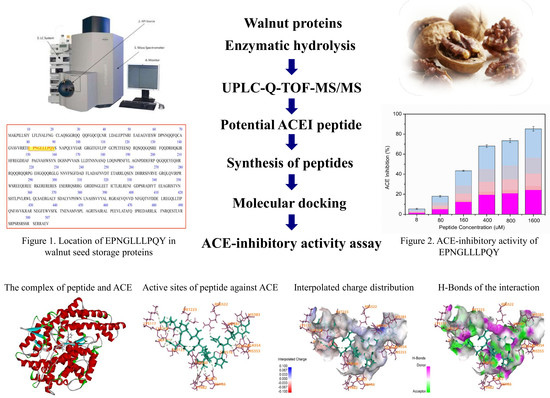
Identification of an ACE-Inhibitory Peptide from Walnut Protein and Its Evaluation of the Inhibitory Mechanism
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Peptides from Walnut Protein Hydrolysates
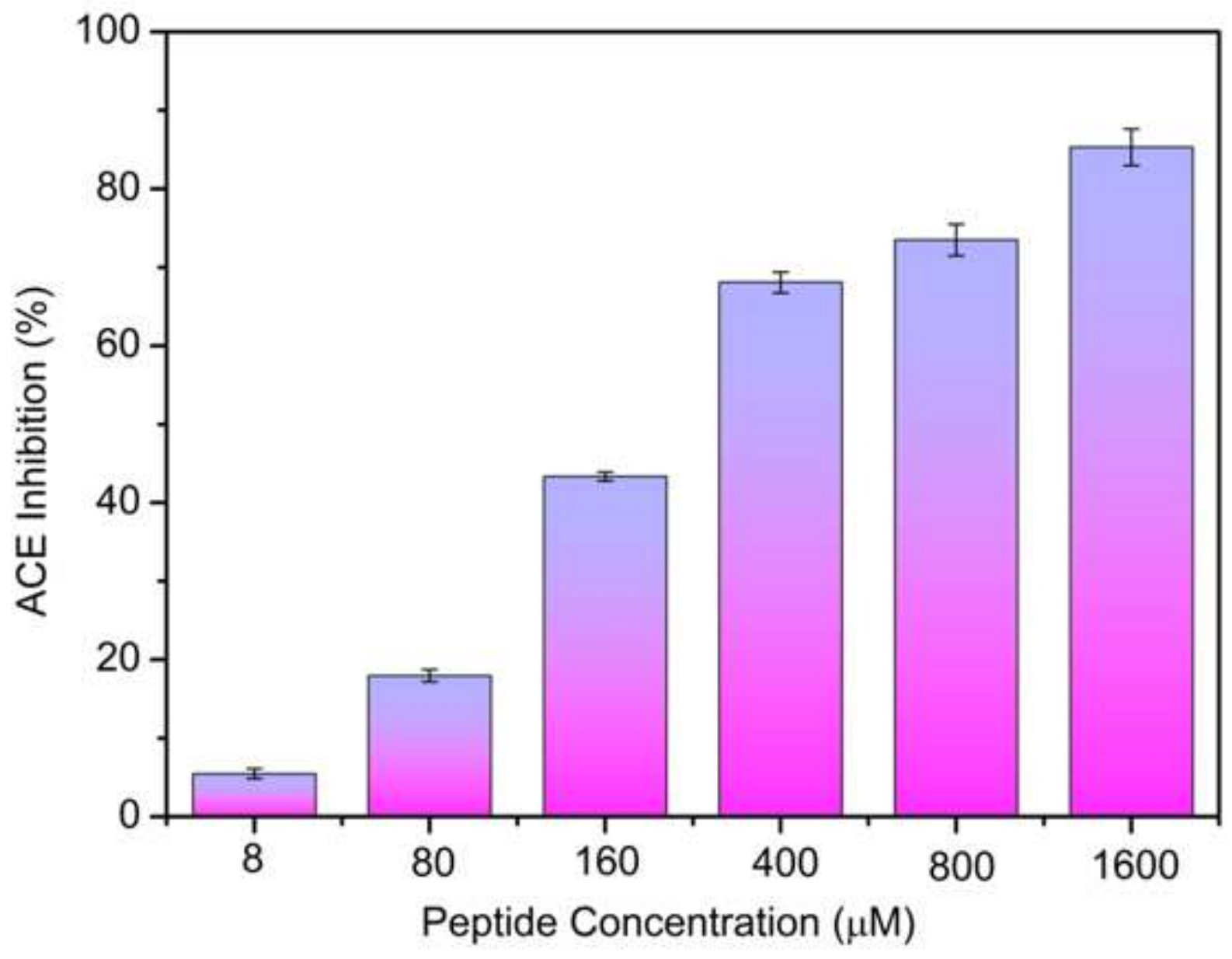
2.2. ACE-Inhibitory Activity Determination
2.3. Inhibition Pattern of the ACE-Inhibitory Peptide
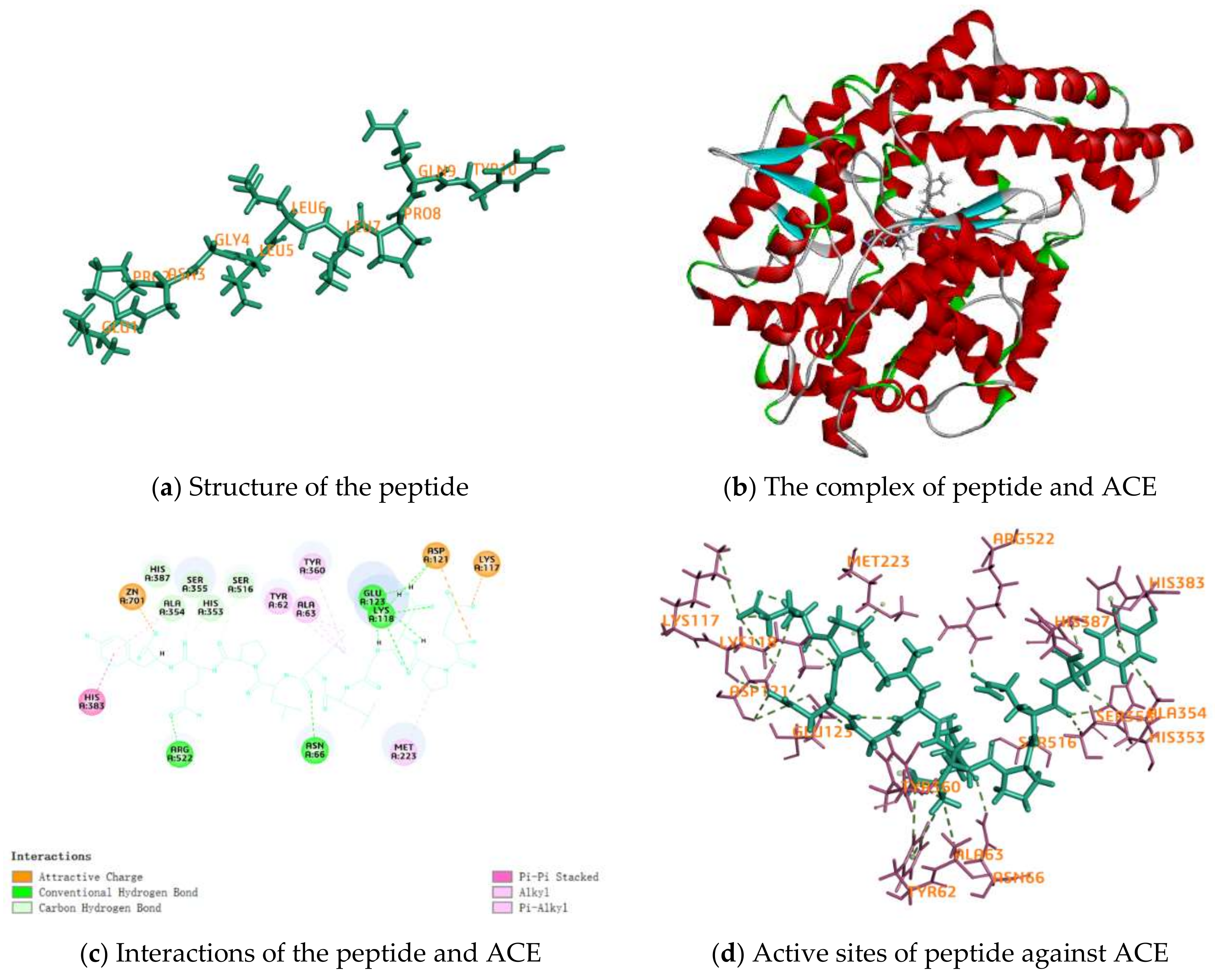
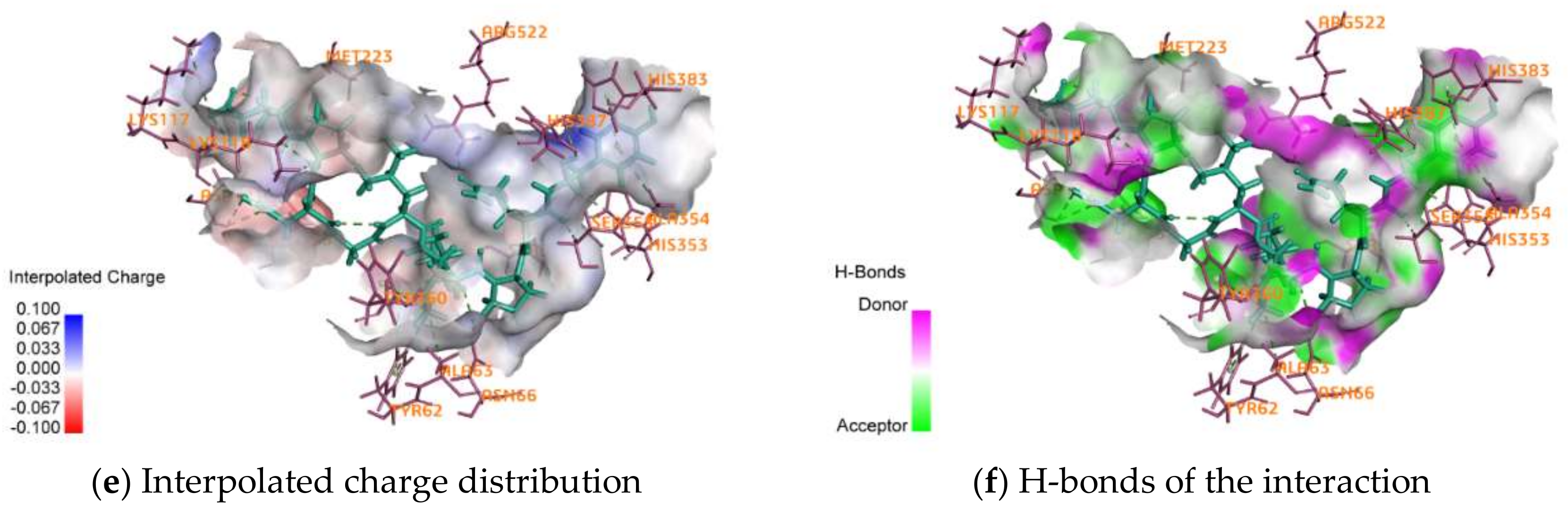
2.4. Activity Mechanism of EPNGLLLPQY
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Enzymatic Hydrolysis of Walnut Proteins
3.3. UPLC-Q-TOF-MS/MS Analysis
3.4. Screening of the Potential ACEI Peptides
3.5. Synthesis of Peptides
3.6. Molecular Docking
3.7. ACE-Inhibitory Activity Assay
3.8. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Cardiovascular Diseases (CVDs) Fact Sheet N 317; Updated January 2015; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Celermajer, D.S.; Chow, C.K.; Marijon, E.; Anstey, N.M.; Woo, K.S. Cardiovascular disease in the developing world. J. Am. Coll. Cardiol. 2012, 60, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Staessen, J.A.; Wang, J.; Bianchi, G.; Birkenhäger, W.H. Essential hypertension. Lancet 2003, 361, 1629–1641. [Google Scholar] [CrossRef]
- Cannon, C.P. Cardiovascular disease and modifiable cardiometabolic risk factors. Clin. Cornerstone 2008, 9, 24–41. [Google Scholar] [CrossRef]
- Gu, Q.; Dillon, C.F.; Burt, V.L.; Gillum, R.F. Association of hypertension treatment and control with all-cause and cardiovascular disease mortality among US adults with hypertension. Am. J. Hypertens. 