide Number | Coverage (%) e | Incidence | Fold f Difference | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ribonuclease/angiogenin inhibitor 1 (RNH1) a | P13489 | 4.5/49 | 4.74/49.4 | 11 | 22.1 | 6/11 | −2.77 | 0.003 |
| 2 | reticulocalbin 1 (RCN1) | Q15293 | 4.6/42.7 | 4.86/38.9 | 3 | 11.8 | 6/11 | −3.74 | 0.04 |
| 3 | galectin 1 (LGALS1) | P09382 | 4.8/16.7 | 5.3/14.7 | 8 | 52.6 | 8/11 | −56.5 | 0.01 |
| 4 | tubulin specific chaperone A (TBCA) | O75347 | 5.3/12.9 | 4.9/17.8 | 8 | 63.8 | 6/11 | +2.92 | 0.07 |
| 5 | ubiquitin conjugating enzyme E2 K (UBE2K) | P61086 | 5/26.1 | 5.33/22.6 | 9 | 46.5 | 6/11 | −3.71 | 0.0005 |
| 6 | polyamine-modulated factor 1 (PMF1) | Q6P1K2 | 5/27.7 | 5.37/23.3 | 5 | 33.7 | 7/11 | −5.1 | 0.00003 |
| 7 | Unidentified | - | - | 5.0/34.0 | - | - | 7/11 | −5.33 | 0.0047 |
| 8 | stathmin 1 (STMN1) | P16949 | 6.1/20.5 | 5.75/17.3 | 17 | 29.5 | 6/11 | −1.88 | 0.008 |
| 9 | toll interacting protein (TOLLIP) | Q9H0E2 | 5.8/33.1 | 5.68/30.3 | 8 | 23.4 | 6/11 | −3.52 | 0.04 |
| 10 | protein CWC15 homolog (CWC15) | Q9P013 | 5.6/26.6 | 5.7/32.3 | 5 | 19 | 6/11 | −3.35 | 0.05 |
| 11 | sorting nexin 9 (SNX9) | Q9Y5X1 | 5.5/83.4 | 5.4/66.6 | 11 | 23 | 6/11 | −2.98 | 0.00004 |
| 12 | scavenger mRNA-decapping enzyme (DCPS) | Q96C86 | 6.5/37.3 | 5.93/38.6 | 12 | 32.3 | 6/11 | −2.82 | 0.028 |
| 13 | fatty acid binding protein 4 (FABP 4) | P15090 | 6.5/16.6 | 7.93/15.2 | 5 | 41.9 | 10/11 | −5.16 | 0.00003 |
| 14 | thioredoxin reductase 1 (TRXR1) | Q16881 | 6.6/58.3 | 6.07/54.4 | 13 | 24.1 | 6/11 | +4.32 | 0.00001 |
| 15 | alcohol dehydrogenase [NADP+] (AKR1A1) | P14550 | 6.9/37.5 | 6.32/36.6 | 14 | 33.2 | 6/11 | −2.24 | 0.005 |
| 16 | N-acetylneuraminate synthase (sialic acid synthase) (NANS) | Q9NR45 | 6.9/37.4 | 6.29/40.3 | 7 | 22.6 | 7/11 | −2.3 | 0.005 |
| 17 | glutamine synthetase (GS) | P15104 | 6.8/41.4 | 6.43/42.1 | 7 | 14.7 | 9/11 | −5.02 | 0.0001 |
| Parameter | Category | Urinary Bladder Urothelial Carcinoma | |||
|---|---|---|---|---|---|
| Case No. | Galectin-1 Expression | p-Value | |||
| Low | High | ||||
| Gender | Male | 216 | 106 | 110 | 0.667 |
| Female | 79 | 41 | 38 | ||
| Age (years) | <65 | 121 | 69 | 52 | 0.039 * |
| ≥65 | 174 | 78 | 96 | ||
| Primary tumor (T) | Ta | 84 | 66 | 18 | <0.001 * |
| T1 | 88 | 51 | 37 | ||
| T2–T4 | 123 | 30 | 93 | ||
| Nodal metastasis | Negative (N0) | 266 | 139 | 127 | 0.012 * |
| Positive (N1-N2) | 29 | 8 | 21 | ||
| Histological grade | Low grade | 56 | 43 | 13 | <0.001 * |
| High grade | 239 | 104 | 135 | ||
| Vascular invasion | Absent | 246 | 138 | 108 | <0.001 * |
| Present | 49 | 9 | 40 | ||
| Perineural invasion | Absent | 275 | 144 | 131 | 0.001 * |
| Present | 20 | 3 | 17 | ||
| Mitotic rate (per 10 high power fields) | <10 | 139 | 84 | 55 | 0.001 * |
| ≥10 | 156 | 63 | 93 | ||
| GS Expression | Low Exp. | 147 | 115 | 32 | <0.001 * |
| High Exp. | 148 | 32 | 116 | ||
| FABP 4 Expression | Low Exp. | 147 | 111 | 36 | <0.001 * |
| High Exp. | 148 | 36 | 112 | ||
| TOLLIP Expression | Low Exp. | 147 | 97 | 50 | <0.001 * |
| High Exp. | 148 | 50 | 98 | ||
| AKR1A1 Expression | Low Exp. | 147 | 111 | 36 | <0.001 * |
| High Exp. | 148 | 36 | 112 | ||
| Parameter | Category | Case No. | Disease-Specific Survival | Metastasis-Free Survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||||||