2010, 23, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Skeggs, L.T.; Kahn, J.R.; Shumway, N.P. The preparation and function of the hypertensin-converting enzyme. J. Exp. Med. 1956, 103, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Erdös, E.; Levin, Y. A dipeptidyl carboxypeptidase that converts angiotensin I and inactivates bradykinin. Biochim. Biophys. Acta Protein Struct. 1970, 214, 374–376. [Google Scholar] [CrossRef]
- Hai-Lun, H.; Xiu-Lan, C.; Cai-Yun, S.; Yu-Zhong, Z.; Bai-Cheng, Z. Analysis of novel angiotensin-I-converting enzyme inhibitory peptides from protease-hydrolyzed marine shrimp Acetes chinensis. J. Pept. Sci. 2006, 12, 726–733. [Google Scholar] [CrossRef] [PubMed]
- López-Fandiño, R.; Otte, J.; Van Camp, J. Physiological, chemical and technological aspects of milk-protein-derived peptides with antihypertensive and ACE-inhibitory activity. Int. Dairy J. 2006, 16, 1277–1293. [Google Scholar] [CrossRef]
- Kim, S.-K.; Wijesekara, I. Development and biological activities of marine-derived bioactive peptides: A review. J. Funct. Foods 2010, 2, 1–9. [Google Scholar] [CrossRef]
- Atkinson, A.; Robertson, J. Captopril in the treatment of clinical hypertension and cardiac failure. Lancet 1979, 314, 836–839. [Google Scholar] [CrossRef]
- Liu, M.; Du, M.; Zhang, Y.; Xu, W.; Wang, C.; Wang, K.; Zhang, L. Purification and identification of an ACE inhibitory peptide from walnut protein. J. Agric. Food Chem. 2013, 61, 4097–4100. [Google Scholar] [CrossRef] [PubMed]
- Pihlanto-Leppälä, A. Bioactive peptides derived from bovine whey proteins: Opioid and ACE-inhibitory peptides. Trends Food Sci. Technol. 2000, 11, 347–356. [Google Scholar] [CrossRef]
- Vercruysse, L.; Van Camp, J.; Smagghe, G. ACE inhibitory peptides derived from enzymatic hydrolysates of animal muscle protein: A review. J. Agric. Food Chem. 2005, 53, 8106–8115. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Ma, H.; Zhao, W.; Wang, Z.; Tian, W.; Luo, L.; He, R. The use of ultrasound for enzymatic preparation of ACE-inhibitory peptides from wheat germ protein. Food Chem. 2010, 119, 336–342. [Google Scholar] [CrossRef]
- Orio, L.P.; Boschin, G.; Recca, T.; Morelli, C.F.; Ragona, L.; Francescato, P.; Arnoldi, A.; Speranza, G. New ACE inhibitory peptides from hemp seed (Cannabis sativa L.) proteins. J. Agric. Food Chem. 2017, 65, 10482–10488. [Google Scholar] [CrossRef] [PubMed]
- Jamdar, S.N.; Rajalakshmi, V.; Pednekar, M.D.; Juan, F.; Yardi, V.; Arun, S. Influence of degree of hydrolysis on functional properties, antioxidant activity and ACE inhibitory activity of peanut protein hydrolysate. Food Chem. 2010, 121, 178–184. [Google Scholar] [CrossRef]
- Wang, C.; Song, W.; Jiang, L.; Du, M. Purification and identification of an ACE-inhibitory peptide from walnut protein hydrolysate. Eur. Food Res. Technol. 2014, 239, 333–338. [Google Scholar]
- Mukuddem-Petersen, J.; Stonehouse, O.W.; Jerling, J.C.; Hanekom, S.M.; White, Z. Effects of a high walnut and high cashew nut diet on selected markers of the metabolic syndrome: A controlled feeding trial. Br. J. Nutr. 2007, 97, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Du, M.; Zheng, D.; Kong, F.; Zu, G.; Feng, Y. Purification and characterization of nattokinase from Bacillus subtilis natto B-12. J. Agric. Food Chem. 2009, 57, 9722–9729. [Google Scholar] [CrossRef] [PubMed]
- Rawendra, R.D.; Chang, C.-I.; Chen, H.-H.; Huang, T.-C.; Hsu, J.-L. A novel angiotensin converting enzyme inhibitory peptide derived from proteolytic digest of Chinese soft-shelled turtle egg white proteins. J. Proteom. 2013, 94, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- Pooja, K.; Rani, S.; Prakash, B. In silico approaches towards the exploration of rice bran proteins-derived angiotensin-I-converting enzyme inhibitory peptides. Int. J. Food Prop. 2017, 20, 2178–2191. [Google Scholar] [CrossRef]