| No. of Event | p-Value | R.R. a | 95% C.I. | p-Value | No. of Event | p-Value | R.R. | 95% C.I. | p-Value | |||
| Gender | Male | 216 | 39 | 0.5392 | - | - | - | 59 | 0.2999 | - | - | - |
| Female | 79 | 11 | - | - | - | 16 | - | - | - | |||
| Age (years) | <65 | 121 | 16 | 0.0972 | - | - | - | 30 | 0.5912 | - | - | - |
| ≥65 | 174 | 34 | - | - | - | 45 | - | - | - | |||
| Primary tumor (T) | Ta | 84 | 1 | <0.0001 * | 1 | - | 0.003 * | 4 | <0.0001 * | 1 | - | 0.033 * |
| T1 | 88 | 9 | 2.732 | 1.232–6.061 | 23 | 4.181 | 1.996–2.591 | |||||
| T2–T4 | 123 | 40 | 9.346 | 1.027–83.333 | 48 | 5.543 | 1.961–23.810 | |||||
| Nodal metastasis | Negative (N0) | 266 | 41 | 0.0037 * | 1 | - | 0.640 | 61 | <0.0001 * | 1 | - | 0.086 |
| Positive (N1-N2) | 29 | 9 | 1.199 | 0.561–2.561 | 14 | 1.746 | 0.924–3.303 | |||||
| Histological grade | Low grade | 56 | 2 | 0.0017 * | 1 | - | 0.703 | 5 | 0.0008 * | 1 | - | 0.807 |
| High grade | 239 | 48 | 1.359 | 0.282–6.558 | 75 | 0.873 | 0.293–2.598 | |||||
| Vascular invasion | Absent | 246 | 36 | 0.0052 * | 1 | - | 0.209 | 54 | 0.0003 * | 1 | - | 0.994 |
| Present | 49 | 14 | 1.616 | 0.765–3.413 | 21 | 0.998 | 0.521–1.911 | |||||
| Perineural invasion | Absent | 275 | 44 | 0.0085 * | 1 | - | 0.353 | 65 | 0.0006 * | 1 | - | 0.202 |
| Present | 20 | 6 | 1.586 | 0.599–4.196 | 10 | 1.646 | 0.765–3.542 | |||||
| Mitotic rate (per 10 high power fields) | <10 | 139 | 12 | <0.0001 * | 1 | - | 0.044 * | 22 | <0.0001 * | 1 | - | 0.021 * |
| ≥10 | 156 | 38 | 2.048 | 1.020–4.109 | 53 | 1.885 | 1.103–3.224 | |||||
| Galectin 1 expression | High Exp. | 147 | 4 | <0.0001 * | 1 | - | 0.007 * | 16 | <0.0001 * | 1 | - | 0.012 |
| Low Exp. | 148 | 46 | 4.628 | 1.5.5–14.225 | 59 | 2.386 | 1.215–4.685 | |||||
| GS expression | Low Exp. | 147 | 11 | 0.0001 * | 1 | - | 0.867 | 27 | 0.0097 * | 1 | - | 0.354 |
| High Exp. | 148 | 39 | 1.066 | 0.504–2.254 | 48 | 0.763 | 0.431–1.351 | |||||
| FABP 4 expression | Low Exp. | 147 | 10 | <0.0001 * | 1 | - | 0.405 | 25 | 0.0002 * | 1 | - | 0.904 |
| High Exp. | 148 | 40 | 1.378 | 0.648–2.927 | 50 | 1.035 | 0.588–1.822 | |||||
| TOLLIP expression | Low Exp. | 147 | 28 | 0.3415 | - | - | - | 34 | 0.5027 | - | - | - |
| High Exp. | 148 | 22 | - | - | - | 41 | - | - | - | |||
| AKR1A1 expression | Low Exp. | 147 | 21 | 0.2467 | - | - | - | 30 | 0.0779 | - | - | - |
| High Exp. | 148 | 29 | - | - | - | 45 | - | - | - | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.-F.; Shen, K.-H.; Chien, L.-H.; Huang, C.-H.; Wu, T.-F.; He, H.-L. Proteomic Identification of the Galectin-1-Involved Molecular Pathways in Urinary Bladder Urothelial Carcinoma. Int. J. Mol. Sci. 2018, 19, 1242. https://doi.org/10.3390/ijms19041242
Li C-F, Shen K-H, Chien L-H, Huang C-H, Wu T-F, He H-L. Proteomic Identification of the Galectin-1-Involved Molecular Pathways in Urinary Bladder Urothelial Carcinoma. International Journal of Molecular Sciences. 2018; 19(4):1242. https://doi.org/10.3390/ijms19041242
Chicago/Turabian StyleLi, Chien-Feng, Kun-Hung Shen, Lan-Hsiang Chien, Cheng-Hao Huang, Ting-Feng Wu, and Hong-Lin He. 2018. "Proteomic Identification of the Galectin-1-Involved Molecular Pathways in Urinary Bladder Urothelial Carcinoma" International Journal of Molecular Sciences 19, no. 4: 1242. https://doi.org/10.3390/ijms19041242