- Iqbal, J.; Li, W.; Ullah, K.; Hasan, M.; Linna, G.; Awan, U.; Zhang, Y.; Batool, S.; Qing, H.; Deng, Y. Study of rat hypothalamic proteome by HPLC/ESI ion trap and HPLC/ESI-Q-TOF MS. Proteomics 2013, 13, 2455–2468. [Google Scholar] [CrossRef] [PubMed]
- Dziuba, J.; Iwaniak, A.; Mine, Y.; Shahidi, F. Database of Protein and Bioactive Peptide Sequences. Nutraceutical Proteins & Peptides in Health & Disease; CRC/Taylor & Francis: Boca Raton, FL, USA, 2006; pp. 543–545. [Google Scholar]
- Forghani, B.; Zarei, M.; Ebrahimpour, A.; Philip, R.; Bakar, J.; Hamid, A.A.; Saari, N. Purification and characterization of angiotensin converting enzyme-inhibitory peptides derived from Stichopus horrens: Stability study against the ACE and inhibition kinetics. J. Funct. Foods 2016, 20, 276–290. [Google Scholar] [CrossRef]
- Fu, Y.; Alashi, A.M.; Young, J.F.; Therkildsen, M.; Aluko, R.E. Enzyme inhibition kinetics and molecular interactions of patatin peptides with angiotensin I-converting enzyme and renin. Int. J. Biol. Macromol. 2017, 101, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Masuyer, G.; Schwager, S.L.; Sturrock, E.D.; Isaac, R.E.; Acharya, K.R. Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides. Sci. Rep. 2012, 2, 717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Y.; Lei, P.; Chen, D.-Q.; Feng, Y.-L.; Bai, X. Renal metabolic profiling of early renal injury and renoprotective effects of Poria cocos epidermis using UPLC Q-TOF/HSMS/MS E. J. Pharm. Biomed. Anal. 2013, 81, 202–209. [Google Scholar] [PubMed]
- Tu, M.; Feng, L.; Wang, Z.; Qiao, M.; Shahidi, F.; Lu, W.; Du, M. Sequence analysis and molecular docking of antithrombotic peptides from casein hydrolysate by trypsin digestion. J. Funct. Foods 2017, 32, 313–323. [Google Scholar] [CrossRef]
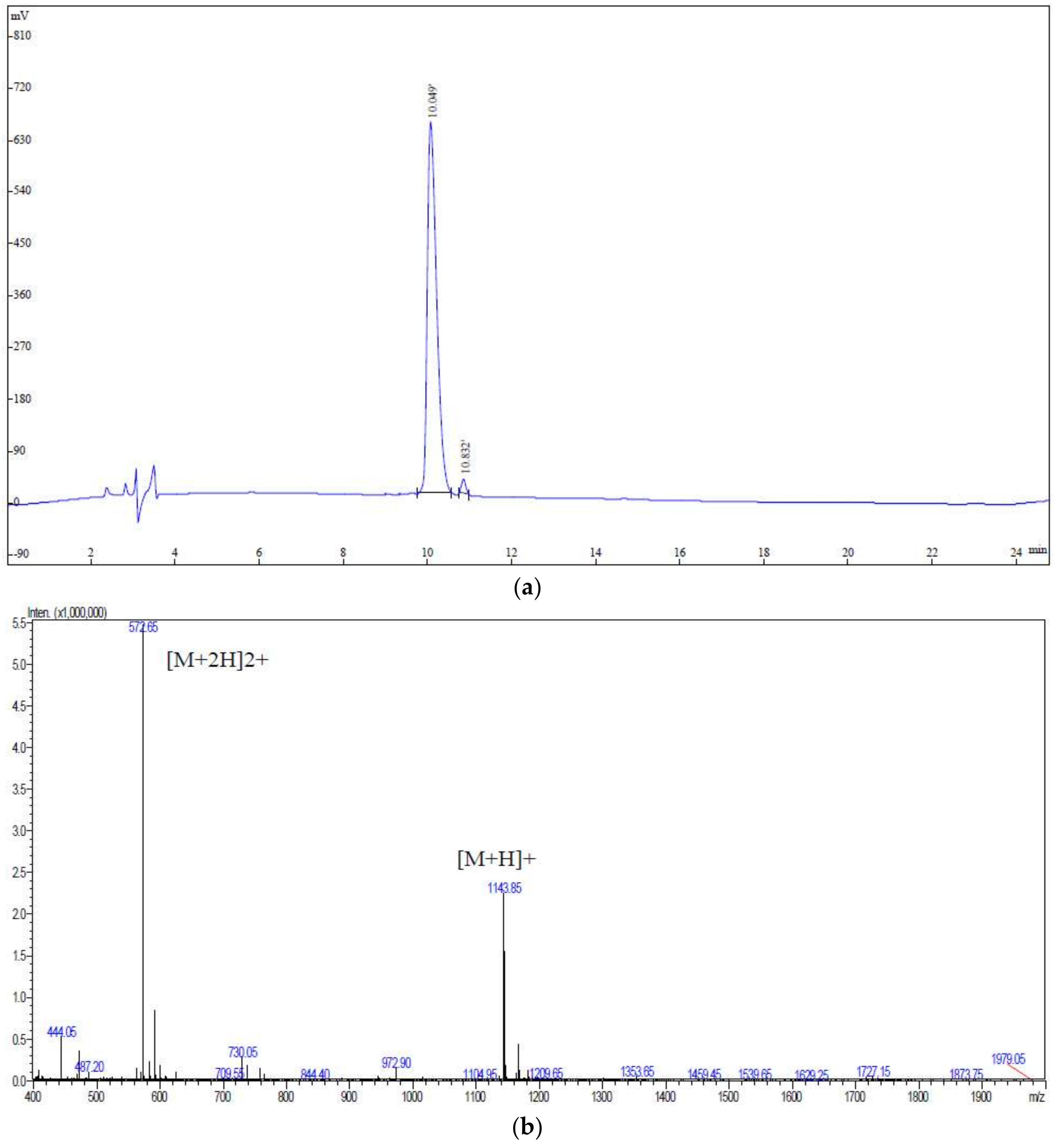


| No. | Protein | m/z Meas. | z | Mr. Calc. | Score | Start | AA Sequence | End | Length |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Histidine kinase OS | 529.3077 | 2 | 1056.593 | 38.4 | L | LQLSALADRA | A | 10 |
| 2 | Legumin OS | 832.4633 | 1 | 831.4491 | 21.37 | L | LTIPQNF | A | 7 |
| 3 | Legumin OS | 570.2736 | 2 | 1138.525 | 27.31 | L | SAERGALYSDA | L | 11 |
| 4 | Legumin OS | 648.374 | 2 | 1294.725 | 33.74 | A | IRALPEEVLANA | L | 12 |
| 5 | Legumin OS | 710.6773 | 3 | 2127.997 | 24.14 | A | VALMDTTNNANQLDQNPRN | F | 19 |
| 6 | Legumin OS | 759.702 | 3 | 2275.065 | 28.2 | A | VALMDTTNNANQLDQNPRNF | Y | 20 |
| 7 | Legumin OS | 864.8346 | 3 | 2590.46 | 29.51 | A | VVRRTIEPNGLLLPQYSNAPQLL | Y | 23 |
| 8 | Legumin OS | 670.3027 | 3 | 2007.871 | 24.94 | L | MDTTNNANQLDQNPRNF | Y | 17 |
| 9 | Multi-sensor signal transduction histidine kinase | 529.3077 | 2 | 1056.593 | 38.4 | P | IQLSAERLQ | F | 9 |
| 10 | Receptor agonist peptide | 520.9299 | 3 | 1558.77 | 21.66 | A | KTPGLPPMPQSDYL | K | 14 |
| 11 | Seed storage protein | 452.7325 | 2 | 903.445 | 28.1 | A | FQIPRED | A | 7 |
| 12 | Seed storage protein | 352.193 | 2 | 702.366 | 29.19 | L | SAERGAL | Y | 7 |
| 13 | Seed storage protein | 520.3148 | 2 | 1038.607 | 33.34 | A | IRALPEEVL | A | 9 |
| 14 | Seed storage protein | 574.2939 | 2 | 1146.567 | 41.27 | L | ATAFQIPRED | A | 10 |
| 15 | Seed storage protein | 555.8338 | 2 | 1109.644 | 22.87 | A | IRALPEEVLA | T | 10 |
| 16 | Seed storage protein | 529.3063 | 2 | 1056.593 | 38.51 | W | LQLSAERGAL | Y | 10 |
| 17 | Seed storage protein | 614.2948 | 3 | 1839.846 | 21.95 | L | LDTNNNANQLDQNPRN | F | 16 |
| 18 | Seed storage protein | 582.9686 | 3 | 1745.873 | 36.22 | F | KTNENAMVSPLAGRTSA | I | 17 |
| 19 | Seed storage protein | 994.97 | 2 | 1986.914 | 26.31 | L | LDTNNNANQLDQNPRNF | Y | 17 |
| 20 | Seed storage protein | 644.6458 | 3 | 1930.903 | 20.57 | P | GEHGQQQRGLGNNVFSGF | D | 18 |
| 21 | Seed storage protein | 921.4422 | 2 | 1839.854 | 62.21 | F | PAGVAHWSYNDGSNPVVA | I | 18 |
| 22 | Seed storage protein | 719.0287 | 3 | 2153.046 | 27.39 | A | ISLLDTNNNANQLDQNPRN | F | 19 |
| 23 | Seed storage protein | 827.139 | 3 | 2477.375 | 29.43 | A | VVRRTIEPNGLLLPQYSNAPQL | V | 22 |
| 24 | Seed storage protein | 526.9371 | 3 | 1577.78 | 26.95 | F | LADAFNVDTETARR | L | 14 |
| 25 | Seed storage protein | 863.9137 | 2 | 1725.803 | 80.77 | L | LDTNNNANQLDQNPR | N | 15 |
| 26 | Seed storage protein | 572.33 | 2 | 1142.60 | 27 | - | EPNGLLLPQY | S | 10 |
| 27 | Seed storage protein | 681.0131 | 3 | 2039.003 | 47.55 | A | ISLLDTNNNANQLDQNPR | N | 18 |
| 28 | Seed storage protein | 711.8505 | 2 | 1421.679 | 26.29 | F | LADAFNVDTETAR | R | 13 |
| 29 | Vicilin-like protein precursor | 432.7356 | 2 | 863.4501 | 25.24 | L | LRGIENY | R | 7 |
| 30 | Vicilin-like protein precursor | 425.7273 | 2 | 849.4345 | 37.36 | Y | LRVFSND | I | 7 |
| 31 | Vicilin-like protein precursor | 482.2702 | 2 | 962.5185 | 28.32 | L | RVFSNDIL | V | 8 |
| 32 | Vicilin-like protein precursor | 538.8131 | 2 | 1075.603 | 35.78 | Y | LRVFSNDIL | V | 9 |
| 33 | Vicilin-like protein precursor | 500.2511 | 3 | 1497.721 | 23.9 | M | ESYFVPTERQSR | R | 12 |
| No. | Peptides | Length | -CDOCKER_Energy Interaction (kcal mol−1) | Peptide Ranker |
|---|---|---|---|---|
| 1 | IRALPEEVL | 9 | 155.937 | 0.14 |
| 2 | LQLSALADRA | 10 | 144.081 | 0.23 |
| 3 | ATAFQIPRED | 10 | 136.401 | 0.18 |
| 4 | EPNGLLLPQY | 10 | 134.600 | 0.37 |
| 5 | LQLSAERGAL | 10 | 130.532 | 0.30 |
| 6 | LRVFSND | 7 | 127.556 | 0.16 |
| 7 | LRVFSNDIL | 9 | 126.776 | 0.30 |
| 8 | IQLSAERLQ | 9 | 123.061 | 0.12 |
| 9 | IRALPEEVLA | 10 | 121.918 | 0.14 |
| 10 | LTIPQNF | 7 | 108.518 | 0.34 |
| 11 | FQIPRED | 7 | 97.6096 | 0.27 |
| 12 | LRGIENY | 7 | 94.0396 | 0.12 |
| 13 | RVFSNDIL | 8 | 85.8807 | 0.31 |
| 14 | SAERGAL | 7 | 68.027 | 0.29 |
| Interactions | Distance | Category | Types |
|---|---|---|---|
| A:LYS117:NZ–EPNGLLLPQY:O15 | 4.63933 | Electrostatic | Attractive Charge |
| A:ZN701:ZN–EPNGLLLPQY:O162 | 2.22675 | Electrostatic | Attractive Charge |
| EPNGLLLPQY:N1–A:ASP121:OD2 | 4.00046 | Electrostatic | Attractive Charge |
| A:ASN66:HD21–EPNGLLLPQY:O90 | 2.53299 | Hydrogen Bond | Conventional Hydrogen Bond |
| A:LYS118:HZ1–EPNGLLLPQY:O40 | 1.79716 | Hydrogen Bond | Conventional Hydrogen Bond |
| A:LYS118:HZ3–EPNGLLLPQY:O31 | 2.13479 | Hydrogen Bond | Conventional Hydrogen Bond |
| A:ARG522:HH12–EPNGLLLPQY:O135 | 1.92374 | Hydrogen Bond | Conventional Hydrogen Bond |
| EPNGLLLPQY:H33–A:GLU123:OE1 | 1.97968 | Hydrogen Bond | Conventional Hydrogen Bond |
| EPNGLLLPQY:H42–A:LYS118:O | 2.29783 | Hydrogen Bond | Conventional Hydrogen Bond |
| EPNGLLLPQY:H42–A:ASP121:O | 2.83625 | Hydrogen Bond | Conventional Hydrogen Bond |
| EPNGLLLPQY:H43–A:ASP121:O | 2.80029 | Hydrogen Bond | Conventional Hydrogen Bond |
| EPNGLLLPQY:H43–A:GLU123:OE2 | 2.2265 | Hydrogen Bond | Conventional Hydrogen Bond |
| EPNGLLLPQY:H47–A:GLU123:OE1 | 2.01368 | Hydrogen Bond | Conventional Hydrogen Bond |
| A:LYS118:HE2–EPNGLLLPQY:O31 | 2.33128 | Hydrogen Bond | Carbon Hydrogen Bond |
| A:HIS353:HD2–EPNGLLLPQY:O140 | 2.40483 | Hydrogen Bond | Carbon Hydrogen Bond |
| A:SER355:HB1–EPNGLLLPQY:O140 | 2.37051 | Hydrogen Bond | Carbon Hydrogen Bond |
| A:HIS387:HD2–EPNGLLLPQY:O162 | 2.74483 | Hydrogen Bond | Carbon Hydrogen Bond |
| A:SER516:HB1–EPNGLLLPQY:O123 | 2.80361 | Hydrogen Bond | Carbon Hydrogen Bond |
| EPNGLLLPQY:H144–A:ALA354:O | 2.26174 | Hydrogen Bond | Carbon Hydrogen Bond |
| A:HIS383–EPNGLLLPQY | 4.51235 | Hydrophobic | Pi-Pi Stacked |
| A:ALA63–EPNGLLLPQY:C81 | 4.15055 | Hydrophobic | Alkyl |
| EPNGLLLPQY–A:MET223 | 4.05541 | Hydrophobic | Alkyl |
| A:TYR62–EPNGLLLPQY:C81 | 5.26359 | Hydrophobic | Pi-Alkyl |
| A:TYR62–EPNGLLLPQY:C85 | 5.3078 | Hydrophobic | Pi-Alkyl |
| A:TYR360–EPNGLLLPQY:C81 | 5.23035 | Hydrophobic | Pi-Alkyl |
| EPNGLLLPQY–A:ALA354 | 4.78628 | Hydrophobic | Pi-Alkyl |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Tu, M.; Wu, D.; Chen, H.; Chen, C.; Wang, Z.; Jiang, L. Identification of an ACE-Inhibitory Peptide from Walnut Protein and Its Evaluation of the Inhibitory Mechanism. Int. J. Mol. Sci. 2018, 19, 1156. https://doi.org/10.3390/ijms19041156
Wang C, Tu M, Wu D, Chen H, Chen C, Wang Z, Jiang L. Identification of an ACE-Inhibitory Peptide from Walnut Protein and Its Evaluation of the Inhibitory Mechanism. International Journal of Molecular Sciences. 2018; 19(4):1156. https://doi.org/10.3390/ijms19041156
Chicago/Turabian StyleWang, Cong, Maolin Tu, Di Wu, Hui Chen, Cheng Chen, Zhenyu Wang, and Lianzhou Jiang. 2018. "Identification of an ACE-Inhibitory Peptide from Walnut Protein and Its Evaluation of the Inhibitory Mechanism" International Journal of Molecular Sciences 19, no. 4: 1156. https://doi.org/10.3390/ijms19041156